Abstract
Coral diseases have been increasingly reported over the past few decades and are a major contributor to coral decline worldwide. The Caribbean, in particular, has been noted as a hotspot for coral disease, and the aptly named white syndromes have caused the decline of the dominant reef building corals throughout their range. White band disease (WBD) has been implicated in the dramatic loss of Acropora cervicornis and Acropora palmata since the 1970s, resulting in both species being listed as critically endangered on the International Union for Conservation of Nature Red list. The causal agent of WBD remains unknown, although recent studies based on challenge experiments with filtrate from infected hosts concluded that the disease is probably caused by bacteria. Here, we report an experiment using four different antibiotic treatments, targeting different members of the disease-associated microbial community. Two antibiotics, ampicillin and paromomycin, arrested the disease completely, and by comparing with community shifts brought about by treatments that did not arrest the disease, we have identified the likely candidate causal agent or agents of WBD. Our interpretation of the experimental treatments is that one or a combination of up to three specific bacterial types, detected consistently in diseased corals but not detectable in healthy corals, are likely causal agents of WBD. In addition, a histophagous ciliate (Philaster lucinda) identical to that found consistently in association with white syndrome in Indo-Pacific acroporas was also consistently detected in all WBD samples and absent in healthy coral. Treatment with metronidazole reduced it to below detection limits, but did not arrest the disease. However, the microscopic disease signs changed, suggesting a secondary role in disease causation for this ciliate. In future studies to identify a causal agent of WBD via tests of Henle–Koch's postulates, it will be vital to experimentally control for populations of the other potential pathogens identified in this study.
Keywords: coral, disease, treatment, antibiotics, Philaster, Vibrio
1. Introduction
Coral reefs and other tropical marine systems have declined in health in recent decades, owing to a variety of local and regional environmental impacts in addition to the effects of climate change. These impacts threaten the fundamental ecological functions of coral reefs [1] as well as the coastal protection, tourism, biodiversity, fisheries production and other ecosystem services that they provide [2].
Many reef coral diseases have emerged in the past 30–40 years, several of which have caused significant regional-scale ecological impacts [3–5]. For example, Acropora species were formerly the dominant ‘bioengineering’ species on shallow and mid-depth zones over most of the Caribbean. Shallow (0–6 m depth) reefs were typically dominated by Acropora palmata, whereas Acropora cervicornis was commonly a dominant species at 6–9 m depth [6]. These species are now relatively rare and recently both species have been listed as critically endangered on the International Union for the Conservation of Nature Red list. In consequence to this loss, the physiographic reef zones they had previously constructed have been eliminated from all but a handful of reefs in the region [7]. It is widely regarded that this decline can be attributed to the emergence of a particularly aggressive coral disease, white band disease (WBD), first observed in the early 1970s [8–10].
As with many coral diseases, the causal agent of WBD has not been definitively identified, and there is some confusion over the two types of WBD that have been described (type I and II). WBD type II is differentiated from WBD type I by a variable band of bleached tissue that precedes the tissue lesion [11]. A causal agent for WBD type II has previously been proposed as Vibrio charchariae, which is a synonym of Vibrio harveyi [11]; however, the reisolation and definitive identification of the proposed agent from experimental inoculations has never been documented. With regard to WBD type I, Pantos & Bythell [12] failed to detect any Vibrios in diseased tissues using culture-independent techniques [12], however, they did show a suite of α-proteobacteria related to Roseobacter to be present in WBD type I, but absent in healthy corals. These were similar to a group of bacteria previously detected exclusively in diseased tissues of other coral diseases (white plague and black band disease). By contrast, Casas et al. [13] failed to detect any specific pathogens in WBD using culture-independent techniques and suggested that there might be a non-pathogenic cause of this disease. However, recently, a study by Kline & Vollmer [14] used homogenates of diseased tissue to demonstrate that WBD is, in fact, a transmissible disease attributable to a 0.22–0.45 µm filterable fraction that was susceptible to antibiotic treatment. They concluded that the causal agent is therefore likely to be a Gram-positive bacterium. However, their study did not involve a comprehensive analysis of the microorganisms involved and therefore the pathogen remains unknown. In this study, a broader set of antibiotics were used to treat WBD type I diseased corals (A. cervicornis) directly, together with a comprehensive screening of the microbial community (bacteria, archaea and ciliates) associated with the coral using culture-independent molecular techniques to detect potential pathogens and narrow the range of potential agents that could be further tested for causality using challenge experiments.
2. Results
A range of antibiotics were used to treat corals showing signs of WBD type I with the aim to detect potential pathogens and reduce the overall range of potential causal agents highlighted in previous studies. We found a significant interaction between time and treatment (repeated-measures ANOVA, F120 = 8.03, p = 0.0001), on lesion progression rate (table 1), indicating that the effect of each antibiotic on WBD progression was different for the period of observation. All apparently healthy, non-diseased (ND) corals survived the 6 days of the experiment and appeared normal at the end of the experiment (figure 1). Disease progression continued in all the diseased corals not treated with antibiotics throughout the duration of the experiment (figure 1 and table 1), and the advance rates of the disease lesion of these untreated corals (from 0.9 ± 0.10 to 3.4 ± 0.40 cm d−1) were within the range reported for WBD in the field (0.2–4 cm d−1) [15]. Initial experiments on healthy corals treated with the four types of antibiotic used in this study showed no visible adverse effects of the treatments.
Table 1.
