Abstract
In convergent evolution, similar phenotypes evolve repeatedly in independent populations, often reflecting adaptation to similar environments. Understanding whether convergent evolution proceeds via similar or different genetic and developmental mechanisms offers insight towards the repeatability and predictability of evolution. Oceanic populations of threespine stickleback fish, Gasterosteus aculeatus, have repeatedly colonized countless freshwater lakes and streams, where new diets lead to morphological adaptations related to feeding. Here, we show that heritable increases in branchial bone length have convergently evolved in two independently derived freshwater stickleback populations. In both populations, an increased bone growth rate in juveniles underlies the convergent adult phenotype, and one population also has a longer cartilage template. Using F2 crosses from these two freshwater populations, we show that two quantitative trait loci (QTL) control branchial bone length at distinct points in development. In both populations, a QTL on chromosome 21 controls bone length throughout juvenile development, and a QTL on chromosome 4 controls bone length only in adults. In addition to these similar developmental profiles, these QTL show similar chromosomal locations in both populations. Our results suggest that sticklebacks have convergently evolved longer branchial bones using similar genetic and developmental programmes in two independently derived populations.
Keywords: convergent evolution, parallel evolution, quantitative trait loci, skeletal development, endochondral bone, stickleback
1. Introduction
Independent populations that converge on similar evolved phenotypes may do so by using similar genetic and developmental mechanisms, suggesting that evolution is, at times, constrained and predictable [1,2]. When convergent phenotypic evolution is caused by parallel genetic mechanisms, the parallelism may occur on a number of different hierarchical levels, including changes in the same nucleotide, gene, genetic pathway or genomic region (reviewed in [2–4]; for examples, see [5–8]). While numerous cases of convergent evolution have been documented across natural and experimental populations of animals, plants and microbes, fewer studies have investigated whether these convergently evolved phenotypes arise in the same way during development [9]. Furthermore, most studies of convergent evolution have focused on traits with a simple genetic architecture, and less is known about whether more complex traits, which are more common in nature, convergently evolve via parallel developmental genetic features.
In vertebrates, the skeleton contributes to organismal form and function, and evolved changes in skeletal elements occur repeatedly as populations adapt to new environments. The skeleton forms largely from two types of bone: endochondral, which develops from a cartilage template, and dermal, which ossifies directly without a cartilage intermediate [10]. Atchley & Hall [11] proposed that skeletal evolution may proceed through a number of cellular mechanisms (e.g. the size of the cartilage template or the rate of bone growth). In support of this proposal, a dramatic difference in jaw size between quails and ducks results from several differences in the specification and morphogenesis of the midbrain and midbrain neural crest cells from which the jaw is derived [12]. In Anolis lizards, however, a smaller number of cellular mechanisms appear to underlie convergent skeletal evolution. On at least four islands of the Caribbean, Anolis limb morphologies have repeatedly evolved in different ecological habitats [13]. While multiple pathways in pre- and post-embryonic development could contribute to differing limb length, increased adult limb size in four different long-limbed species arose from an increase in the size of the embryonic limb template, followed by growth rates equal to those in shorter-limbed species [14]. Whether this mechanism of evolved bone length differences is seen in other convergently evolved skeletal changes is largely unknown.
The threespine stickleback (Gasterosteus aculeatus) provides an excellent model system for studying both the developmental and genetic basis of convergent skeletal evolution. Ancestral marine sticklebacks have colonized thousands of freshwater environments throughout the Northern Hemisphere and have evolved numerous adaptations to these new freshwater environments [15]. For example, freshwater sticklebacks have repeatedly evolved changes to their head skeletons to improve feeding efficiency on new foods in freshwater environments, including convergent decreases in gill raker number [16–18], as well as increases in jaw width [17,19] and suction feeding index [20].
Here, we hypothesized that other trophic skeletal elements may also differ between marine and freshwater sticklebacks. The branchial skeleton (figure 1, adapted from [21]) is primarily made up of bilateral, segmentally reiterated bones: five ventral pairs (ceratobranchials, CB1–CB5) and four dorsal pairs (epibranchials, EB1–EB4). In fish, these bones arise from neural crest cells in the pharyngeal arches during development, and the dorsal and ventral bones are segmental homologues of the upper and lower jaw, respectively [22,23]. These long dorsal and ventral bones of the branchial skeleton are endochondral and resemble mammalian long bones (e.g. the femur) in appearance [24]. In fish, the branchial cartilages start to form late in embryogenesis, just before hatching [23,25]. As development continues, this cartilage is mostly replaced with bone deposited by osteoblasts that originate both outside and within the cartilage template [26]. The bones then elongate as the fish grows larger [24]. Thus, two key developmental processes contribute to the length of the bone: the establishment of the cartilage template early in embryonic development and the rate of subsequent bone growth.
Figure 1.
