Abstract
In response to DNA damage the E2F1 transcription factor is phosphorylated at serine 31 (serine 29 in mouse) by the ATM or ATR kinases, which promotes E2F1 protein stabilization. Phosphorylation of E2F1 also leads to the recruitment of E2F1 to sites of DNA damage where it functions to enhance DNA repair. To study the role of this E2F1 phosphorylation event in vivo, a knock-in mouse model was generated in which serine 29 was mutated to alanine. The S29A mutation impairs E2F1 stabilization in response to ultraviolet (UV) radiation and doxorubicin treatment but has little effect on the expression of E2F target genes. The apoptotic and proliferative responses to acute UV radiation exposure is also similar between wild type and E2F1S29A/S29A mice. As expected, the S29A mutation prevents E2F1 association with damaged DNA and reduces DNA repair efficiency. Moreover, E2f1S29A/S29A mice display increased sensitivity to UV-induced skin carcinogenesis. This knock-in mouse model thus links the ability of E2F1 to directly promote DNA repair with the suppression of tumor development.
Keywords: E2F1, ATR, GCN5, nucleotide excision repair, skin carcinogenesis
Introduction
Members of the E2F family of transcription factors regulate the expression of genes involved in a variety of processes, including cell cycle progression, DNA replication, differentiation, and apoptosis (1). The transcriptional activity of the E2F family is regulated through association with the retinoblastoma (Rb) tumor suppressor and related proteins, p107 and p130. The generation of mouse models inactivated for individual E2F genes has revealed the complexity and overlapping functions of the E2F family (1, 2). For example, mice lacking E2F1 are viable and fertile but display cell type-specific defects in some tissues, including defective negative selection of T cells in younger mice and exocrine gland hyperplasia and testicular atrophy in older mice (3, 4). E2f1-/- mice are also mildly tumor prone, developing an unusual array of spontaneous tumors after one year of age (4). Moreover, inactivation of E2f1 in transgenic mice expressing Myc under the control of a keratin 5 promoter significantly accelerates tumor development (5). In sharp contrast, inactivation of E2f1 in Rb+/- mice significantly reduces spontaneous tumor development in that model (6). Taken together, these findings demonstrate that endogenous E2F1 can either enhance or inhibit tumorigenesis depending on the experimental context.
The mechanism by which E2F1 suppresses tumor development is unclear but may be related to its role in the DNA damage response (7-11). The ATM and ATR kinases phosphorylate human E2F1 at serine 31, a site not conserved in other E2F family members (12). E2F1 phosphorylation at this site creates a binding motif for 14-3-3τ, which contributes to E2F1 stabilization (13). E2F1 is also acetylated on several lysine residues in response to agents that cause DNA double-strand breaks, which enhances the ability of E2F1 to bind and activate some pro-apoptotic genes, such as p73, and induce apoptosis (14-18). However, E2F1 is not acetylated in response to UV radiation so E2F1 does not transcriptionally activate p73 or participate in UV-induced apoptosis (18). In fact, the absence of E2F1 appears to increase the apoptotic response to UV radiation (19, 20).
Phosphorylation of E2F1 at serine 31 also creates a binding motif for a BRCT domain in the TopBP1 protein and this interaction represses E2F1 transcriptional activity independent of Rb (21, 22). Phosphorylation of E2F1 and binding to TopBP1 also recruits E2F1 to sites of DNA double-strand breaks where it forms foci that co-localize with BRCA1 (21). Moreover, cells lacking E2F1 are impaired for the recruitment of some DNA repair factors to sites of double-strand breaks and display genome instability (23). E2F1 also accumulates at sites of UV-induced DNA damage dependent on ATR and serine 31 of E2F1 (24). E2F1 was shown to stimulate nucleotide excision repair (NER) dependent on serine 31 but independent of its DNA binding or transactivation domains. The ability of E2F1 to enhance NER correlated with E2F1-dependent recruitment of the GCN5 histone acetyltransferase to sites of UV-induced DNA damage, increased H3K9 acetylation, and enhanced co-localization of NER factors with damaged DNA (25). Taken together, these findings suggest that E2F1 stimulates the repair of several types of DNA damage and that E2F1 phosphorylation by ATM/ATR is critical for this transcription-independent function.