Mean lesion progression rate (cm d−1) of white band diseased coral nubbins during the antibiotic experiment. (Time 0 represents lesion progression rate in the 12 h prior to the start of the experiment confirming that all lesions were advancing prior to treatment. ND, non-diseased corals kept under experimental conditions as controls. No disease lesions developed in these controls. WBD, white band diseased corals untreated with antibiotic. These lesions continued to progress throughout the experiment and samples were collected before the whole coral nubbin had died (see + for collection points). Amp, ampicillin-treated white band diseased corals. Lesion progression was immediately halted under this treatment. Gent, gentamicin-treated white band diseased corals. Lesions in this treatment continued to progress throughout the experiment, but all nubbins survived to the end of the experiment and were sampled at 144 h. Met, metronidazole-treated white band diseased corals. Lesions in this treatment continued to progress throughout the experiment, and samples were collected before the nubbins were completely killed (see + for collection points). Para, paromomycin-treated white band diseased corals. Lesion progression halted after 24 h in this treatment. Plus symbols show time points when an individual nubbin was sampled, either when <1 cm of tissue was left on the coral nubbin or at the end of the experiment. Data are means ± s.e. (n = 6 initially).)
| time (h) | ND | WBD | amp | gent | met | para |
|---|---|---|---|---|---|---|
| 0 | 0 | 0.9 ± 0.10 | 0.9 ± 0.10 | 0.8 ± 0.23 | 0.9 ± 0.20 | 0.9 ± 0.2 |
| 24 | 0 | 0.9 ± 0.07 | 0 | 0.6 ± 0.19 | 0.6 ± 0.19 | 0.7 ± 0.45 |
| 48 | 0 | 1.2 ± 0.08 | 0 | 0.8 ± 0.28 | 1.4 ± 0.3 | 0 |
| 72 | 0 | 1.5 ± 0.20 | 0 | 1.4 ± 0.53 | 1.8 ± 0.55 ++ | 0 |
| 96 | 0 | 3.4 ± 0.40 ++++ | 0 | 1.7 ± 0.7 | 2.3 ± 0.7 | 0 |
| 120 | 0 | ++ | 0 | 1.9 ± 0.8 | 1.9 ± 0.1 +++ | 0 |
| 144 | 0 | 0 | 1 ± 0.4 | ++ | 0 | |
| ++++++ | ++++++ | ++++++ | ++++++ |
Figure 1.
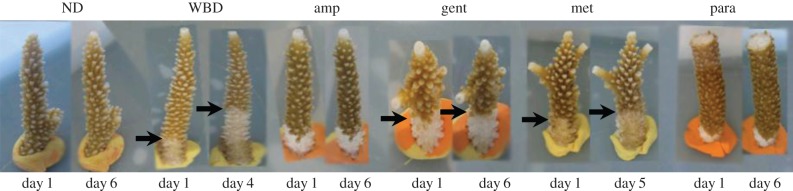
Photographs of healthy, non-diseased (ND) corals and those with signs of white band disease (WBD) both controls and treated corals. The experiment was run for 6 days, during which the healthy corals showed no signs of lesion development or other visible signs of stress. The lesion of untreated corals showing signs of white band disease Type 1 (WBD) continued to progress, while those treated with ampicillin (amp) and paromomycin sulfate (para) arrested lesion progression. (Online version in colour.)
(a). Identification of potential pathogens
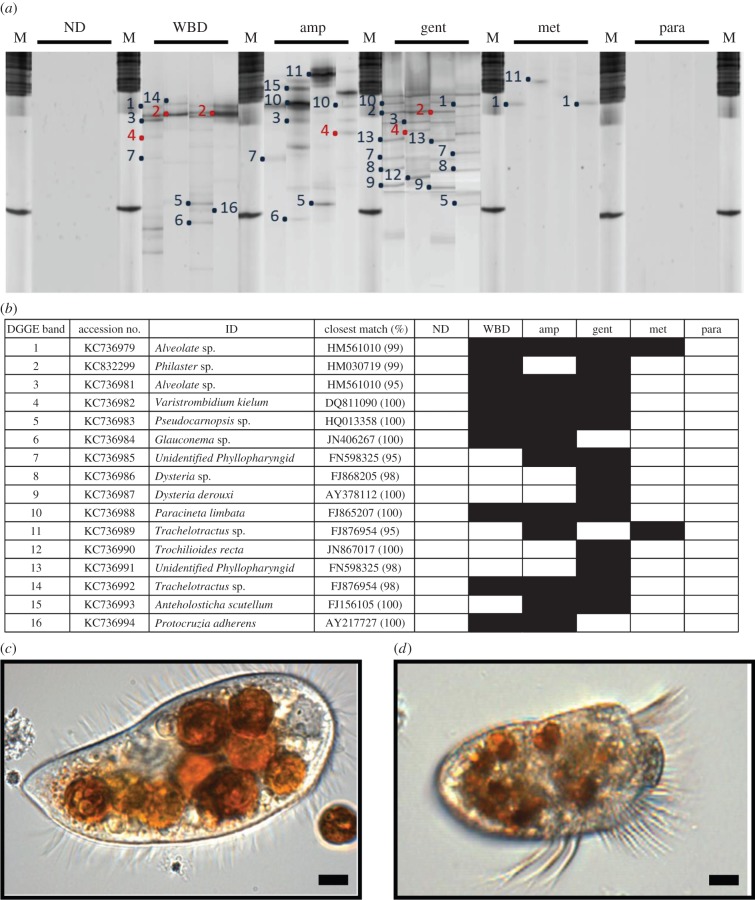
There was little variation apparent in the bacterial diversity of replicate samples of the same treatment, as determined by denaturing gradient gel electrophoresis (DGGE) analysis (electronic supplementary material, figure S1); therefore, only a subset (n = 3) of randomly selected samples were used for detailed clone library analysis. There were significant differences in 16S rRNA gene bacterial diversity in both the clone libraries (n = 3 per treatment) and the DGGE (n = 6 per treatment; analysis of similarity (ANOSIM R = 1, p = 0.029 and R = 1, p = 0.001, respectively) between all treatments (electronic supplementary material, table S1). Despite metronidazole-targeting protozoans, this treatment also had a significant effect on the bacterial communities (electronic supplementary material, table S1).