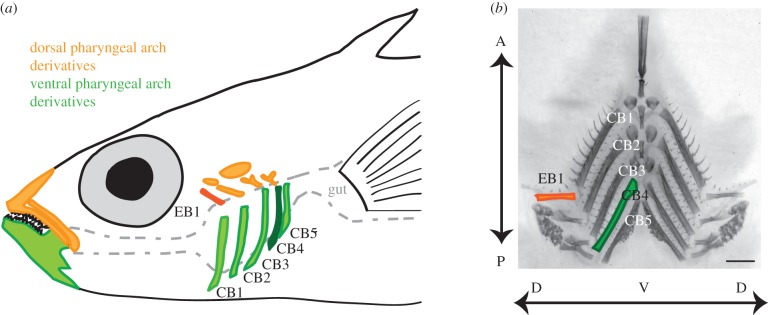
Anatomy of stickleback branchial bones. (a) Fish ingest food into the buccal cavity, which is flanked bilaterally by dorsal (epibranchial, EB; orange) and ventral (ceratobranchial, CB; green) pharyngeal arch bones between the mouth and the gut. Also shown are the upper and lower oral jaw, segmental homologues of the branchial bones in the first pharyngeal arch. In some experiments, we focused on the highlighted bones: EB1 and CB4, the dorsal and ventral bones with the strongest detected genetic effects. Some bones have been omitted for clarity. (b) Dissected and flat-mounted Alizarin red-stained branchial skeleton with dorsal (EB1) and ventral (CB1–CB5) bones indicated. Dorsal–ventral (DV) and anterior–posterior (AP) axes are labelled with arrows. Scale bar, 1 mm.
A previous genome-wide linkage mapping study of the genetic basis of skeletal variation in sticklebacks identified 14 quantitative trait loci (QTL) with significant effects on the length of branchial bones in a marine-by-freshwater F2 cross, including two QTL on chromosomes 4 and 21 with large effects [27]. Combined, these two QTL explain approximately 27% of the variance in length of the dorsal EB1 and approximately 25% of variance in length of the ventral bones. Most (11) of these QTL, including both of the large-effect QTL, had effects in the same direction, with freshwater alleles conferring longer bones [27]. However, this study did not measure the bone length phenotypes of the parental populations. Orr [28] proposed that a concerted sign of QTL effect indicates a trait is under natural selection, as similar directions of effect would be unlikely to be observed by chance. Here, we test the hypothesis that the two previously identified large-effect QTL on chromosomes 4 and 21 are used in a second independently derived freshwater population. By studying the developmental trajectories of evolved increases in bone length, as well as the developmental timing of two bone length QTL, we also test whether similar developmental and genetic effects contribute to these evolved increases in bone length.
2. Material and methods
(a). Wild collections
Wild anadromous marine fish were collected from the Little Campbell River (LITC) in British Columbia under a fish collection permit from the British Columbia Ministry of Environment (permit #SU08-44549). Wild freshwater fish were collected from Fishtrap Creek (FTC) in Washington under fish scientific collection from the Washington Department of Fish and Wildlife (permit #08-284). Wild sticklebacks were collected in the summer of 2008. All wild and laboratory-reared fish were euthanized with 0.08% Tricaine and stored in 100% ethanol until staining and dissection.
(b). Fish husbandry and crosses
For the FTC × LITC cross, a wild male FTC fish was crossed to a wild female LITC. For the Paxton Benthic (PAXB, British Columbia, Canada) × LITC cross, a laboratory-reared male offspring of wild PAXB fish was crossed to two wild LITC females. Adult F1 fish were then intercrossed to their siblings to create F2 families, which were grown to ages of 20, 40 and 80 days post-fertilization (dpf), or adults (see the electronic supplementary material, table S1). All fish were raised in 3 ppt salinity (approx. 10% seawater) at 18°C in 110 l (29 gallon) tanks. Fish were fed a diet of live Artemia as young fry, live Artemia and frozen Daphnia as juveniles, and frozen bloodworms and Mysis shrimp as adults.
(c). Phenotyping, genotyping and quantitative trait loci analysis
Detailed descriptions of phenotyping, genotyping and QTL analysis can be found in the electronic supplementary material.
(d). Statistical analysis
All statistical analyses were performed using the R statistical software package (www.r-project.org). QTL analysis was performed using R/qtl (www.rqtl.org).
3. Results
(a). Population differences in bone length
To test the hypothesis that wild marine and freshwater fish differ in branchial bone length, we analysed wild-caught marine (LITC) and freshwater (FTC) sticklebacks for differences in length of the dorsal (epibranchial, EB1) and ventral (ceratobranchial, CB1–CB5) branchial long bones (figure 1). All six branchial long bones differed significantly in length (figure 2a), with freshwater fish having longer bones relative to standard length than marine fish (ventral bones were 8.8–17.1% longer; dorsal bone was 23.8% longer in 60 mm fish). Because a strong genetic component of bone length was previously observed in a large F2 cross [27], we next hypothesized that these differences in bone length were heritable in multiple freshwater populations. We tested these hypotheses by raising adult marine and freshwater fish under common laboratory-reared conditions. Supporting our hypotheses, fish from both FTC and a second freshwater population (PAXB) raised in the laboratory had significantly longer branchial bones than marine LITC fish (figure 2b; electronic supplementary material, figure S1). FTC bones were consistently slightly longer than PAXB bones, and the increase in FTC over LITC bone length ranged from 7.6 to 19.7% for ventral bones and was 33.6% for EB1.
Figure 2.