Here we describe the generation of a knock-in mouse model in which E2F1 serine 29 (equivalent to human serine 31) is mutated to alanine (E2f1S29A/S29A mice). As expected, E2F1 stabilization in response to UV radiation and doxorubicin treatment was impaired by the E2F1 S29A mutation but the expression of several E2F target genes and the apoptotic and proliferative responses to UV were similar between E2f1S29A/S29A and wild type mice. E2F1 was unable to associate with DNA containing UV photoproducts in cells from E2f1S29A/S29A mice and this correlated with decreased association of GCN5, acetylated H3K9, and NER factors with damaged DNA. Consistent with these findings, the S29A knock-in mutation reduced DNA repair efficiency and enhanced sensitivity to UV-induced skin carcinogenesis. This mouse model highlights the importance of E2F1 as a downstream target of ATR for enhancing NER in the context of chromatin and suppressing skin tumor development.
Materials and Methods
Generation of E2f1S29A/S29A knock-in mouse model
Genomic DNA containing E2f1 exon 1 was amplified by PCR and cloned using standard procedures. Site directed mutagenesis was used to create a two base pair substitution that resulted in a silent mutation in codon 28 (a serine) and altering codon 29 from a serine to an alanine. This mutation also created an AviII site, which can be used for genotyping purposes to identify the knock-in allele. The targeting vector as shown in Figure 1A was electroporated into mouse embryonic stem (ES) cells and colonies were selected in G418 at the University of Texas MD Anderson Cancer Center Genetically Engineered Mouse Facility. Southern blot analysis was performed on genomic DNA isolated from ES cell clones and digested with BamHI and AviII using standard procedures to identify correctly targeted ES clones. Chimeric mice were developed using two positive clones. Chimeric mice were crossed with FVB mice to produce F1 generation of heterozygous mice. One heterozygous mouse was crossed with FLPer mice to excise the Neo-cassette from the targeted allele. For UV carcinogenesis experiments, mice containing the S29A knock-in allele were backcrossed seven times to the FVB strain before mating heterozygous mice to produce homozygous knock-in and wild type sibling control mice.
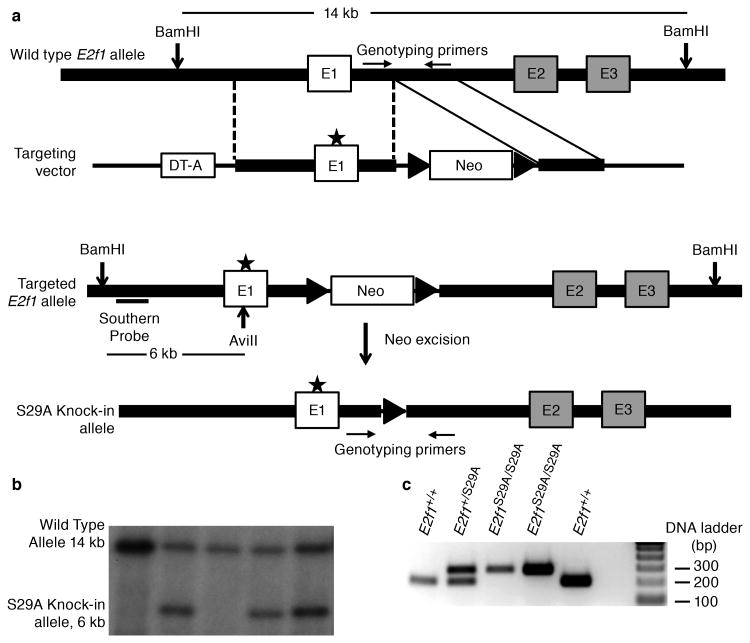
Figure 1. Generation of an E2f1S29A/S29A knock-in mouse model.
(a) Schematic of gene targeting construct and knock-in strategy (b) ES cell clones with the correctly targeted E2f1 allele were detected by Southern blot analysis of genomic DNA digested with BamHI and AviII. The wild type allele produces a 14 kb band while the knock-in allele produces a 6 kb band (lanes 2, 4 and 5). (c) Genotyping of mouse litters by PCR using primers across the remaining Frt site. The wild type allele without Frt produces an approximately 200 bp band and the knock-in allele produces a band of approximately 300 bp.
UV irradiation
UVB treatment of mice was performed using a panel of FS20 sunlamps in an irradiation chamber as previously described (19). For UVB-induced skin carcinogenesis, the dorsal skin of 4-5 week old mice was shaved and 24 h later mice were exposed to 337 J/m2 of UVB. This treatment continued three times per week for up to 48 weeks or until tumors reached approximately 1 cm in size. Histological examination confirmed that the tumors were squamous cell carcinoma (SSC).