Fifteen prokaryote (bacteria and archaea) ribotypes were identified in this study as potential pathogens of WBD, being consistently present in all samples of corals displaying WBD, but absent in ND samples. These were ribotypes related to: Oceanicola (KC736995), Sandarakinotalea (KC736996), Anaeroplasma bactoclasticum (KC736998), Parachlamydia acanthamoebae (KC737010), Photobacterium aplysia (KC737015), Comamonas (KC737017), Halobacteria (KC737020), Asteroleplasma (KC737022), Vi. charchariae (KC737024), Lactobacillus suebicus (KC737026), Roseovarius crassostreae (KC737031), Bacillus (KC737032), Cyclobacterium (KC737035) and an unidentified β-proteobacteria (KC737036; electronic supplementary material, table S1). An additional ribotype, an archaea, Pyrobaculum (KC737045) was present only in low frequency in one of the three samples of ND corals and frequent in all three diseased samples and was therefore also considered a potential pathogen. The Vi. charchariae ribotype has previously been identified (100% sequence similarity) as a pathogen in WBD type II [11], the Ri. crassostreae ribotype (100% similarity) was highlighted as a potential pathogen of WBD type I [12] and the Bacillus species (100% similarity) was identified as a possible pathogen of WBD in acroporas from Indonesia [16].
There was a diverse community of ciliates associated with WBD samples, but ciliates were not detected in any of the ND samples (figure 2). Ciliate ribotypes present in samples with WBD were related to: two Alveolate sp. (KC736981 and KC736979), a Pseudocarnopsis sp. (KC736983), a Glauconema sp. (KC736984), Paracineta limbata (KC736988), a Trachelotractus sp. (KC736992), Protocruzia adherens (AY217727), Philaster lucinda (KC832299) and Varistrombidium kielum (KC736982). The latter two histophagous ciliates have previously been found associated with diseased corals (white syndrome; WS) in the Pacific [17]. Philaster lucinda (99% sequence similarity) was consistently present in all samples of WBD and has similarly been observed consistently in WS in the Pacific [17]. As previously observed in WS, this highly active ciliate was present in dense, mobile population masses and observed to burrow under the lesion boundary and consume intact coral tissues in all WBD samples analysed in this study (electronic supplementary material, video S1). The Varistrombidium species (100% sequence similarity to that present in WS in the Pacific) is also histophagous (observed ingesting the coral tissues and engulfing the symbiotic algae), but was not consistently observed in all diseased samples. No ciliate other than Phi. lucinda was consistently observed in all samples of the disease, and it is therefore likely that the other species observed are secondary invaders.
Figure 2.
(a) Representative denaturing gradient gel electrophoresis (DGGE) profiles obtained using ciliate-specific primers of: non-diseased (ND); white band diseased (WBD); ampicillin treated (amp); gentamicin-treated (gent); metronidazole-treated (met) and paromomycin-treated (para) corals; (b) summary table showing presence/absence of specific ciliates in the different samples, (c) light micrograph of the histophagous ciliate Phialster lucinda (KC832299); and (d) light micrograph of the other histophagous ciliate Varistrombidium kielum (KC736982). Ingested coral symbiotic algae clearly visible. Scale bars represent 10 µm. (Online version in colour.)
(b). Identification of candidate primary pathogens by experimental antibiotic treatment
In specimens initially displaying an actively progressing WBD lesion, two antibiotic treatments, ampicillin and paromomycin sulfate, arrested the advance of the lesion in all samples (figure 1 and table 1; n = 6 coral fragments per treatment). The disease lesion continued to progress in diseased corals treated with gentamicin and metronidazole (figure 1 and table 1). Of the 15 prokaryote candidate pathogens identified above, seven were reduced to undetectable levels by the gentamicin treatment that failed to arrest the progression of the WBD lesion; (Oceanicola (KC736995), Sandarakinotalea (KC736996), Pa. acanthamoebae (KC737010), Pho. aplysia (KC737015), Comamonas (KC737017), Cyclobacterium (KC737035) and the β-proteobacteria (KC737036)). Similarly, the ciliate Philaster lucinda was reduced to undetectable levels by the metronidazole treatment that failed to arrest the lesion progression. It was conversely eliminated in both the ampicillin and paromomycin treatments. These eight candidate pathogens are therefore unlikely to be primary pathogens. Of the eight remaining candidate pathogens, five were not eliminated either by the ampicillin treatment, the paromomycin sulfate treatment, or both, which arrested the WBD lesion progression (Anaeroplasma bactoclasticum (KC736998), Halobacteria (KC737020), Asteroleplasma (KC737022), R. crassostreae (KC737031) and Pyrobaculum (KC737045)). Thus, three of the potential pathogens identified in this study: V. charchariae (KC737024), L. suebicus (KC737026) and the Bacillus sp. (KC737032) remain as potential primary pathogens of WBD.