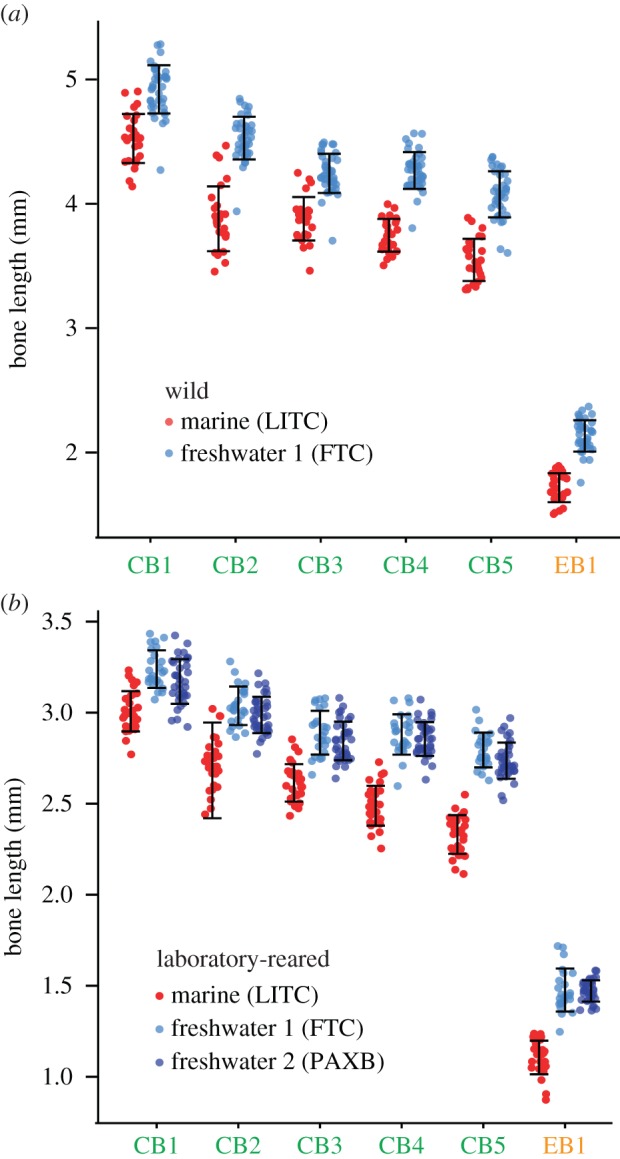
Heritable increases in branchial bone lengths in freshwater sticklebacks. (a) Wild Fishtrap Creek (FTC) fish have significantly longer dorsal (EB1) and ventral (CB1–CB5) branchial bones relative to size-matched wild marine Little Campbell (LITC) fish. All six bones are significantly longer in FTC fish than LITC fish (p < 10−10 for all bones, nLITC = 27, nFTC = 40, Welch's T-test). (b) Increased bone lengths are heritable in adult laboratory-reared fish, and longer bones are also found in a second laboratory-reared freshwater population, Paxton Benthic (PAXB). All bones are significantly longer in each freshwater population relative to LITC marine (Tukey HSD: p < 10−5, nLITC = 32, nFTC = 25, nPAXB = 36). PAXB and FTC bone lengths do not significantly differ (p > 0.05). Error bars = standard deviation of the mean. Red, LITC; light blue, FTC; dark blue, PAXB.
In wild fish, LITC bones were sexually dimorphic, with males having longer bones than females, but FTC bone lengths did not differ significantly between sexes (electronic supplementary material, table S2). In laboratory-reared fish, LITC branchial bones were all sexually dimorphic; some ventral FTC bones (CB1, CB2 and CB4) were sexually dimorphic, while no PAXB bones were significantly sexually dimorphic. This observation matches previous findings for sticklebacks: marine fish are sexually dimorphic for body shape and feeding kinematic phenotypes, while freshwater fish have lost this sexual dimorphism, with both sexes having an overall phenotype more similar to marine males [29,30].
(b). Developmental basis of bone length differences
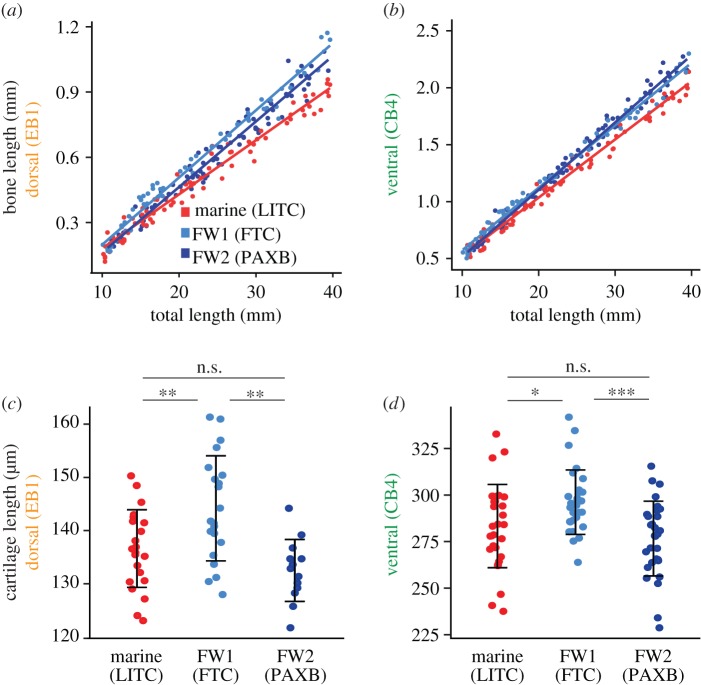
We hypothesized that stickleback bone length differences, like evolved Anolis limb length [14], would manifest during development as transposition of the y-intercept, but not slope, of a regression of bone length against standard length. We collected laboratory-reared fish from each population at regular developmental time points, resulting in fish varying in total length from 10 to 40 mm. We looked for differences in bone growth rate and initial bone size using an ANCOVA with standard length as the covariate and population as an interacting factor. Contrary to our prediction, we observed significant differences in the slopes (population × standard length interaction term) of dorsal and posterior ventral bone lengths relative to standard length between marine and freshwater fish, suggesting that freshwater bones grow more rapidly relative to body size (figure 3a,b; electronic supplementary material, figure S2 and table S5). Thus, unlike in Anolis lizards, the convergent evolution of increased bone length in two derived freshwater stickleback populations appears to use a similar faster bone growth rate in both populations.
