Abstract
Microbial community structure on coral reefs is strongly influenced by coral–algae interactions; however, the extent to which this influence is mediated by fishes is unknown. By excluding fleshy macroalgae, cultivating palatable filamentous algae and engaging in frequent aggression to protect resources, territorial damselfish (f. Pomacentridae), such as Stegastes, mediate macro-benthic dynamics on coral reefs and may significantly influence microbial communities. To elucidate how Stegastes apicalis and Stegastes nigricans may alter benthic microbial assemblages and coral health, we determined the benthic community composition (epilithic algal matrix and prokaryotes) and coral disease prevalence inside and outside of damselfish territories in the Great Barrier Reef, Australia. 16S rDNA sequencing revealed distinct bacterial communities associated with turf algae and a two to three times greater relative abundance of phylotypes with high sequence similarity to potential coral pathogens inside Stegastes's territories. These potentially pathogenic phylotypes (totalling 30.04% of the community) were found to have high sequence similarity to those amplified from black band disease (BBD) and disease affected corals worldwide. Disease surveys further revealed a significantly higher occurrence of BBD inside S. nigricans's territories. These findings demonstrate the first link between fish behaviour, reservoirs of potential coral disease pathogens and the prevalence of coral disease.
Keywords: microbes, territorial grazers, coral reefs, benthic dynamics, epilithic algal matrix, coral disease
1. Introduction
Microbes are abundant across terrestrial and marine environments, and have prominent roles in community dynamics, yet there remains considerable unexplored complexity within microbial communities, particularly within marine environments [1,2]. The forefront of marine microbial ecology emphasizes the role of microbial processes on ecosystem functioning and the ecology of microbial diseases [3]. Moreover, marine benthic microbes are known to be important drivers of environmental change, so an understanding of benthic microbial community structure on coral reefs has the potential to provide forewarning of macro-ecological change [4]. Patterns of microbial communities on coral reefs are strongly influenced by the presence, physiological activity and ecological interactions of corals and algae [5,6]; however, it is largely unknown how fishes mediate these links between microbes and the macro-benthos. There is strong evidence that territorial grazers, particularly territorial damselfishes, play a key role in benthic dynamics on coral reefs [7–9]. Territorial grazers engineer benthic structure in their territories by grazing turf algae, coral-pecking (biting coral polyps at the base of coral colonies to further propagate turf algae) and weeding undesirable species of turf algae and fleshy macroalgae [10,11]. They also engage in frequent and sustained aggression towards intruders to defend food resources within their territories [12,13], which are composed of microbes, detritus, filamentous algae, corals and macroinvertebrates [14–16]. Along with shaping benthic community structure, territorial grazers are abundant and widespread, and it is estimated that up to 77% of the substratum of shallow reef flats (approx. 2 m depth) may be covered with damselfish territories [15,17,18]. Here, we aim to determine the role of territorial grazers in structuring coral–algal–bacteria linkages on the reef benthos.
Territorial grazers' behaviour markedly increases the productivity of palatable filamentous algae and dramatically decelerates the succession of macroalgae on coral reefs [19,20]. Among territorial grazers, there is a spectrum of guilds from extensive to intensive grazers [21]. Extensive grazers have large territories with unclear or overlapping boundaries, highly diverse algae turfs and low levels of territorial aggression. In contrast, intensive grazers have more distinct territorial boundaries, monocultures or low-diversity algae turfs with higher algal biomass per unit area and prompt aggressive responses to intruding species [7,22–24]. Within intensive grazers, there are large discrepancies among territory characteristics, as each damselfish species farms a unique turf assemblage with highly variable turf biomass per unit area [25]. However, the consequences of territorial variations on coral, algal and microbial assemblages within the defined guilds are largely unknown.
The dominant benthic component of damselfish territories is the epilithic algal matrix (EAM), which is composed of a conglomeration of living and non-living components, including filamentous turf algae, juvenile macroalgae, cyanobacteria, detritus, invertebrates and a consortium of microbes [5,16,26,27]. The effects of turf algae on corals are largely detrimental, and many of these interactions are attributed to the microbiota associated with turfs. In addition to inhibiting coral recruitment [28], retarding coral growth [29,30] and stressing coral physiology [31,32], turf algae also enhance microbial activity and coral mortality by releasing dissolved compounds that are harmful to corals [6,33,34]. Previously, both macroalgae and benthic turf algae have been shown to harbour pathogens that are associated with coral disease [35,36]. Yet the microbial communities of damselfish territories are virtually unexplored. While select communities of turf algae may harbour potential pathogens, the indirect effects of territorial grazers on benthic microbial communities, including the prevalence of potentially pathogenic bacteria and the consequential manifestation of coral disease, remain unresolved.
This paper aims to determine how the cultivation of turf algae-dominated territories by intensive territorial grazers in the genus Stegastes influences the structure of the microbial community and the prevalence of coral disease. Specifically, we characterized the algal assemblages inside and outside Stegastes apicalis's and Stegastes nigricans's territories to determine which algae were cultivated or excluded by these fish species, assessed and compared associated differences of microbial communities in the EAM inside and outside of Stegastes's territories, and ran coral disease surveys inside and outside of S. nigricans's territories. Our results reveal that microbial assemblages and coral disease prevalence are considerably different inside Stegastes's territories and have substantial implications for coral health in reef systems, elucidating an important ecological link between microbes and macro-organisms in the marine environment.
2. Material and methods
(a). Study site and species
This study took place at Lizard Island in the northern Great Barrier Reef (GBR), Australia (14°41′5″ S, 145°26′55″ E) from February to August of 2013. The main study site was on the back reef in the lagoon between Palfrey and South Island (Palfrey; electronic supplementary material, figure S1), at a depth of 1–3 m. For the coral disease surveys, there were four study sites on the back reef in the lagoon around Lizard Island (Palfrey, Bird, Loomis and Horseshoe; electronic supplementary material, figure S1).