(c). Association of potential pathogens with tissue pathogenesis
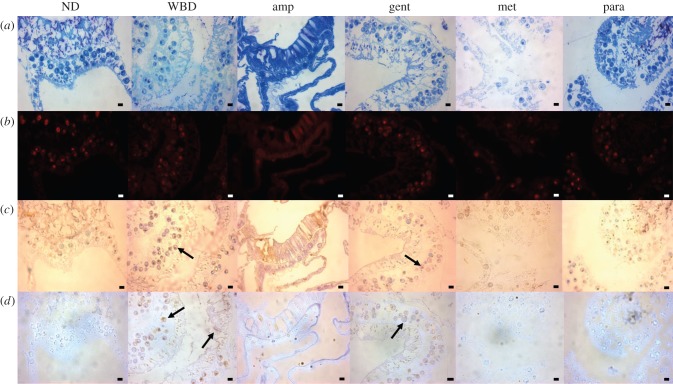
There was a significant difference in bacterial abundance between tissues (ANOVA R = 0.87 p = 0.001; figure 3). Specifically, there was a significant increase in total bacterial abundance between ND and WBD diseased tissues (p < 0.0001; figure 3). However, there was no large bacterial mass or evidence of widespread tissue necrosis and/or apoptosis in histological sections (figure 4), indicating that the bacterial population build-up probably occurred in the surface mucus layer, external to the tissues, which would be lost during routine histological processing. However, sections stained with nigrosin indicated some cellular necrosis within both the host tissues and symbiotic algae of WBD samples (figure 4). Necrosis was present at low levels in the peripheral surface of the epidermis and in localized deeper pockets (figure 4). Furthermore, a similar positive staining was associated with tissues stained with in situ end labelling (ISEL) indicating simultaneous apoptosis occurring within the necrotic tissues (figure 4). In corals treated with ampicillin, gentamicin and paromomycin sulfate, the tissues appeared normal under the general stain toluidine blue (figure 4), whereas corals treated with metronidazole showed extensive tissue fragmentation, but without any evidence for cellular necrosis or apoptosis (figure 4). Cellular necrosis, as detected by nigrosin staining, and apoptosis, as detected by ISEL, were observed in the gentamicin treatments, following a similar pattern to that of the untreated WBD tissues with low levels of staining in the peripheral epidermis.
Figure 3.
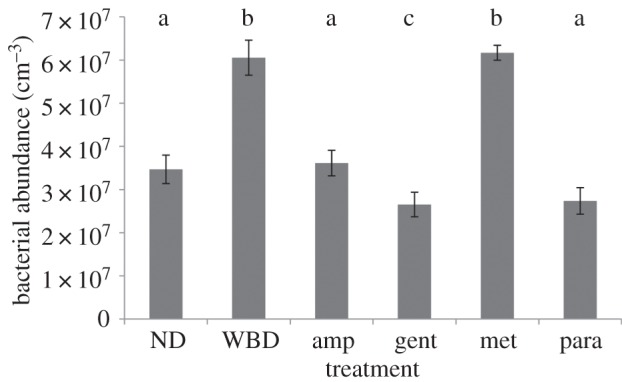
Total bacterial abundance of non-diseased tissues (ND), corals with white band disease (WBD) and diseased corals treated with paromomycin sulfate (para), ampicilin (amp), gentamicin (gent) and metronidazole (met). Letters above the bars (a, b and c) show which treatments showed significant differences (Tukey's HSD post hoc tests).
Figure 4.
Representative histological sections from each treatment at day 6 of the experiment for non-diseased tissues (ND) and those treated with paromomycin sulfate (para), ampicilin (amp) and gentamicin (gent). The white band diseased (WBD) tissues and those treated with metronidazole (met) were sampled before complete tissue loss occurred, on days 2, 3, 4 and 5 (table 1). In all cases (except that of ND tissues), the samples were taken at the disease lesion interface, within 1 mm of the lesion boundary. (a) Sections stained with Toluidine blue. Tissues appear indistinguishable from healthy samples in the diseased (WBD) samples and in all treatments except for the Met treatment, in which extensive tissue fragmentation can be seen. (b) Fluorescence in situ hybridization (FISH) probed with EUBMIX, giving a comprehensive ‘eubacterial’ detection as visualized by red (CY3) fluorescence. In this case, few if any bacterial-sized fluorescent particles can be identified in the sections, with red fluorescence attributed to autofluorescence of symbiotic algae and nematocysts. (c) In situ end labelling (ISEL), programmed cell death assay. Little or no probe binding (indicated by red-brown staining) could be detected in ND tissues and those treated with amp and para. Positive ISEL staining was observed in diseased tissues (WBD) and gent-treated samples (arrows). (d) Nigrosin (Nig) staining, which targets necrotic tissues. Brown-stained (necrotic) host tissues and symbiotic algae were observed in diseased (WBD) and gent treatments (arrows), but not in any other samples. Para- and amp-treated tissues appeared indistinguishable from healthy tissue sections in all cases. Scale bars represent 10 µm. (Online version in colour.)
Bacterial abundance in corals treated with ampicillin and paromomycin sulfate (3.61 × 107 cm−3 and 2.73 × 107 cm−3, respectively) returned to similar levels recorded in non-diseased tissues (3.46 × 107 cm−3; figure 3). Furthermore, there was no significant difference in bacterial abundance between WBD tissues and those treated with metronidazole (p = 0.12; figure 3), although bacterial species composition was affected (electronic supplementary material, table S1).
3. Discussion
We show here that WBD is amenable to treatment with either ampicillin or paromomycin sulfate and therefore support previous studies [14] that have used the application of filtrate to conclude that WBD is caused by microorganisms rather than by physiological stress. This study also shows that WBD, as previously shown in this disease [12], and other diseases such as WS [17–19], white plague [20–25] and black band disease [26,27] is a polymicrobial disease associated with a number of specific microorganisms that are consistently associated with diseased samples but absent, or undetectable, in healthy ones. We identify a suite of 16 such specific associates (14 bacteria, one archaea and one ciliate) as potential pathogens of WBD. One of these, the ciliate Phi. lucinda (KC832299), has recently been shown to be consistently associated with the coral disease WS in the Pacific [17] and within aquaria [28], which all have identical visible and histopathological disease signs, namely the advancing band of cleared skeleton immediately adjacent to visibly normal tissues (table 2). In all cases, the ciliate has been observed to ingest visibly intact coral tissues and to engulf and digest the coral endosymbionts. Furthermore, what was probably the same ciliate species, has also been observed to digest entire coral spats [29], although at the time it was misidentified as Helicostoma nonatum. This histophagous activity and the consistent, specific association of Phi. lucinda with the two diseases indicates that: (i) the ciliate is an active pathogen, and (ii) that the two diseases are probably synonymous. However, selective elimination of this pathogen using the antibiotic metronidazole failed to arrest disease lesion progression in controlled experiments, indicating that the Philaster (KC832299) ciliate is unlikely to be a primary pathogen of WBD. However, histological analysis showed that the histopathology of the disease changed in the metronidazole treatment. This result is consistent with this ciliate being a secondary pathogen which nonetheless contributes to the typical pathogenesis of WBD and WS [17]. We also show that another histophagous ciliate, V. kielum (KC736982), also associated with WS in Pacific corals, is present in WBD, but is not consistently associated with all cases of the disease. This and a diverse community of other ciliates are non-specific associates, likely to be secondary invaders of WBD. Observations from these studies have led us to the conclusion that the visible disease signs of WBD and WS [17], can largely be attributed to this ciliate assemblage and other protozoans and micro-metazoans feeding on both the intact tissues (Philaster lucinda and Va. kielum) and tissue detritus at the edge of the lesion (many species; figure 2).