Figure 3.
Developmental basis of dorsal and ventral bone length differences. (a,b) Developmental time course of (a) dorsal (EB1) and (b) ventral (CB4) bone lengths plotted against total length of laboratory-reared fish under five months of age. Both bones show statistically significant differences in slope (bone growth rate) as well as y-intercept between marine and freshwater. ANCOVA statistics are shown in the electronic supplementary material, table S5; additional bones are shown in the electronic supplementary material, figure S2. Red, LITC; light blue, FTC; dark blue, PAXB. (c) EB1 and (d) CB4 cartilages are longer in FTC relative to LITC and PAXB fry (Tukey HSD p < 0.05 for LITC-FTC and PAXB-FTC comparisons of both cartilages). In (c), the FTC fish were slightly shorter in total length than the LITC and PAXB fish (Tukey HSD test p < 0.05), which makes the cartilage size increase even greater relative to body size. Error bars = standard deviation. Asterisks indicate Tukey HSD p-values: n.s., not significant; *p < 0.05, **p < 0.01, ***p < 0.001.
The significant differences in y-intercepts in the bone development time courses (electronic supplementary material, table S5) led us to hypothesize that the cartilage templates that prefigure branchial bones may be larger in freshwater fish relative to marine fish. For ventral cartilages, we focused on CB4 because it had a large marine–freshwater difference and had strong genetic effects in a previous cross [27]. We raised FTC and LITC fry to stage 26 (approx. 10 dpf) [31] to measure the CB4 cartilage and stage 28 (approx. 13–14 dpf) to measure EB1 cartilage. We found that both cartilage templates were longer in FTC relative to both LITC and PAXB (figure 3c,d). Thus, despite the convergent increased bone growth rates, one unique developmental difference contributes to the convergent evolution, with one freshwater population (FTC) but not a second (PAXB) evolving a longer cartilage template early in development.
(c). Genetic basis of bone length differences
QTL mapping provides a powerful first test of possible parallel genetic mechanisms underlying convergent evolution. We hypothesized that previously identified bone length QTL might be re-used in multiple freshwater stickleback populations due to extensive sharing of the genetic basis of evolved traits in stickleback populations [32,33], and the similar increased bone growth rates in FTC and PAXB. Because there are probably multiple developmental mechanisms that can be altered to change bone length [11], we further predicted that these QTL might exert different effects at specific points in development.
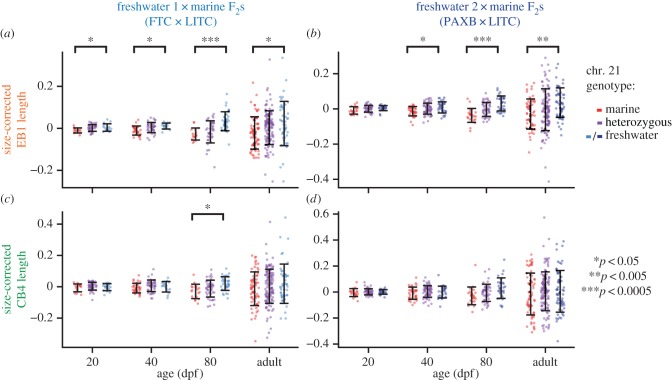
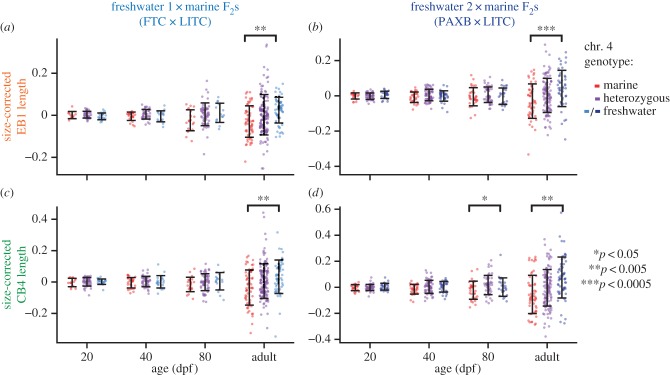
We focused on the two largest-effect QTL controlling adult bone length in a previous cross (chromosomes 4 and 21 [27]) and observed strikingly similar developmental profiles of these QTL in our two crosses. We raised F2 fish to four time points (20, 40 and 80 dpf and adults, see the electronic supplementary material, table S1), and tested for the effect of chromosomes 4 and 21 at each time point using the peak markers from the previous cross [27]. We found that freshwater alleles of chromosome 21 increased dorsal bone length at all stages in the FTC × LITC cross and at all stages except 20 dpf in the PAXB × LITC cross (figure 4; electronic supplementary material, table S6; ANOVA p < 0.05 for marker Stn489 on chromosome 21), suggesting that the freshwater allele of this QTL acts relatively early in development to increase bone size. The 20 dpf time point in the PAXB × LITC cross strongly trends in the same direction (p = 0.06). The dorsal effect of chromosome 21 was strongest at 80 dpf in both crosses. Chromosome 21 controlled ventral (CB4) bone length at the 80 dpf timepoint in the FTC × LITC cross and was nearly significant (p = 0.056) in the PAXB × LITC cross at 80 dpf. These results suggest that the freshwater allele may specifically increase both dorsal and ventral bone growth rates during juvenile stages. By contrast, chromosome 4 exhibited effects on dorsal and ventral bones only in adults of the two crosses (figure 5; electronic supplementary material, table S6; ANOVA p < 0.05), suggesting that this QTL acts later in bone development. In both crosses, the effect of the chromosome 4 QTL was greatest on CB4 (electronic supplementary material, figure S3).