Stegastes nigricans and Stegastes apicalis (f. Pomacentridae), two intensive territorial grazers [7,21,25], were our study species. Stegastes apicalis occurs at a depth of 1–15 m and reaches up to 15 cm (total length). Stegastes nigricans occurs at the depth of 1–12 m and reaches up to 14 cm (total length). Both species form social groups (termed ‘colonies’) made up of several contiguous territories, each territory belonging to an individual adult damselfish [37]. Stegastes apicalis form colonies on the reef flat in association with crevices, coral rubble, sparse acroporids and soft corals, whereas S. nigricans form colonies in staghorn coral outcrops dominated by Acropora muricata (J. M. Casey 2013, personal observation). Both damselfish species are aggressive territorial grazers that are not easily perturbed by human observers [13].
All algal composition surveys and EAM sampling were conducted on two colonies of S. apicalis (comprised 12 and 20 territories, respectively) and two colonies of S. nigricans (comprised 30 and 38 territories, respectively). We mapped each colony, and we used the minimum convex polygon method to estimate the territory size of each individual fish [38]. We observed each individual for a 5 min period then placed four flagged fishing weights around the extremities of the individual's territory. The longest and shortest diameters were measured to the nearest centimetre of the elliptical territory, and the average diameter was used to calculate territory area.
(b). Algal composition
To assess the algal communities inside S. apicalis's and S. nigricans's territories, we surveyed 20 territories from each study species. After estimating territory size, percentage coverage of algae in each territory was assessed visually and photographically. To assess the exclusion of macroalgae from Stegastes' territories, ten 1 m2 quadrats were placed on the benthos to identify the macroalgae taxa that did not appear in damselfish territories. Algae were identified to genus level and, when possible, to species level [39,40]. For the two damselfish species' territories, we quantified differences in algal species richness, evenness and Shannon diversity.
(c). Epilithic algal matrix microbial communities
To determine the impact of territorial damselfish on microbial communities, we examined the bacterial composition of the EAM inside and outside damselfish territories. In the field, ten 50 ml EAM samples were collected from control plots outside of Stegastes's territories, inside S. apicalis's territories and inside S. nigricans's territories. For the collections from S. apicalis's and S. nigricans's territories, samples were taken across two damselfish colonies, and each sample came from a different damselfish territory. Samples were immediately snap-frozen in liquid nitrogen, stored at −80°C [41] and transported to James Cook University for processing and DNA extraction.
Samples were homogenized under liquid nitrogen, DNA was isolated using a PowerPlant DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA) following the manufacturer's instructions and DNA quality was checked using a nanodrop. 27f and 519r universal reverse primers and the V1–V3 region of the 16S rDNA gene were used for amplification with a single-step 30-cycle PCR using a HotStarTaq Plus Master Mix Kit (Qiagen, Valencia, CA). MR DNA, a next generation sequencing and bioinformatics provider (Shallowater, TX), performed the PCR (under the following conditions: 94°C for 3 min, 28 cycles of 94°C for 30 s, 53°C for 40 s and 72°C for 1 min; 72°C for 5 min). All amplicon products were mixed in equal concentrations and purified using Agencourt ampure beads (Agencourt Bioscience Corporation, MA) and sequenced with Roche 454 FLX titanium instruments and reagents according to manufacturer's guidelines. The sequence data were processed using a proprietary analysis pipeline at MR DNA and further analysed with Quantitative Insights Into Microbial Ecology (QIIME) [42]. Sequences were depleted of barcodes and primers, and short sequences (less than 200 bp) and sequences with homopolymer runs exceeding 6 bp were removed. The average read length was 431 bp after primer and barcode removal. The sequences were denoised and chimeras were removed using Acacia [43]. After normalization, operational taxonomic units (OTUs) were defined with clustering at 3% divergence (97% similarity) [44–50]. Taxonomy was assigned to OTUs in QIIME, using SILVA [51] and BLASTN [52] against a curated GreenGenes database [53].
We assessed the beta-diversity of the EAM microbial communities inside and outside of damselfish territories with QIIME using a weighted UniFrac analysis. An unweighted pair group method using average linkages (UPGMA) clustering and a principal coordinates analysis (PCoA) were generated from the UniFrac distances [54]. The PCoA was generated from weighted UniFrac distances and plotted in two dimensions. Individual OTUs (generated using the proprietary pipeline analysis) were then assigned into three categories—autotrophs, heterotrophs and potential pathogens—based on literature reviews (electronic supplementary material, table S1). All genera with a less than 2% relative abundance were excluded from the analyses. Our data included five genera that are considered potential pathogens for a broad array of hosts; however, among these five genera, only two have previously been linked to coral disease. These two coral-specific potential pathogens made up 95% of the potential pathogen category, so we only included coral-specific potential pathogens in the analyses. Kruskal–Wallis one-way analyses of variance were used to analyse the differences in the relative abundances of these components of microbial communities in the EAM outside of Stegastes's territories as compared with inside S. apicalis's territories and S. nigricans's territories.
(d). Coral disease surveys
To assess the prevalence of coral disease inside and outside of damselfish territories, we determined coral disease presence in A. muricata outcrops, a common staghorn coral in the lagoon of Lizard Island. We ran six 10 m transects at each of the four study sites: Palfrey, Bird, Loomis and Horseshoe (electronic supplementary material, figure S1), for a total of 24 transects. At each transect, a 1 m2 quadrat was attached to the transect every 2 m, for a total of five quadrats per transect. Of the six transects at each site, three transects were in A. muricata outcrops that were occupied by S. nigricans, and three transects were in A. muricata outcrops with no territorial damselfish. As A. muricata is common in the lagoon of Lizard Island and S. nigricans typically form colonies around A. muricata outcrops, it was straightforward to survey comparable A. muricata outcrops outside damselfish territories. In contrast, S. apicalis's territories and their associated habitat have very heterogeneous coral assemblages. Since the prevalence of coral disease varies among coral taxa [55] and there is evidence that coral composition differs inside and outside damselfish territories [8], this substantially complicates the attribution of differences in coral disease prevalence to the behaviour of S. apicalis's rather than differences in coral species composition. Therefore, S. apicalis territories were not included in this component of the study.