Table 2.
Summary showing the main effects of treatments and associated potentially pathogenic bacteria (Vibrio carchariae, Lactobacillus sp., Roseovarius crassostreae and a Bacillus sp.) and histophagous ciliates (Philaster lucinda and Varistrombidium sp.) identified within the study.
| ND | WBD | amp | gent | met | para | |
|---|---|---|---|---|---|---|
| progressive lesion | − | + | − | + | + | − |
| tissue necrosis | − | + | − | + | − | − |
| apoptosis | − | + | − | + | − | − |
| tissue fragmentation | − | − | − | − | + | − |
| Vibrio carchariae | − | + | − | + | + | − |
| Lactobacillus sp. | − | + | − | + | + | − |
| Roseovarius crassostreae | − | + | + | + | + | − |
| Bacillus sp. | − | + | − | + | + | − |
| Philaster lucinda | − | + | − | + | − | − |
| Varistrombidium sp. | − | + | + | + | − | − |
| in vitro effect on ciliates | NA | NA | − | − | + | + |
Fifteen potential prokaryote (bacterial and archaeal) pathogens were also identified in this study that were consistently associated with all samples of disease and absent from ND samples. The selective elimination of different groups of these prokaryotes by antibiotic treatment indicated that most of these are likely to be secondary pathogens or secondary invaders, despite their specific and consistent association with the disease. Three of the identified candidate bacterial pathogens, Vi. charchariae (KC737024), L. suebicus (KC737026) and the Bacillus sp. (KC737032) could not be eliminated by selective antibiotic treatment and remain as potential primary pathogens of WBD. Two of these have previously been implicated as causal agents in this and similar ‘white diseases’ in other locations and host species, including Vi. charchariae in WBD type II [11], and the Bacillus sp. in WBD in Indonesian acroporas [16].
By combining a culture-independent analysis with selective antibiotic treatment, we were able to identify a diverse pool of candidate pathogens involved in WBD. While several authors [30,31] have suggested that a wide range of normally commensal, non-specific microorganisms may become pathogenic under the right conditions, such as environmental stress-related reductions in immunity [32], these candidate pathogens were not detected in ND tissues (using the specific techniques in this study), suggesting that these potential pathogens are not a normal part of the commensal community. Most of these candidate pathogens appear to have no significant role in pathogenesis, whereas three potential bacterial primary pathogens and one specific ciliate secondary pathogen were identified. A key question is whether a combination of more than one of these three potential primary bacterial pathogens is required to maintain the disease state, although all three plus the ciliate consistently co-occurred in the disease in nature. Regardless, results from this study strongly suggest that at least one from this candidate group cause a systemic infection resulting in compromised health indicated by limited cell necrosis and increased apoptosis (both in the host tissues and the symbiotic algae) in advance of the disease lesion (figure 3). Interestingly, an increase in apoptosis has been identified previously in corals showing signs of WS [33], again highlighting similarities between these two diseases. The ciliate Phi. lucinda contributes to the pathology of the disease yet appears not to be a primary causal agent. Although these results highlight potential pathogens of WBD, challenge experiments such as Henle–Koch's postulates would be required to definitively prove causation. However, such challenge experiments must also control for the effects of the treatments on naturally occurring populations of these and other potential pathogens of WBD.
Although antibiotic treatments could be used as a potential cure for WBD in the field, extreme care would need to be taken as many microbes are known to develop resistance to antibiotics [34,35]. Furthermore, such treatments might have unwarranted effects on other host–microbe interactions in the natural environment. It would be unfeasible and unethical to apply antibiotic treatment at the regional and global scale of coral disease zoonoses, but it would probably be effective to use ampicillin or paromomycin sulfate treatment in specific circumstances where collateral effects could be minimized, for example in aquarium treatments.
4. Methods
Acropora cervicornis showing signs of WBD (type I) were monitored to show signs of progression. Only those with advancing lesions were used in the experiment. N = 2, 5 cm2 coral nubbins showing signs of WBD (type I) were placed in three individual aquaria per treatment. Therefore, n = 6 replicate coral nubbins were used for each treatment. All corals were maintained in the aquarium for 24 h prior to the start of the experiment to allow for acclimatization and to confirm lesion progression in diseased nubbins. Four types of antibiotics were used in treatments to determine their effects on the diseased corals; ampicillin, gentamicin, metronidazole and paromomycin sulfate. 100 µg ml−1 was used for all four antibiotics after preliminary laboratory trials on both bacteria and healthy corals. The antibiotics were added directly into tanks filled with 3 l of seawater collected from the original location of the corals. Repeat dosage was dissolved in 1.5 l of seawater every 12 h, and half the water in the experimental tanks was replaced with the new water. n = 6 corals were used per treatment (n = 2 per tank, three tanks). One set with WBD were left untreated in the tanks and sampled before all the tissue had been lost. Healthy corals were also collected and held in the aquaria for the duration of the experiment to address any tank effects on the health of the corals. Prior to the onset of the main experiment, healthy corals were also treated with the antibiotics at the same dose rates used within the experiment to ensure that the antibiotics were having no adverse effect on the corals. All these treatments survived to the end of the experiment with no visual appearance of tissue deterioration or discoloration.