Figure 4.
Similar developmental effects of chromosome 21 QTL in two independently derived freshwater populations. F2 fish from two marine × freshwater F2 crosses ((a,c) FTC × LITC; (b,d) PAXB × LITC) were raised to four time points (given in days post-fertilization, dpf; or adults > 150 dpf) and tested for effects of chromosome 21 genotype on size-corrected bone length residuals. See the electronic supplementary material, table S1 for a summary of the fish included in each time point. (a,b) Chromosome 21 controlled dorsal (EB1) bones at all time points in the FTC cross and at all time points except 20 dpf in the PAXB cross, and the effect was strongest at 80 dpf in both crosses. (c,d) The effect of chromosome 21 on ventral bone length (CB4) was significant at 80 dpf in the FTC cross and nearly significant at 80 dpf in the PAXB cross. ANOVA p-values for the marker Stn489 are indicated with asterisks when significant (p < 0.05; see the electronic supplementary material, table S6 for complete listing of ANOVA results for all branchial bones). Error bars = standard deviation.
Figure 5.
Similar developmental effects of chromosome 4 QTL in two independently derived freshwater populations. F2 fish from two crosses ((a,c) FTC × LITC; (b,d) PAXB × LITC) were raised to four time points (given in dpf for sub-adults) and tested for effects of chromosome 4 genotype on size-corrected bone length residuals. In both crosses, a chromosome 4 QTL was only significant in adults. ANOVA p-values for the marker Stn382 are indicated with asterisks when significant (p < 0.05; see the electronic supplementary material, table S6 for complete listing of ANOVA results). Error bars = standard deviation.
Finally, to test whether the QTL overlap in the two crosses, we genotyped markers across each chromosome and tested for association with bone length, focusing on the ventral effect of chromosome 4 and dorsal effect of chromosome 21. In support of a parallel genetic basis of these QTL, we saw similar localization of the two QTL in the two crosses (figure 6). The chromosome 4 peak marker from the PAXB × LITC cross (Stn382) was identical to the previously identified peak marker [27] and was within 3 cM (6 Mb) of the peak marker of the FTC × LITC cross (Stn42). The peak chromosome 21 marker from the PAXB × LITC cross (Stn491) was only 0.9 cM (0.9 Mb) away from the peak marker of the FTC × LITC cross (Stn489), which was the peak marker in the previous study. Furthermore, the dorsal chromosome 21 and ventral chromosome 4 QTL are additive in both crosses (dominances between −0.15 and 0.21; electronic supplementary material, table S7). Combined with the QTL developmental profiles, these localization and dominance data suggest that FTC and PAXB share several parallel genetic features for evolved bone length gain, including overlapping QTL on chromosomes 4 and 21.
Figure 6.
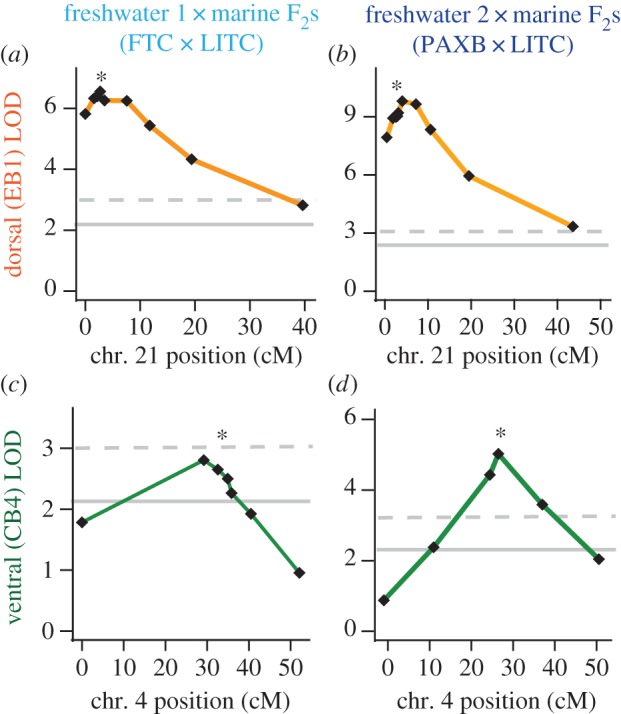
Similar localization of chromosome 21 dorsal bone QTL and chromosome 4 ventral bone QTL in two crosses. The chromosome 21 QTL was mapped in fish from the 40 dpf, 80 dpf and adult time points in each cross, while the chromosome 4 QTL was mapped in adults only. The significance threshold of α = 0.05, calculated based on 10 000 permutations of the phenotype data, is shown as a solid grey line. The 0.01 significance threshold is shown as a dashed line. The 0.001 significance thresholds were 4.20, 3.92, 4.10 and 4.21 for panels (a)–(d), respectively. Genetic markers (black diamonds) from left to right: (a) chr21_0049, Stn487, Stn423, Stn489, Stn491, Stn424, Stn425, chr21_0991, chr21_1170; (b) chr21_0049, chr21_0163, Stn487, Stn423, Stn489, Stn491, Stn424, Stn425, chr21_0997, chr21_1136; (c) Stn38, Stn42, Stn382, Stn46, Stn266, Stn253, chr4_280; (d) Stn38, Stn40, Gac4147, Stn382, Stn253, chr4_280. Marker Stn489 is asterisked for chromosome 21 and marker Stn382 is asterisked for chromosome 4 (see the electronic supplementary material, table S3 for a linkage map of each cross).