To analyse the effect of damselfish territories on coral disease presence, we used a generalized linear mixed model (GLMM) with a binomial error distribution and logit-link function. The response variable was binary (coral disease present in quadrat/absent in quadrat). The fixed effect was the presence or absence of S. nigricans's territories. We included transect nested within site (Palfrey, Bird, Loomis and Horseshoe) as random factors. This analysis was conducted with the packages lme4 [56] and arm [57] using the software program R [58].
3. Results
(a). Algal composition
In the northern GBR around Lizard Island, S. apicalis's territories have a more diverse assemblage of turf algae than S. nigricans's territories (10 versus five species of turf algae; electronic supplementary material, table S2). The species richness, evenness and Shannon diversity index indicate that the algal community diversity of S. apicalis's territories is significantly higher than S. nigricans's territories (electronic supplementary material, table S3). The three dominant turf algae genera in S. apicalis's and S. nigricans's territories are Polysiphonia, Amphiroa and Ceramium, all of which are rhodophytes. Polysiphonia sp. is the most abundant turf in both S. apicalis's and S. nigricans's territories; it comprises over 50% of turf algae coverage across all territories. Both damselfish species actively exclude macroalgae (except for Halimeda, which occurs in very low frequencies in S. apicalis's territories) that commonly occur in the lagoon around Lizard Island and were observed in our control plots. Despite the more diverse algal assemblage in S. apicalis's territories, S. apicalis's territories have a thinner layer of turf algae cultivated on barren flat regions and coral rubble on the benthos compared with S. nigricans's territories, which are characterized by a thicker turf (largely composed of Polysiphonia sp.) cultivated on the branches of acroporids (figure 1). The thicker turf algae in S. nigricans's territories may be attributable to their more intensive farming behaviours (electronic supplementary material, figure S2 and table S4) and greater aggression than S. apicalis (electronic supplementary material, figures S3 and S4).
Figure 1.

The differential effects of (a) S. apicalis and (b) S. nigricans on the EAM. Stegastes apicalis cultivates a thin layer of turf algae on barren regions and coral rubble on the benthos, whereas S. nigricans cultivates a thick turf on the branches of acroporids. (Online version in colour.)
(b). Epilithic algal matrix microbial communities
Bacterial 16S rDNA gene amplicon sequencing retrieved 102 038 high-quality sequence reads from 24 EAM samples. Sequence reads were normalized to 1050 reads per sample to allow for comparison between samples and bacterial community patterns. All sequences were submitted to the NCBI Sequence Read Archive (SAMN02808132–SAMN02808155).
The microbial community of the EAM is distinct among S. apicalis's territories, S. nigricans's territories and control plots outside of damselfish territories (figure 2). There is some overlap between S. apicalis's and S. nigricans's territories, suggesting that these two damselfish territories have similar microbial communities. Polysiphonia sp. is the dominant taxon in both damselfish species' territories, so this may explain the clustering between S. apicalis's and S. nigricans's territories (figure 2; electronic supplementary material, figure S5). Further, there is some similarity between control plots outside of damselfish territories and S. apicalis's territories. Since S. apicalis cultivates algae on flattened regions of the benthos, which is a similar substrate to our control plots, whereas S. nigricans cultivates algae on the branches of acroporids (figure 1), microbial assemblages within control plots outside of damselfish territories cluster with S. apicalis's territories (figure 2; electronic supplementary material, figure S5).
Figure 2.
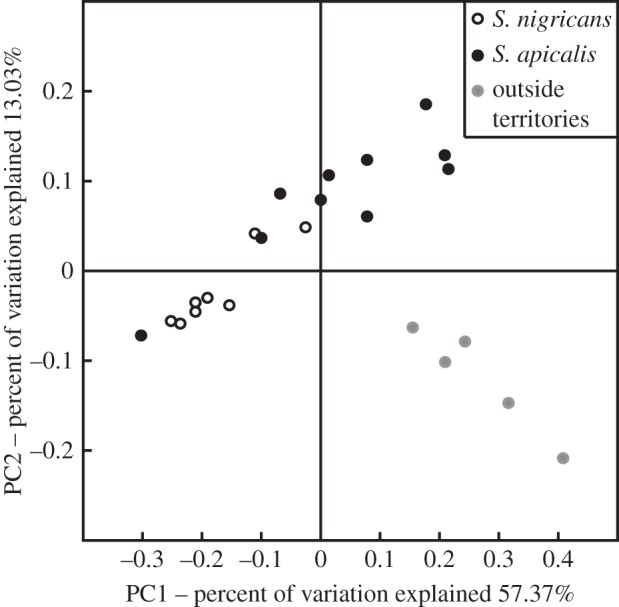
PCoA plot showing the per cent of variation in the microbial community of the EAM explained among S. nigricans's territories, S. apicalis's territories and outside of Stegastes's territories.
Assigning bacterial phylotypes into broad groupings of autotrophs, heterotrophs and potential coral pathogens (electronic supplementary material, table S1) further reveals that damselfish territories have distinct microbial consortia (figure 3). The relative abundance of autotrophs is significantly lower inside S. apicalis's territories (Kruskal–Wallis, χ2 = 6.615, p = 0.010), as is the relative abundance of heterotrophs inside S. apicalis's territories (Kruskal–Wallis, χ2 = 3.84, p = 0.050) and S. nigricans's territories (Kruskal–Wallis, χ2 = 8.218, p = 0.004) as opposed to outside Stegastes's territories (figure 3; electronic supplementary material, table S5). The relative abundances of potential coral pathogens is two to three times greater inside Stegastes's territories as opposed to outside Stegastes's territories (figure 3); there are significantly higher relative abundances of potential opportunistic coral pathogens inside of S. apicalis's (Kruskal–Wallis, χ2 = 4.335, p = 0.037) and S. nigricans's (Kruskal–Wallis, χ2 = 7.471, p = 0.006) territories than outside of Stegastes's territories (electronic supplementary material, table S4). The coral-specific potential pathogen communities are composed of the genera Leptolyngbya and Oscillatoria, key cyanobacteria associated with the pathogenicity of black band disease (BBD) [59].
Figure 3.