Ampicillin belongs to the penicillin group of beta-lactam antibiotics, it is able to penetrate Gram-positive and some Gram-negative bacteria. It acts as a competitive inhibitor of the enzyme transpeptidase, which is needed by bacteria to make their cell walls. Inhibition of cell wall synthesis ultimately leads to cell lysis. Gentamicin targets mainly Gram-negative bacterium and inhibits protein synthesis. Metronidazole is a nitroimidazole antibiotic used particularly for anaerobic bacteria (particularly from the genus clostridium and bacteroides) and protozoa. Once metronidazole is taken up by the microorganisms, it is non-enzymatically reduced by reacting with reduced ferredoxin, which is generated by pyruvate oxidoreductase. Many of the reduced nitroso intermediates will form sulfinamides and thioether linkages with cysteine-bearing enzymes, thereby deactivating these critical enzymes. Paromomycin sulfate is a protein synthesis inhibitor and binds directly to the 16S rRNA. It is a broad spectrum antibiotic which is soluble in water.
Rates of application of ampicillin, gentamicin and metronidazole were measured twice daily at 10.00 and 16.00 for 6 days until the end of the experiment. Paromomycin sulfate was applied only twice on the first day owing to the cost of this antibiotic.
Sterile surgical gloves were worn at all times to avoid contamination. Samples were taken either when the corals had less than half their remaining tissue on the nubbins or at the end of the experiment. Samples were placed in Falcon tubes underwater and sealed. The water was then replaced with 100% ethanol and stored at −20°C until extraction.
(a). PCR and denaturing gradient gel electrophoresis
Extraction, PCR and denaturing gradient gel electrophoresis were undertaken as described in [16]. DNA was extracted from all samples using QIAGEN DNeasy blood and tissue kits, and bacterial 16S rRNA gene diversity was amplified using primers 357F and 518R. PCR protocol was as in [17]. Ciliate 18S rRNA gene diversity was amplified using primers CilF and CilDGGE-r. PCR protocol was as in [17]. For each of the above primer pairs, 30 µl PCR mixtures containing 1.5 mM MgCl2, 0.2 mM dNTP (Promega), 0.5 mM of each primer, 2.5 Ul of Taq DNA polymerase (QBiogene), incubation buffer and 20 ng of template DNA [17]. DGGE was performed as in [17] using the D-Code universal mutation detection system (Bio-Rad). PCR products were resolved on 10% (w/v) polyacrylamide gels for bacterial 16S rRNA gene diversity and 8% (w/v) for ciliate diversity. Bands of interest (those which explained the greatest differences/similarities between samples) were excised from DGGE gels, re-amplified with the same original primers, labelled using a big dye (Applied Biosystems) transformation sequence kit and sent to Genevision (Newcastle University, UK) for sequencing.
(b). Clone libraries and amplified ribosomal DNA restriction analysis screening
As all replicates within samples showed no significant differences in relation to their DGGE profiles, a random subset of only n = 3 replicates per treatment were further analysed using Clone Libraries. Almost complete 16S rRNA gene fragments were amplified from the DNA extracted using the ‘universal’ eubacterial 16S rRNA gene primers 27F and 1542R. PCR protocols were as in [27,36]. Amplified products, purified using the Qiagen PCR purification kit, were inserted into the pGEM-T vector system (Promega) and transformed into Escherichia coli JM109 cells. A total of 392 clones containing the 16S rRNA gene inserts were randomly selected from each sample/replicate, and boiled lysates were prepared as in [17]. PCR protocols were as in [17]. The products were then digested with the restriction enzymes HaeIII and RsaI (Promega; 4 µg of PCR product, 2 µl of restriction buffer, 0.2 µl of bovine serum albumin, 0.07 µl of HaeIII, 0.1 µl of RsaI and made up to 20 µl with sigma water for 2 h at 37°C then 10 min at 67°C). Restriction fragments were resolved by 3% agarose gel electrophoresis, visualized using a ultraviolet transilluminator and grouped based on restriction patterns. n = 10 representatives from each group were sequenced. Closest match of retrieved sequences was determined by RDP II similarity matching [37]. All sequences in this study have been deposited in GenBank and their unique accession numbers reported in the text.
(c). Total bacterial abundance
To estimate bacterial abundance, 1000 µl of tissue slurry was filtered through a 0.22 µm black polycarbonate filter and fixed with 100 µl of paraformaldehyde until analysis. These filters were stained with 100 μl DAPI solution (final concentration 5 μg ml−1) for 10 min, rinsed with phosphate-buffered solution, and viewed under epifluorescence microscopy (Nikon UK Ltd, Surrey, UK) at 1000× magnification using a DAPI-specific filter set. Counts on 50 fields of view were taken using an automatic cell counter (Cell C). The parameters were set to exclude any objects smaller than 0.03 μm2 and anything larger than 0.7 μm2. Counts were scaled up to the total area of the filter and calculated to give total bacterial abundance per volume of tissue on the diseased corals (cells cm3). Total amount of diseased tissue rather than complete coral nubbin surface area was used to account for the varying amount of tissue on the diseased samples as this could not be standardized at time of collection. All six nubbins were sampled for bacterial abundance with n = 3 subsamples taken from each coral and averaged to provide a cell density per sample.