4. Discussion
(a). A heritable increase in branchial bone length in two freshwater stickleback populations is likely to be a trophic adaptation
A previous QTL mapping study found that most (11/14) freshwater alleles controlling stickleback branchial bone length produced longer bones [27], suggesting increased branchial bone length is under natural selection in freshwater environments. Supporting this prediction, we show that marine and freshwater bone lengths differ in the wild, and that two populations of freshwater stickleback show strongly heritable increases in branchial bone length. This elongation of branchial bones may facilitate the processing of larger prey items in freshwater by providing a larger buccal cavity for food to pass through, generating greater crushing force and/or offering increased muscle attachment area for the crushing of freshwater prey. While many studies have focused on evolutionary loss, these evolved increases in bone length demonstrate that despite the predictable loss of several skeletal elements (including gill rakers, dorsal spines and armour plates) in freshwater environments [18,32,34], other parts of the skeleton (i.e. the branchial bones) increase in size despite the much lower environmental calcium concentration in freshwater. In both freshwater populations studied here, the increased bone length differences are most pronounced in the dorsal (EB1) and posterior ventral bones (CB4 and CB5 demonstrate larger marine–freshwater differences than the more anterior three CBs). These findings suggest that the entire branchial skeleton is not uniformly enlarged relative to standard length in freshwater fish, but rather that independent genetic and developmental mechanisms have led to modular changes in the relative sizes of bones in the branchial skeleton. Heritable and similarly modular increases in bone length in two independent freshwater populations suggest that this trait may be adaptive in these environments [35].
(b). A convergent increase in bone growth rate underlies bone elongation in freshwater sticklebacks
Here, we find two developmental mechanisms of evolved bone elongation (increased cartilage template size and bone growth rate) are at work in freshwater stickleback populations. Relative to marine fish, both freshwater populations have evolved an increased bone growth rate. All PAXB branchial bones and dorsal and posterior ventral FTC branchial bones have an increased growth rate relative to marine bones. Early cartilage template size is also increased in FTC freshwater fish. Therefore, the convergent evolution of these independent stickleback populations uses one shared developmental feature (increased bone growth rate) as well as at least one unique feature (increased cartilage template size in only one freshwater population). Differences in juvenile bone growth rates have been observed in the limbs of large and small mouse strains [36], and the elongated craniofacial bones of needlefish [37]. Multiple aspects of chondrocyte hypertrophy (the enlargement of chondrocytes that promotes bone growth) are altered to produce elongated digits in bats [38] and elongated limbs in jerboas [39]. Thus, developmental modulation of bone growth rates seems to be a shared mechanism of altering skeletal proportions in multiple taxa, including sticklebacks.
(c). Shared quantitative trait loci on chromosomes 4 and 21 suggest a parallel developmental genetic basis for freshwater bone length increase
Consistent with the convergent increased bone growth rate in two freshwater populations, these populations also share two overlapping QTL with strong effects on bone length at various stages of development. These two QTL, initially identified in the PAXB freshwater population [27], were successfully replicated here by crossing a different PAXB fish to a different marine background, and also were observed in a second freshwater population, Fishtrap Creek. The developmental profiles of the QTL are remarkably similar between the two crosses. The effect of chromosome 4 is only seen in adult bones in both crosses. This QTL may only act late in development, or its earlier effects may only be apparent when fish reach a larger size. By contrast, chromosome 21 seems to exert its effects earlier than chromosome 4 in both crosses. Thus, similar developmental genetic features underlie the convergent evolution of longer branchial bones, suggesting that even complex traits can have at least partially predictable genetic bases.
Although QTL in vertebrates are typically mapped in adults, a handful of studies have linked adult phenotypes to changes in juveniles. QTL for stickleback juvenile pigmentation and standard length have been identified [40]. In a finding similar to ours, a QTL controls shank growth rate in chickens during a specific time period of juvenile development [41]. Additionally, a QTL controlling adult hair colour in beach mice can be traced to differential expression of the Agouti gene early in development [42]. These studies demonstrate that genetic changes that manifest at specific developmental stages can contribute to differences in final adult phenotype.
Future fine-mapping work will determine whether FTC and PAXB share the alleles on chromosomes 4 and 21 that control bone size. Since freshwater stickleback populations are derived from a large oceanic ancestral population and often share alleles controlling evolved morphological changes [32,33,43,44], we parsimoniously hypothesize that the same alleles are at work in the FTC and PAXB populations. This recycling of ancestral alleles to produce a convergent phenotype has been called ‘collateral’ evolution [3]. This hypothesis is supported by the similar developmental profiles of the QTL, and the similarities in chromosomal location and dominances of the QTL in each cross. However, sticklebacks have also been shown to independently evolve alleles in multiple populations in the case of pelvic reduction [45], so the possibility remains that unique alleles have evolved in each population.