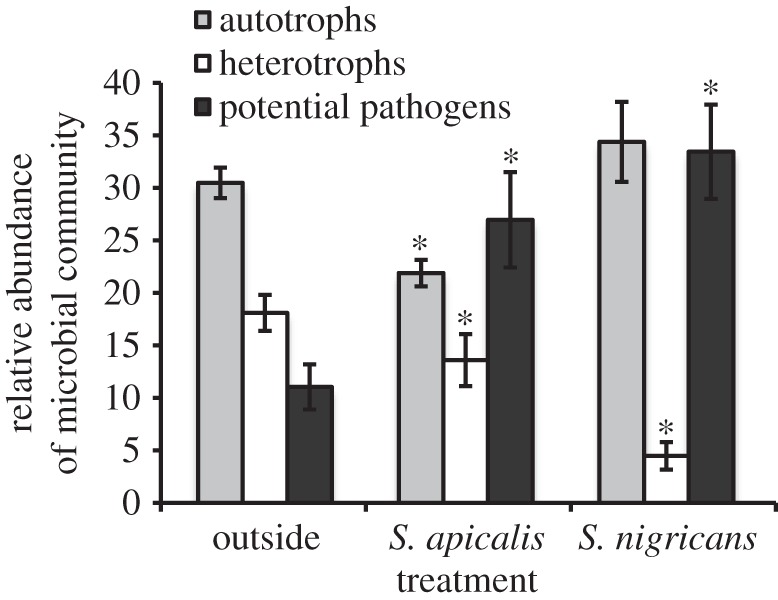
Relative abundances of autotrophs, heterotrophs and coral-specific potential pathogens in the EAM in control plots outside of Stegastes's territories, inside S. apicalis's territories and inside S. nigricans's territories. Asterisks represent significant differences (p < 0.05) between the control plot (outside Stegastes's territories) and respective Stegastes's territories (Kruskal–Wallis one-way analysis of variance; see the electronic supplementary material, table S5).
We compared the identified OTUs with high sequence similarity to potential coral pathogens (cyanobacterial genera, Leptolyngbya and Oscillatoria) to the entire BLAST nucleotide collection [50]. The highest abundant Oscillatoria sequences from the current study yielded 34 BLAST hits (more than 95% sequence identity) from two previous studies of BBD [60,61]. Within the overlapping region of the amplified sequence, the most abundant Leptolyngbya sequences from the current study showed 97% sequence similarity to a Leptolyngbya sequence reported in a previous BBD study [62] and to uncultured bacterial clones amplified from BBD corals (electronic supplementary material, table S6). The taxonomy of these cyanobacterial potential coral pathogens is probably unresolved, which is reflective of the complex phylogeneny of the cyanoprokaryote and supports previous calls for a re-evaluation of the taxonomy of the genus Leptolyngbya [63]. We also aligned our four most abundant uncultured cyanobacterial potential pathogen sequences to the unidentified bacterium clone sequence associated with the highest similarity from the BLAST results, which was the top match for 46.61% of our OTUs and associated with BBD affected corals (electronic supplementary material, table S6 and figure S6).
(c). Coral disease surveys
In A. muricata outcrops inside of S. nigricans's territories, we detected an average of 3.17 ± 1.41 occurrences of BBD per transect, whereas in A. muricata outcrops outside of S. nigricans's territories, there were zero occurrences of BBD (figure 4). The GLMM revealed that A. muricata inside S. nigricans's territories have a significantly higher occurrence of BBD than A. muricata outside of S. nigricans's territories (z = 2.670, p = 0.008; electronic supplementary material, table S7; figure 4).
Figure 4.
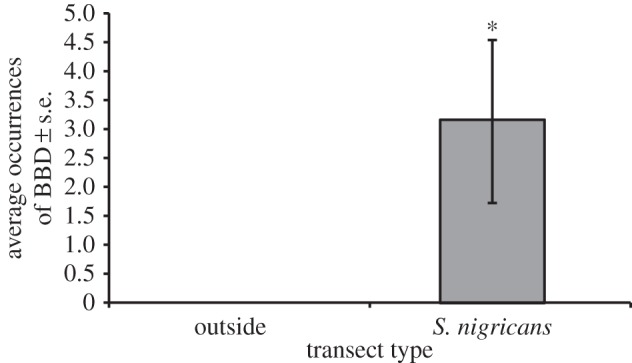
Average occurrences of BBD in A. muricata outcrops outside damselfish territories (n = 60) and inside S. nigricans's territories (n = 60; GLMM: p = 0.008). Asterisk represents a significant difference (p < 0.05).
4. Discussion
This study shows how the modification of benthic habitat by a macro-organism can influence microbial assemblage structure, with potentially broad implications for ecosystem health. Specifically, on coral reefs, territorial grazers strongly influence both algal and microbial community structure, with notable implications for the dynamics of coral disease. Intensive territorial grazers in the genus Stegastes demonstrably shape turf algae communities by excluding macroalgae and cultivating low-diversity, high-density communities of rhodophytes [11,19]. These behaviours also result in a shift of the microbial communities associated with the EAM in damselfish territories to communities with a high prevalence of potential opportunistic pathogens linked to coral disease. Moreover, at least for S. nigricans, these differences in microbial community structure are associated with a higher occurence of coral disease relative to corals outside of damselfish territories. This provides the crucial first link between fish behaviour, reservoirs of potential coral disease pathogens and the occurrence of coral disease. In light of territorial grazers' extensive space occupation on many shallow coral reefs [17,18], territorial damselfish may have a large role in coral disease dynamics via their extensive manipulation of the marine benthos.
Although both study species' behaviour appears to induce turf-dominant territories, we found considerable differences between our study species. Stegastes nigricans have a less diverse assemblage of turf algae than S. apicalis, suggesting that S. nigricans should be classified as a more intensive territorial grazer than S. apicalis. This inference is supported by direct behavioural observations, which indicate more active cultivation of algae and greater aggression in S. nigricans as opposed to S. apicalis (see the electronic supplementary material, figure S2). It is likely that S. nigricans has a less diverse assemblage of turf algae because they cultivate a thick mat of Polysiphonia sp. (figure 1), a highly filamentous turf that readily retains detritus and is one of the main food sources of Stegastes [64–68]. When cultivated to reach high densities, such as inside S. nigricans's territories, Polysiphonia sp. may outcompete some of the more delicate turf algae present in S. apicalis's territories (electronic supplementary material, table S2) due to Polysiphonia's advantageous lateral vegetative propagation and resilience to disturbance [69,70].