(d). Histology
Samples were collected as for microbial analysis; however, tissue samples were preserved with 5% paraformaldehyde for 24 h then stored in 100% EtOH until resin embedding in LR white (r). Survey sections of each tissue type were stained with the general DNA stain toluidine blue. The location of bacteria was recorded using fluorescent in situ hybridization (FISH). For FISH, samples were stained and sectioned following the protocols in [38]. Oligonucleotide probes were purchased from Interactiva (http://www.interactiva.de) with an aminolink C6/MMT at the 5′ end. Four probes were used: the ‘universal’ eubacterial probes EUB338, EUB338-II, EUB338-III and the ‘non-sense probe’ NONEUB. The three eubacterial probes were used in an equimolar mix (EUBMIX), and the NONEUB probe was used singly. Samples of pure cultured E. coli were run alongside each section and for each staining protocol as a positive stain. Sections were viewed under epiflourescence microscopy with an FITC-specific filter block (Nikon UK Ltd, Surrey, UK) and images recorded using an integrating camera (model JVC KY-SSSB: Foster Findlay and Associates, Newcastle upon Tyne, UK). Further histological samples were analysed for signs of apoptosis using ISEL of fragmented DNA (in situ apoptosis detection kit S7101 Chemicon International, Inc. USA) as per [33], whereby apoptotic cells are brown. The stain nigrosin [39] was used for evaluating the extent of mass tissue necrosis, necrotic cells appear black/brown in coloration.
(e). Statistical analysis
Because the same experimental subjects (coral nubbins) were followed throughout the experiment, a two-way repeated-measure ANOVA was conducted to test the effect of each drug on WBD progression rates over time. The analysis was conducted with Statistica for Windows v. 6. An ANOSIM was conducted to test differences in 16S rRNA gene bacterial assemblage and 18S rRNA gene ciliate assemblage. Percentage similarity (SIMPER) was performed to determine which ribotypes better explained differences and/or similarities between sample types. Patterns of the 16S rRNA gene bacterial assemblages were represented on a multidimensional scaling plot.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Deborah Burn and Juan Jose Cruz-Motta for their assistance in the field and the reviewers who have greatly improved this paper throughout the process.
Funding statement
The work was supported by a grant from the Natural Environmental Research Council, UK (NE/E006949).
References
- 1.Wild C, et al. 2011. Climate change impedes scleractinian corals as primary reef ecosystem engineers. Mar. Freshw. Res. 62, 205–215. ( 10.1071/MF10254) [DOI] [Google Scholar]
- 2.Costanza R, et al. 1997. The value of the world's ecosystem services and natural capital. Nature 387, 253–260. ( 10.1038/387253a0) [DOI] [Google Scholar]
- 3.Hayes RL, Goreau NI. 1998. The significance of emerging diseases in the tropical coral reef ecosystem. Rev. Biol. Trop. 46, 173–185. [Google Scholar]
- 4.Rogers CS, Miller J. 2006. Permanent ‘phase shifts’ or reversible declines in coral cover? Lack of recovery of two coral reefs in St. John, US Virgin Islands. Mar. Ecol. Prog. Ser. 306, 103–114. ( 10.3354/meps306103) [DOI] [Google Scholar]
- 5.Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. 2007. The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. ( 10.1038/nrmicro1635) [DOI] [PubMed] [Google Scholar]
- 6.Geister J. 1977. The influence of wave exposure on the ecological zonation of Caribbean coral reefs. Proc. Int. Coral Reef Symp. 3, 23–29. [Google Scholar]
- 7.Lirman D, Bowden-kerby A, Schopmeyer S, Huntington B, Thyberg T, Gough M, Gough T, Gough R, Gough Y. 2010. A window to the past: documenting the status of one of the last remaining ‘megapopulations’ of the threatened staghorn coral Acropora cervicornis in the Dominican Republic. Aquat. Conserv. Mar. Freshw. Ecosyst. 20, 773–781. ( 10.1002/aqc.1146) [DOI] [Google Scholar]
- 8.Aronson RB, Precht WF. 2001. White-band disease and the changing face of Caribbean coral reefs. Hydrobiologia 460, 25–38. ( 10.1023/A:1013103928980) [DOI] [Google Scholar]
- 9.Bythell J, Sheppard C. 1993. Mass mortality of caribbean shallow corals. Mar. Pollut. Bull. 26, 296–297. [Google Scholar]
- 10.Gladfelter WB. 1982. White-band disease in Acropora palmate: implications for the structure and growth of shallow reefs. Bull. Mar. Sci. 32, 639–643. [Google Scholar]
- 11.Gil-Agudelo DL, Smith GW, Weil E. 2006. The white band disease type II pathogen in Puerto Rico. Rev. Biol. Trop. 54(Suppl. 3), 59–67.18457175 [Google Scholar]
- 12.Pantos O, Bythell JC. 2006. Bacterial community structure associated with white band disease in the elkhorn coral Acropora palmata determined using culture-independent 16S rRNA techniques. Dis. Aquat. Organ. 69, 79–88. ( 10.3354/dao069079) [DOI] [PubMed] [Google Scholar]
- 13.Casas V, Kline DI, Wegley L, Yu Y, Breitbart M, Rohwer F. 2004. Widespread association of a Rickettsiales-like bacterium with reef-building corals. Environ. Microbiol. 6, 1137–1148. ( 10.1111/j.1462-2920.2004.00647.x) [DOI] [PubMed] [Google Scholar]
- 14.Kline DI, Vollmer SV. 2011. White band disease (type I) of endangered Caribbean acroporid corals is caused by pathogenic bacteria. Sci. Rep. 1, 1–7. ( 10.1038/srep00007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams DE, Miller MW. 2005. Coral disease outbreak: pattern, prevalence and transmission in Acropora cervicornis. Mar. Ecol. Progr. Ser. 301, 119–128. ( 10.3354/meps301119) [DOI] [Google Scholar]
- 16.Hakim H, et al. 2012. Causative agents of white band disease from culturable bacterial community associated with healthy and diseased corals Acropora humilis and Acropora tortuosa from Karimunjawa Islands, Indonesia. Ecologia 2, 52 ( 10.3923/ecologia.2012.52.59) [DOI] [Google Scholar]
- 17.Sweet MJ, Bythell J. 2012. Ciliate and bacterial communities associated with white syndrome and brown band disease in reef building corals. Environ. Microbiol. 14, 2184–2199. ( 10.1111/j.1462-2920.2012.02746.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luna GM, Bongiorni L, Gili C, Biavasco F, Danovaro R. 2010. Vibrio harveyi as a causative agent of the white syndrome in tropical stony corals. Environ. Microbiol. Rep. 2, 120–127. ( 10.1111/j.1758-2229.2009.00114.x) [DOI] [PubMed] [Google Scholar]
- 19.Sussman M, Willis BL, Victor S, Bourne DG. 2008. Coral pathogens identified for white syndrome (WS) epizootics in the Indo-Pacific. PLoS ONE 3, e2393 ( 10.1371/journal.pone.0002393) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atad I, Zvuloni A, Loya Y, Rosenberg E. 2012. Phage therapy of the white plague-like disease of Favia favus in the Red Sea. Coral Reefs 31, 665–670. [Google Scholar]
- 21.Barash Y, et al. 2005. Bacterial strain BA-3 and a filterable factor cause a white plague-like disease in corals from the Eilat coral reef. Aquat. Microb. Ecol. 40, 183–189. ( 10.3354/ame040183) [DOI] [Google Scholar]
- 22.Bythell J, Pantos O, Richardson L. 2004. White plague, white band, and other “white” diseases. In Coral Health and Disease (eds E Rosenberg, Y Loya), p. 351–365. Berlin, Germany: Springer.