In conclusion, we find evidence of similar genetic and developmental properties underlying evolved increases in bone length in two independently derived freshwater stickleback populations. Both derived freshwater populations share an increased rate of growth of some bones relative to the bones of their marine counterparts, and the two QTL on chromosomes 4 and 21 demonstrate strikingly similar effects throughout development in crosses of each population. Our developmental genetic evidence supports a model that the same chromosome 4 and 21 genomic regions were selected independently in two freshwater populations to produce quantitative changes in a convergently evolved trophic phenotype. Future studies of other freshwater populations and crosses will test whether this evolved gain trait and the use of bone length QTL on chromosomes 4 and 21 are predictable features of freshwater adaptation.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Monica Jimenez, Nihar Patel, Brittany Bartolome, Jessica Grindheim and Jiyeon Baek for assistance in genotyping and phenotyping; Anthony Lee and Patrick Lee for expertise in fish husbandry; and David Kingsley, Dolph Schluter, and Alex Pollen for help and support in wild fish collection.
All animal work was approved by the Institutional Animal Care and Use Committees of the University of California-Berkeley or Stanford University (protocol numbers R330 and 13834).
Data accessibility
All data are available as an electronic supplementary material file.
Funding statement
This work was supported in part by the NIH (R01-DE021475), a March of Dimes Basil O'Connor Starter Scholar Award, a Hellman Family Faculty award (C.T.M.), an NIH Predoctoral Training grant (5T32GM007127) to P.A.E., A.M.G. and P.A.C., and National Science Foundation Graduate Research Fellowships (A.M.G. and P.A.C.).
References
- 1.Losos JB. 2011. Convergence, adaptation, and constraint . Evolution 65, 1827–1840. ( 10.1111/j.1558-5646.2011.01289.x) [DOI] [PubMed] [Google Scholar]
- 2.Stern DL, Orgogozo V. 2008. The loci of evolution: how predictable is genetic evolution? Evolution 62, 2155–2177. ( 10.1111/j.1558-5646.2008.00450.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stern DL. 2013. The genetic causes of convergent evolution . Nat. Rev. Genet. 14, 751–764. ( 10.1038/nrg3483) [DOI] [PubMed] [Google Scholar]
- 4.Martin A, Orgogozo V. 2013. The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation . Evolution 67, 1235–1250. ( 10.1111/evo.12081) [DOI] [PubMed] [Google Scholar]
- 5.Jones FC, et al. 2012. The genomic basis of adaptive evolution in threespine sticklebacks . Nature 484, 55–61. ( 10.1038/nature10944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steiner CC, Weber JN, Hoekstra HE. 2007. Adaptive variation in beach mice produced by two interacting pigmentation genes . PLoS Biol. 5, e219 ( 10.1371/journal.pbio.0050219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldman CR, Brodie ED, Brodie ED, Pfrender ME. 2012. Constraint shapes convergence in tetrodotoxin-resistant sodium channels of snakes . Proc. Natl Acad. Sci. USA 109, 4556–4561. ( 10.1073/pnas.1113468109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tishkoff SA, et al. 2007. Convergent adaptation of human lactase persistence in Africa and Europe . Nat. Genet. 39, 31–40. ( 10.1038/ng1946) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall BK. 2012. Parallelism, deep homology, and evo-devo . Evol. Dev. 14, 29–33. [DOI] [PubMed] [Google Scholar]
- 10.Hall BK. 2005. Bones and cartilage: developmental and evolutionary skeletal biology. San Diego, CA: Academic Press. [Google Scholar]
- 11.Atchley WR, Hall BK. 1991. A model for development and evolution of complex morphological structures . Biol. Rev. 66, 101–157. ( 10.1111/j.1469-185X.1991.tb01138.x) [DOI] [PubMed] [Google Scholar]
- 12.Fish JL, Sklar RS, Woronowicz KC, Schneider RA. 2014. Multiple developmental mechanisms regulate species-specific jaw size . Development 141, 674–684. ( 10.1242/dev.100107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Losos J. 2011. Lizards in an evolutionary tree: ecology and adaptive radiation of anoles. Berkeley, CA: University of California Press. [Google Scholar]
- 14.Sanger TJ, Revell LJ, Gibson-Brown JJ, Losos JB. 2012. Repeated modification of early limb morphogenesis programmes underlies the convergence of relative limb length in Anolis lizards . Proc. R. Soc. B 279, 739–748. ( 10.1098/rspb.2011.0840) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster SA, Bell MA. 1994. The evolutionary biology of the threespine stickleback. New York, NY: Oxford University Press. [Google Scholar]
- 16.Hagen D, Gilbertson L. 1972. Geographic variation and environmental selection in Gasterosteus aculeatus L. in the Pacific Northwest, America . Evolution 26, 32–51. ( 10.2307/2406981) [DOI] [PubMed] [Google Scholar]
- 17.Schluter D, McPhail JD. 1992. Ecological character displacement and speciation in sticklebacks . Am. Nat. 140, 85–108. ( 10.1086/285404) [DOI] [PubMed] [Google Scholar]
- 18.Gross HP, Anderson J. 1984. Geographic variation in the gillrakers and diet of European threespine sticklebacks, Gasterosteus aculeatus . Copeia 1, 87–97. ( 10.2307/1445038) [DOI] [Google Scholar]
- 19.Lavin P, McPhail J. 1986. Adaptive divergence of trophic phenotype among freshwater populations of the threespine stickleback (Gasterosteus aculeatus) . Can. J. Fish. Aquat. Sci. 43, 2455–2463. ( 10.1139/f86-305) [DOI] [Google Scholar]