Our most striking finding is that damselfish have a marked effect on benthic microbial communities and increase the prevalence of BBD, at least in S. nigricans's territories. We found that Oscillatoria, an opportunistic pathogen associated with BBD [59], and uncultured cyanobacterial potential pathogens with homology to those isolated from BBD were strongly associated with the EAM inside damselfish territories. Oscillatoria and our uncultured cyanobacterial potential pathogens are both cyanobacteria, which is consistent with previous studies that show that diseased corals, particularly those with BBD, have elevated levels of cyanobacteria [71–73]. The prevalence of these phylotypes has major implications for coral reefs, suggesting that through altering benthic structure and preventing the establishment of macroalgae, territorial grazers may increase the incidence of microbes associated with coral disease. Intensive territorial grazers cultivate turf algae and detritus as food resources [16], which appear to harbour potential pathogens linked to BBD and may act as a coral disease pathogen reservoir. Turf algae have previously been shown to harbour pathogens that are associated with coral disease [30,36]. However, fish behaviour has not previously been directly linked to reservoirs of potential pathogens that may cause coral disease or the actual increased prevalence of coral disease.
Identifying reservoirs for marine disease is a major priority within the realm of ocean disease research because knowledge of the rates of spread and modes of transmission of pathogens are limited [74–76], yet no study has closely examined the role of coral reef fishes in the transmission of coral disease. A previous study argued for a negative link between reef fish diversity and coral disease, and this was attributed to the fact that chaetodontids, which are typically not fished, remain on reefs disturbed by fishing and may play a role in the transmission of disease [77]. If a link between fish diversity and coral disease does exist, it requires a more focused analysis of reef fish activities and associated disease prevalence. As our results suggest, coral reef fishes, especially herbivores and detritivores that strongly impact benthic dynamics, may play an important role in the formation of reservoirs of coral disease due to their feeding behaviours and territoriality. Consequently, we need to further examine widespread and abundant groups with strong benthic interactions, such as territorial pomacentrids, to determine their relationship to coral disease.
By actively cultivating benthic assemblages and within their territories, territorial pomacentrids function as ecosystem engineers [78]. Our study demonstrates that in the course of structuring habitat and modulating the availability of resources, ecosystem engineers may induce substantial alterations to microbial assemblages, with repercussions for microbial processes that influence disease ecology. On coral reefs, connections between fish populations, behaviour, disease reservoirs and coral health are of potentially substantial importance in regions where damselfish abundances are increasing, a phenomenon that may be associated with overfishing. Recent studies have reported up to a 60-fold increase in damselfish abundances across a gradient of human impact (e.g. Hawaiian Islands [79]; Line Islands [80]). Although large-scale patterns in fish community structure are influenced by numerous factors, including historical biogeography, reef geomorphology and human activities unrelated to fishing [81,82], a global meta-analysis of local-scale studies has found a positive relationship between fishing pressure and damselfish abundance that is consistent with the earlier studies [83]. Regardless of the specific mechanism that underlies this relationship, our study indicates that an increase in intensive territorial grazers will be likely to have substantial implications for the structure of the benthic microbial community on coral reefs, potentially increasing reservoirs of opportunistic pathogens linked to coral disease as well as the occurrence of coral disease. Consequently, understanding the mechanistic links among fishing pressure, damselfish abundances, shifts in microbial assemblages and the dynamics of associated coral communities may be important for anticipating and managing ongoing changes to the structure and functioning of coral reef ecosystems.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Lizard Island Research Station, K. D. Anderson, S. A. Blowes and C. Heckathorn for field support. We also thank D. R. Bellwood for conceptual advice and the Ecological Modelling Group at James Cook University, S. A. Blowes, S. J. Brandl and two anonymous reviewers for helpful comments that improved the manuscript.
All work was carried out under the Great Barrier Reef Marine Parks Permit no. G11/34774.1.
Data accessibility
DNA sequences: SRA accessions SAMN02808132–SAMN02808155. All other data have been uploaded as the electronic supplemental material.
Funding statement
Funds were provided by the Australian Research Council Centre of Excellence for Coral Reef Studies and the Great Barrier Reef Marine Park Authority Science for Management Award.