- 23.Denner EBM, et al. 2003. Aurantimonas coralicida gen. nov., sp nov., the causative agent of white plague type II on Caribbean scleractinian corals. Int. J. Syst. Evol. Microbiol. 53, 1115–1122. ( 10.1099/ijs.0.02359-0) [DOI] [PubMed] [Google Scholar]
- 24.Efrony R, Atad I, Rosenberg E. 2009. Phage therapy of coral white plague disease: properties of phage BA3. Curr. Microbiol. 58, 139–145. ( 10.1007/s00284-008-9290-x) [DOI] [PubMed] [Google Scholar]
- 25.Pantos O, Cooney RP, Le Tissier MDA, Barer MR, O'Donnell AG, Bythell JC. 2003. The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis. Environ. Microbiol. 5, 370–382. ( 10.1046/j.1462-2920.2003.00427.x) [DOI] [PubMed] [Google Scholar]
- 26.Frias-Lopez J, Bonheyo GT, Jin Q, Fouke BW. 2003. Cyanobacteria associated with coral black band disease in Caribbean and Indo-Pacific reefs. Appl. Environ. Microbiol. 69, 2409–2413. ( 10.1128/AEM.69.4.2409-2413.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooney RP, Pantos O, Le Tissier MDA, Barer MR, O'Donnell AG, Bythell JC. 2002. Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ. Microbiol. 4, 401–413. ( 10.1046/j.1462-2920.2002.00308.x) [DOI] [PubMed] [Google Scholar]
- 28.Sweet MJ, et al. 2013. Assessment of the microbial communities associated with white syndrome and brown jelly syndrome in aquarium corals. J. Zoo Aquar. Res. 1, 21–27. [Google Scholar]
- 29.Cooper W, Lirman D, Schmale M, Lipscomb D. 2007. Consumption of coral spat by histophagic ciliates. Coral Reefs 26, 249–250. ( 10.1007/s00338-007-0196-z) [DOI] [Google Scholar]
- 30.Casadevall A, Pirofski LA. 2003. The damage-response framework of microbial pathogenesis. Nat. Rev. Microbiol. 1, 17–24. ( 10.1038/nrmicro732) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pirofski L-A, Casadevall A. 2008. The damage-response framework of microbial pathogenesis and infectious diseases. Gi Microbiota Regul. Immune Syst. 635, 135–146. ( 10.1007/978-0-387-09550-9_11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer CV, Bythell JC, Willis BL. 2010. Levels of immunity parameters underpin bleaching and disease susceptibility of reef corals. FASEB J. 24, 1935–1946. ( 10.1096/fj.09-152447) [DOI] [PubMed] [Google Scholar]
- 33.Ainsworth TD, Kvennefors EC, Blackall LL, Fine M, Hoegh-Guldberg O. 2007. disease and cell death in white syndrome of acroporid corals on the great barrier reef. Mar. Biol. 151, 19–29. ( 10.1007/s00227-006-0449-3) [DOI] [Google Scholar]
- 34.Al-Bahry SN, Al-Zadjali MA, Mahmoud IY, Elshafie AE. 2012. Biomonitoring marine habitats in reference to antibiotic resistant bacteria and ampicillin resistance determinants from oviductal fluid of the nesting green sea turtle, Chelonia mydas. Chemosphere 87, 1308–1315. ( 10.1016/j.chemosphere.2012.01.051) [DOI] [PubMed] [Google Scholar]
- 35.Eisenberg JNS, et al. 2012. In-roads to the spread of antibiotic resistance: regional patterns of microbial transmission in northern coastal Ecuador. J. R. Soc. Interface 9, 1029–1039. ( 10.1098/rsif.2011.0499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galkiewicz JP, Kellogg CA. 2008. Cross-kingdom amplification using bacteria-specific primers: complications for studies of coral microbial ecology. Appl. Environ. Microbiol. 74, 7828–7831. ( 10.1128/AEM.01303-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cole JR, et al. 2009. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37(Suppl. 1), D141–D145. ( 10.1093/nar/gkn879) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bythell JC, et al. 2002. Histopathological methods for the investigation of microbial communities associated with disease lesions in reef corals. Lett. Appl. Microbiol. 34, 359–364. ( 10.1046/j.1472-765X.2002.01097.x) [DOI] [PubMed] [Google Scholar]
- 39.Bjorndahl L, Soderlund I, Kvist U. 2003. Evaluation of the one-step eosin-nigrosin staining technique for human sperm vitality assessment. Hum. Reproduct. 18, 813–816. ( 10.1093/humrep/deg199) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.