- 20.McGee MD, Wainwright PC. 2013. Convergent evolution as a generator of phenotypic diversity in threespine stickleback . Evolution 67, 1204–1208. ( 10.1111/j.1558-5646.2012.01839.x) [DOI] [PubMed] [Google Scholar]
- 21.Anker G. 1974. Morphology and kinetics of the head of the stickleback, Gasterosteus aculeatus . Trans. Zool. Soc. Lond. 32, 311–416. ( 10.1111/j.1096-3642.1974.tb00030.x) [DOI] [Google Scholar]
- 22.Schilling TF, Kimmel CB. 1994. Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo . Development 120, 483–494. [DOI] [PubMed] [Google Scholar]
- 23.Schilling TF, Kimmel CB. 1997. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo . Development 124, 2945–2960. [DOI] [PubMed] [Google Scholar]
- 24.Haines R. 1934. Epiphysial growth in the branchial skeleton of fishes . Q. J. Microsc. Sci. 77, 77–97. [Google Scholar]
- 25.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. 1995. Stages of embryonic development of the zebrafish . Dev. Dyn. 203, 253–310. ( 10.1002/aja.1002030302) [DOI] [PubMed] [Google Scholar]
- 26.Hammond CL, Schulte-Merker S. 2009. Two populations of endochondral osteoblasts with differential sensitivity to Hedgehog signalling . Development 136, 3991–4000. ( 10.1242/dev.042150) [DOI] [PubMed] [Google Scholar]
- 27.Miller CT, et al. 2014. Modular skeletal evolution in sticklebacks is controlled by additive and clustered quantitative trait loci . Genetics 197, 405–420. ( 10.1534/genetics.114.162420) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orr HA. 1998. Testing natural selection versus genetic drift in phenotypic evolution using quantitative trait locus data. Genetics 149, 2099–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albert AY, Sawaya S, Vines TH, Knecht AK, Miller CT, Summers BR, Balabhadra S, Kingsley DM, Schluter D. 2008. The genetics of adaptive shape shift in stickleback: pleiotropy and effect size . Evolution 62, 76–85. [DOI] [PubMed] [Google Scholar]
- 30.McGee MD, Wainwright PC. 2013. Sexual dimorphism in the feeding mechanism of threespine stickleback . J. Exp. Biol. 216, 835–840. ( 10.1242/jeb.074948) [DOI] [PubMed] [Google Scholar]
- 31.Swarup H. 1958. Stages in the development of the stickleback Gasterosteus aculeatus (L.) . J. Embryol. Exp. Morphol. 6, 373–383. [PubMed] [Google Scholar]
- 32.Colosimo PF, et al. 2005. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles . Science 307, 1928–1933. ( 10.1126/science.1107239) [DOI] [PubMed] [Google Scholar]
- 33.Bell MA, Aguirre WE. 2013. Contemporary evolution, allelic recycling, and adaptive radiation of the threespine stickleback . Evol. Ecol. Res. 15, 377–411. [Google Scholar]
- 34.Gross HP. 1978. Natural selection by predators on the defensive apparatus of the three-spined stickleback, Gasterosteus aculeatus L. Can. J. Zool. 56, 398–413. ( 10.1139/z78-058) [DOI] [Google Scholar]
- 35.Schluter D. 2000. The ecology of adaptive radiation. New York, NY: Oxford University Press. [Google Scholar]
- 36.Sanger TJ, Norgard EA, Pletscher LS, Bevilacqua M, Brooks VR, Sandell LJ, Cheverud JM. 2011. Developmental and genetic origins of murine long bone length variation . J. Exp. Zool. B Mol. Dev. Evol. 316, 146–161. ( 10.1002/jez.b.21388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gunter HM, Koppermann C, Meyer A. 2014. Revisiting de Beer's textbook example of heterochrony and jaw elongation in fish: calmodulin expression reflects heterochronic growth, and underlies morphological innovation in the jaws of belonoid fishes . EvoDevo 5, 8 ( 10.1186/2041-9139-5-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farnum C, Tinsley M, Hermanson J. 2008. Forelimb versus hindlimb skeletal development in the big brown bat, Eptesicus fuscus: functional divergence is reflected in chondrocytic performance in autopodial growth plates . Cells Tissues Organs (Print) 187, 35–47. ( 10.1159/000109962) [DOI] [PubMed] [Google Scholar]
- 39.Cooper KL, Oh S, Sung Y, Dasari RR, Kirschner MW, Tabin CJ. 2013. Multiple phases of chondrocyte enlargement underlie differences in skeletal proportions . Nature 495, 375–378. ( 10.1038/nature11940) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greenwood AK, et al. 2011. The genetic basis of divergent pigment patterns in juvenile threespine sticklebacks . Heredity 107, 155–166. ( 10.1038/hdy.2011.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao Y, Du Z, Feng C, Deng X, Li N, Da Y, Hu X. 2010. Identification of quantitative trait loci for shank length and growth at different development stages in chicken . Anim. Genet. 41, 101–104. ( 10.1111/j.1365-2052.2009.01962.x) [DOI] [PubMed] [Google Scholar]
- 42.Manceau M, Domingues VS, Mallarino R, Hoekstra HE. 2011. The developmental role of Agouti in color pattern evolution . Science 331, 1062–1065. ( 10.1126/science.1200684) [DOI] [PubMed] [Google Scholar]
- 43.Schluter D, Conte GL. 2009. Genetics and ecological speciation . Proc. Natl Acad. Sci. USA 106, 9955–9962. ( 10.1073/pnas.0901264106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller CT, Beleza S, Pollen AA, Schluter D, Kittles RA, Shriver MD, Kingsley DM. 2007. cis-regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans . Cell 131, 1179–1189. ( 10.1016/j.cell.2007.10.055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan YF, et al. 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer . Science 327, 302–305. ( 10.1126/science.1182213) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available as an electronic supplementary material file.