References
- 1.Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proc. Natl Acad. Sci. USA 95, 6578–6583. ( 10.1073/pnas.95.12.6578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrigo KR. 2005. Marine microorganisms and global nutrient cycles. Nature 437, 349–355. ( 10.1038/nature04159) [DOI] [PubMed] [Google Scholar]
- 3.Ainsworth TD, Thurber RV, Gates RD. 2009. The future of coral reefs: a microbial perspective. Trends Ecol. Evol. 25, 233–240. ( 10.1016/j.tree.2009.11.001) [DOI] [PubMed] [Google Scholar]
- 4.Garren M, Azam F. 2012. New directions in coral reef microbial ecology. Environ. Microbiol. 44, 833 ( 10.1111/j.1462-2920.2011.02597.x) [DOI] [PubMed] [Google Scholar]
- 5.Barott KL, Rohwer FL. 2012. Unseen players shape benthic competition on coral reefs. Trends Microbiol. 20, 621–628. ( 10.1016/j.tim.2012.08.004) [DOI] [PubMed] [Google Scholar]
- 6.Vega Thurber R, Burkepile DE, Correa AMS, Thurber AR, Shantz AA, Welsh R, Pritchard C, Rosales S. 2012. Macroalgae decrease growth and alter microbial community structure of the reef-building coral, Pories astreoides. PLoS ONE 7, e44246 ( 10.1371/journal.pone.0044246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hata H, Kato M. 2004. Monoculture and mixed-species algal farms on a coral reef are maintained through intensive and extensive management by damselfishes. J. Exp. Mar. Biol. Ecol. 313, 285–296. ( 10.1016/j.jembe.2004.08.009) [DOI] [Google Scholar]
- 8.Ceccarelli DM, Jones GP, McCook LJ. 2001. Territorial damselfishes as determinants of the structure of benthic communities on coral reefs. Oceanogr. Mar. Biol. Annu. Rev. 39, 355–389. [Google Scholar]
- 9.Ceccarelli DM, Hughes TP, McCook LJ. 2006. Impacts of simulated overfishing on the territoriality of coral reef damselfish. Mar. Ecol. Prog. Ser. 309, 255–262. ( 10.3354/meps309255) [DOI] [Google Scholar]
- 10.Hinds PA, Ballantine DL. 1987. Effects of the Caribbean threespot damselfish, Stegastes planifrons (Cuvier), on algal lawn composition. Aquat. Bot. 27, 299–308. ( 10.1016/0304-3770(87)90070-2) [DOI] [Google Scholar]
- 11.Klumpp DW, Polunin NVC. 1989. Partitioning among grazers of food resources within damselfish territories on a coral reef. J. Exp. Mar. Biol. Ecol. 125, 145–169. ( 10.1016/0022-0981(89)90040-3) [DOI] [Google Scholar]
- 12.Letourneur Y, Galzin R, Harmelin-Vivien M. 1997. Temporal variations in the diet of the damselfish Stegastes nigricans (Lacepede) on a Réunion fringing reef. J. Exp. Mar. Biol. Ecol. 217, 1–18. ( 10.1016/S0022-0981(96)02730-X) [DOI] [Google Scholar]
- 13.Newton AA. 1994. Ranching versus farming in Stegastes nigricans Lacapede (PISCES: Pomacentridae), a territorial herbivore, p. 78 Honours thesis James Cook University, Townsville, Australia. [Google Scholar]
- 14.Horn MH. 1989. Biology of marine herbivorous fishes. Oceanogr. Mar. Biol. Annu. Rev. 27, 167–172. [Google Scholar]
- 15.Choat JH. 1991. The biology of herbivorous fishes on coral reefs. In The ecology of fishes on coral reefs (ed. Sale P.), pp. 120–155. San Diego, CA: Academic Press. [Google Scholar]
- 16.Wilson S, Bellwood DR. 1997. Cryptic dietary components of territorial damselfishes (Pomacentridae, Labroidei). Mar. Ecol. Prog. Ser. 153, 299–310. ( 10.3354/meps153299) [DOI] [Google Scholar]
- 17.Klumpp DW, McKinnon D, Daniel P. 1987. Damselfish territories: zones of high productivity on coral reefs. Mar. Ecol. Prog. Ser. 40, 41–51. ( 10.3354/meps040041) [DOI] [Google Scholar]
- 18.Meekan MG, Steven ADL, Fortin MJ. 1995. Spatial patterns in the distribution of damselfishes on a fringing coral reef. Coral Reefs 14, 151–161. ( 10.1007/BF00367233) [DOI] [Google Scholar]
- 19.Ceccarelli DM, Jones GP, McCook LJ. 2005. Foragers versus farmers: contrasting effects of two behavioiural groups of herbivores on coral reefs. Oecologia 145, 445–453. ( 10.1007/s00442-005-0144-y) [DOI] [PubMed] [Google Scholar]
- 20.Ceccarelli DM, Jones GP, McCook LJ. 2011. Interactions between herbivorous fish guilds and their influence on algal succession on a coastal coral reef. J. Exp. Mar. Biol. Ecol. 399, 60–67. ( 10.1016/j.jembe.2011.01.019) [DOI] [Google Scholar]
- 21.Emslie MJ, Logan M, Ceccarelli DM, Cheal AJ, Hoey AS, Miller I, Sweatman HPA. 2012. Regional-scale variation in the distribution and abundance of farming damselfishes on Australia's Great Barrier Reef. Mar. Biol. 159, 1293–1304. ( 10.1007/s00227-012-1910-0) [DOI] [Google Scholar]
- 22.Hata H, Kato M. 2002. Weeding by the herbivorous damselfish Stegastes nigricans in nearly monocultural algae farms. Mar. Ecol. Prog. Ser. 237, 227–231. ( 10.3354/meps237227) [DOI] [Google Scholar]
- 23.Hata H, Nishihira M, Kamura S. 2002. Effects of habitat-conditioning by the damselfish Stegastes nigricans (Lacepede) on the community structure of benthic algae. J. Exp. Mar. Biol. Ecol. 280, 95–116. ( 10.1016/S0022-0981(02)00395-7) [DOI] [Google Scholar]
- 24.Hoey AS, Bellwood DR. 2010. Damselfish territories as a refuge for macroalgae on coral reefs. Coral Reefs 29, 107–118. ( 10.1007/s00338-009-0567-8) [DOI] [Google Scholar]
- 25.Ceccarelli DM. 2007. Modification of benthic communities by territorial damselfish: a multi-species comparison. Coral Reefs 26, 853–866. ( 10.1007/s00338-007-0275-1) [DOI] [Google Scholar]
- 26.Wilson SK, Bellwood DR, Choat JH, Furnas MJ. 2003. Detritus in the epilithic algal matrix and its use by coral reef fishes. Ocean. Mar. Biol. Annu. Rev. 41, 279–309. [Google Scholar]
- 27.Fricke A, Teichberg M, Beilfuss S, Bischof K. 2011. Succession patterns in algal turf vegetation on a Caribbean coral reef. Bot. Mar. 54, 111–126. ( 10.1515/bot.2011.021) [DOI] [Google Scholar]
- 28.Birrell CL, McCook LJ, Willis BL. 2005. Effects of algal turfs and sediment on coral settlement. Mar. Pollut. Bull. 51, 408–414. ( 10.1016/j.marpolbul.2004.10.022) [DOI] [PubMed] [Google Scholar]
- 29.Quan-Young LI, Espinoza-Avalos J. 2006. Reduction of zooxanthellae density, chlorophyll a concentration, and tissue thickness of the coral Montastraea faveolata (Scleractinia) when competing with mixed turf algae. Limnol. Oceanogr. 51, 1159–1166. ( 10.4319/lo.2006.51.2.1159) [DOI] [Google Scholar]
- 30.Barott KL, Rodriguez-Brito B, Janouskovec J, Marhaver KL, Smith JE, Keeling P, Rohwer FL. 2011. Microbial diversity associated with four functional groups of benthic reef algae and the reef building coral Montastraea annularis. Appl. Environ. Microbiol. 13, 1192–1204. ( 10.1111/j.1462-2920.2010.02419.x) [DOI] [PubMed] [Google Scholar]
- 31.Vermeij MJA, van Moorselaar I, Engelhard S, Hornlein C, Vonk SM, Visser PM. 2010. The effects of nutrient enrichment and herbivore abundance on the ability of turf algae to overgrow coral in the Caribbean. PLoS ONE 5, e14312 ( 10.1371/journal.pone.0014312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wangpraseurt D, Weber M, Roy H, Polerecky L, de Beer D, Nugues MM. 2012. In situ dynamics in coral–algal interactions. PLoS ONE 7, e31192 ( 10.1371/journal.pone.0031192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith JE, et al. 2006. Indirect effects of algae on coral: algae-mediated, microbe-induced coral mortality. Ecol. Lett. 9, 835–845. ( 10.1111/j.1461-0248.2006.00937.x) [DOI] [PubMed] [Google Scholar]
- 34.Haas AF, Nelson CE, Kelly LW, Carlson CA, Rohwer F, Leichter JJ, Wyatt A, Smith JE. 2011. Effects of coral reef benthic primary producers on dissolved organic carbon and microbial activity. PLoS ONE 6, e27973 ( 10.1371/journal.pone.0027973) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nugues MM, Smith GW, Van Hooidonk RJ, Seabra MI, Bak RPM. 2004. Algal contact as a trigger for coral disease. Ecol. Lett. 7, 919–923. ( 10.1111/j.1461-0248.2004.00651.x) [DOI] [Google Scholar]
- 36.Sweet MJ, Bythell JC, Nugues MM. 2013. Algae as reservoirs for coral pathogens. PLoS ONE 8, e69717 ( 10.1371/journal.pone.0069717) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Randall JR, Allen GR, Steene RC. 1991. Fishes of the Great Barrier Reef and coral sea. Bathurst, Australia: Crawford House Press. [Google Scholar]
- 38.Mohr CO. 1947. Table of equivalent populations of North American mammals. Am. Midland Nat. 37, 223–249. ( 10.2307/2421652) [DOI] [Google Scholar]
- 39.Price IR. 1992. The turf algal flora of the Great Barrier Reef: Rhodophyta. Townsville, Australia: James Cook University, Botany Department. [Google Scholar]
- 40.Cribb AB. 1996. Seaweeds of Queensland: a naturalist's guide. Brisbane, Australia: The Queensland Naturalist's Club. [Google Scholar]
- 41.Witt V, Wild C, Anthony KRN, Diaz-Pulido G, Uthicke S. 2011. Effects of ocean acidification on microbial community composition of, and oxygen fluxes through, biofilms from the Great Barrier Reef. Environ. Microbiol. 13, 1–14. ( 10.1111/j.1462-2920.2011.02571.x) [DOI] [PubMed] [Google Scholar]
- 42.Caporaso JG, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. ( 10.1038/nmeth.f.303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW. 2012. Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat. Methods 9, 425–426. ( 10.1038/nmeth.1990) [DOI] [PubMed] [Google Scholar]
- 44.Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS. 2008. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 8, 125 ( 10.1186/1471-2180-8-125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dowd SE, Sun Y, Wolcott RD, Domingo A, Carroll JA. 2008. Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodbourne Pathog. Dis. 5, 459–472. ( 10.1089/fpd.2008.0107) [DOI] [PubMed] [Google Scholar]
- 46.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. ( 10.1093/bioinformatics/btq461) [DOI] [PubMed] [Google Scholar]
- 47.Capone KA, Dowd SE, Stamatas GN, Nikolovski J. 2011. Diversity of the human skin microbiome early in life. J Invest. Dermatol. 131, 2026–2032. ( 10.1038/jid.2011.168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dowd SE, et al. 2011. Survey of fungi and yeast in polymicrobial infections in chronic wounds. J. Wound Care 20, 40–47. ( 10.12968/jowc.2011.20.1.40) [DOI] [PubMed] [Google Scholar]
- 49.Eren AM, Zozaya M, Taylor CM, Dowd SE, Martin DH, Ferris MJ. 2011. Exploring the diversty of Gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation. PLoS ONE 6, e26732 ( 10.1371/journal.pone.0026732) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanson KS, et al. 2011. Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME 5, 639–649. ( 10.1038/ismej/2010.162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. ( 10.1093/nar/gks1219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Z, Schwartz S, Wagner L, Miller W. 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7, 203–214. ( 10.1089/10665270050081478) [DOI] [PubMed] [Google Scholar]
- 53.DeSantis TZ, Hugenhotlz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Anderson GL. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34, W394–W399. ( 10.1093/nar/gkl244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. ( 10.1128/AEM.71.12.8228-8235.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carpenter KE, et al. 2008. One-third of reef-building corals face elevated extinction risk from climate change and local impacts. Science 321, 560–563. ( 10.11126/science.1159196) [DOI] [PubMed] [Google Scholar]
- 56.Bates D, Maechler M, Bolker B. 2012. lme4: linear mixed-effects models using S4 classes. R package v. 0.999999–0 See http://CRAN.R-project.org/package=lme4.
- 57.Gelman A, Su Y. 2013. arm: Data analysis using regression and multilevel/hierarchical models. R package v. 1.6–0.5 See http://CRAN.R-project.org/package=arm.
- 58.R Core Team. 2012. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org/. [Google Scholar]
- 59.Myers JL, Sekar R, Richardson LL. 2007. Molecular detection and ecological significance of the cyanobacterial genera Geitlerinema and Leptolyngbya in black band disease of corals. Appl. Environ. Microbiol. 73, 5173–5182. ( 10.1128/AEM.00900-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klaus JS, Janse I, Fouke BW. 2011. Coral black band disease microbial communities and genotypic variability of the dominant cyanobacteria (CD1C11). Bull. Mar. Sci. 87, 795–821. ( 10.5343/bms.2010.1050) [DOI] [Google Scholar]
- 61.Sekar R, Kaczmarsky LT, Richardson LL. 2008. Microbial community composition of black band disease on the coral host Siderastrea siderea from three regions of the wider Caribbean. Mar. Ecol. Prog. Ser. 362, 85–98. ( 10.3354/meps07496) [DOI] [Google Scholar]
- 62.Gantar M, Sekar R, Richardson LL. 2009. Cyanotoxins from black band disease of corals and from other coral reef environments. Microb. Ecol. 58, 856–864. ( 10.1007/s00248-009-9540-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stoyanov P, Moten D, Mladenov R, Dzhambazov B, Teneva I. 2014. Phylogenetic relationships of some cyanoprokaryotic species. Evol. Bioinform. 10, 39–49. ( 10.4137/EBO.S13748) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Galetto MJ, Bellwood DR. 1994. Digestion of algae by Stegastes nigricans and Amphiprion akindynos (Pisces: Pomacentridae), with an evaluation of methods used in digestibility studies. J. Fish Biol. 44, 415–428. ( 10.1111/j.1095-8649.1994.tb01222.x) [DOI] [Google Scholar]
- 65.Ceccarelli DM, Jones GP, McCook LJ. 2005. Effects of territorial damselfish on an algal-dominated coastal coral reef. Coral Reefs 24, 606–620. ( 10.1007/s00338-005-0035-z) [DOI] [Google Scholar]
- 66.Jones GP, Santana L, McCook LJ, McCormick MI. 2006. Resource use and impact of three herbivorous damselfishes on coral reef communities. Mar. Ecol. Prog. Ser. 328, 215–224. ( 10.3354/meps328215) [DOI] [Google Scholar]
- 67.Hata H, Kato M. 2006. A novel obligate cultivation mutualism between damselfish and Polysiphonia algae. Biol. Lett. 2, 593–596. ( 10.1098/rsbl.2006.0528) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hata H, Umezawa Y. 2011. Food habits of the farmer damselfish Stegastes nigricans inferred by stomach content, stable isotope, and fatty acid composition analyses. Ecol. Res. 26, 809–818. ( 10.1007/s11284-011-0840-5) [DOI] [Google Scholar]
- 69.Airoldi L. 2000. Effects of disturbance, life histories, and overgrowth on coexistence of algal crusts and turfs. Ecology 81, 798–814. ( 10.1890/0012-9658(2000)081[0798:EODLHA]2.0.CO;2) [DOI] [Google Scholar]
- 70.Airoldi L. 1998. Roles of disturbance, sediment stress, and substratum retention of spatial dominance in algal turf. Ecology 79, 2759–2770. ( 10.1890/0012-9658(1998)079[2759:RODSSA]2.0.CO;2) [DOI] [Google Scholar]
- 71.Frias-Lopez J, Klaus JS, Bonheyo T, Fouke BW. 2004. Bacterial community associated with black band disease in corals. Appl. Environ. Microbiol. 70, 5955–5962. ( 10.1128/AEM.70.10.5955-5962.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sato Y, Bourne DG, Willis BL. 2009. Dynamics of seasonal outbreaks of black band disease in an assemblage of Montipora species at Pelorus Island (Great Barrier Reef, Australia). Proc. R. Soc. B 276, 2795–2803. ( 10.1098/rspb.2009.0481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mouchka ME, Hewson I, Harvell CD. 2010. Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr. Comp. Biol. 50, 662–674. ( 10.1093/icb/icq061) [DOI] [PubMed] [Google Scholar]
- 74.Harvell CD, et al. 1999. Emerging marine diseases—climate links and anthropogenic factors. Science 285, 1505–1510. ( 10.1126/science.285.5433.1505) [DOI] [PubMed] [Google Scholar]
- 75.Harvell CD, et al. 2004. The rising tide of ocean diseases: unsolved problems and research priorities. Front. Ecol. Environ. 2, 375–382. ( 10.1890/1540-9295(2004)002[0375:TRTOOD]2.0.CO;2) [DOI] [Google Scholar]
- 76.Bourne DG, Garren M, Work TM, Rosenberg E, Harvell CD. 2009. Microbial disease and the coral holobiont. Trends Microbiol. 17, 554–562. ( 10.1016/j.tim.2009.09.004) [DOI] [PubMed] [Google Scholar]
- 77.Raymundo LJ, Halford AR, Maypa AP, Kerr AM. 2009. Functionally diverse reef-fish communities ameliorate coral disease. Proc. Natl Acad. Sci. USA 106, 17 067–17 070. ( 10.1073/pnas.0900365106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jones CG, Lawton JH, Shachak M. 1994. Organisms as ecosystem engineers. Oikos 69, 373–386. ( 10.2307/3545850) [DOI] [Google Scholar]
- 79.Friedlander AM, DeMartini EE. 2002. Contrasts in density, size, and biomass of reef fishes between the northwestern and the main Hawaiian Islands: the effects of fishing down apex predators. Mar. Ecol. Prog. Ser. 230, 253–264. ( 10.3354/meps230253) [DOI] [Google Scholar]
- 80.Sandin SA, et al. 2008. Baselines and degradation of coral reefs in the Northern Line Islands. PLoS ONE 3, e1548 ( 10.1371/journal.pone.0001548) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pinca S, Kronen M, Magron F, McArdle B, Vigliola L, Kulbicki M, Andrefouet S. 2011. Relative importance of habitat and fishing in influencing reef fish communities across seventeen Pacific Island Countries and Territories. Fish Fish. 13, 361–379. ( 10.111/j.1467-2979.2011.00425.x) [DOI] [Google Scholar]
- 82.Taylor BM. 2014. Divers of protogynous sex change differ across spatial scales. Proc. R. Soc. B 281, 20132423 ( 10.1098/rspb/2013.2423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Edwards CB, et al. 2014. Global assessment of the status of coral reef herbivorous fishes: evidence for fishing effects. Proc. R. Soc. B 281, 20131835 ( 10.1098/rspb.2013.1835) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences: SRA accessions SAMN02808132–SAMN02808155. All other data have been uploaded as the electronic supplemental material.