Cells and antibodies
Primary mouse embryonic fibroblasts (MEFs) were isolated from 13.5 days old embryos derived from crossing heterozygous mice following standard procedures and maintained in DMEM supplemented with 15% FBS, penicillin-streptomycin and 100 μm β-mercaptoethanol. Mouse primary keratinocytes were isolated from 2 day old mice following previously described protocol (26) and cultured in defined keratinocyte-SFM (Life Technologies Cat. No. 10744019). The following antibodies were used in the study: anti-E2F1 (C20, Santa Cruz, Cat. No. sc-193), anti-CPD (Cosmo Bio, clone TDM-2, Cat. No. CAC-MN-DND-001), anti-6-4PP (Cosmo Bio, clone 64M-2, Cat. No. CAC-MN-DND-002), anti-XPC (Bioss USA antibodies Cat. No. bs-6634R), anti-XPA (sc-853), anti-GCN5 (Cell signaling Cat. No. 3305S), anti-H3K9ac (Cell Signaling Cat. No. 9649S), anti-p53 (Novus Biologicals Cat. No. NB200-103), GAPDH (GenScript Cat. No. A00191) and normal rabbit IgG (Cell Signaling Cat. No. 2729).
Western Blot Analysis
Western blotting was performed according to previously described procedure (24). For detecting E2F1, whole cell lysates were resolved on SDS-PAGE and transferred onto PVDF membrane. The blots were incubated with anti-E2F1 at 1:500 dilution overnight at 4°C with gentle shaking.
Gene expression profiling of E2F1 target genes
RNA was isolated from primary keratinocytes exposed to UVB or MEFs treated with doxorubicin using Qiagen RNeasy mini kit (Cat. No. 74104) and RNA was converted to cDNA using the Invitrogen SuperScript® First-Strand Synthesis System (Cat. No. 11904-018). Quantitative real-time PCR for select E2F target genes was performed in the Molecular Biology Core at the University of Texas MD Anderson Cancer Center, Science Park. Each sample was analyzed in triplicate (p73) or duplicate and GAPDH was used as an internal control.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed using the SimpleChIP® Enzymatic Chromatin IP Kit (Cell Signaling; Cat. No. 9003) following manufacturer's instructions. Briefly, cells were cross-linked with 1% formaldehyde (final) for 10 min at room temperature with gentle rocking. Cross-linking was blocked by addition of glycine and incubating for 5 minutes at room temperature. Nuclei were isolated and incubated with micrococcal nuclease and sonicated in 1X ChIP buffer containing a protease inhibitor cocktail (Roche) and a phosphatase inhibitor cocktail (Roche) to produce chromatin fragments ranging from 200-1000 base pairs. An ELISA was used to measure the amount of CPD in the immunoprecipitated DNA. Alternatively, primers specific for sequences flanking an I-PpoI cut site on mouse chromosome 10 were used to examine enrichment at a DNA double-strand break.
Enzyme-linked immunosorbent assay (ELISA) for DNA photoproducts
An ELISA was used to measure the amount of CPD and 6-4PP present in UV damaged DNA using anti-CPD and anti-6-4PP antibodies (Cosmo Bio Co. Ltd., Japan) following manufacturer's instructions. Briefly, 96 well polyvinylchloride plates (Thermo Cat. No. 2801) were coated with 50 μl of 0.003% protamine sulfate overnight at 37°C. Plates were washed with 100 μl distilled water and stored in the dark until used. Plates were coated with DNA obtained from ChIP or directly isolated from UV treated cells (0.02 μg/ml for CPD and 2 μg/ml for 6-4PP) for 30 min at 37°C. Plates were washed 5 times with PBS-T (0.05% Tween-20 in PBS), blocked with 2% FBS in PBS, incubated with appropriate dilutions of anti-CPD and anti-6-4PP antibodies, detected with biotin-F (ab)2 fragment of anti-mouse IgG (H+L) and peroxidase-streptavidin using o-phenylene diamine as a substrate.
UV repair assay
Repair of UVB-induced damage was determined by measuring the amount of 6-4PP and CPD remaining in genomic DNA over time as previously described (19). Briefly, the dorsal skins of mice were shaved 24 h prior to a single UVB dose of 1,000 J/m2. At various time points post-UVB, epidermis from the exposed skin was collected and genomic DNA was isolated using Sigma GenElute mammalian genomic DNA isolation Kit. Equal amounts of DNA were coated onto 96 well microtiter plate pre-coated with protamine sulfate. The DNA photoproducts CPD and 6-4PP were detected by ELISA using anti-CPD and anti-6-4PP antibodies (Cosmo Bio Co. Ltd., Japan) following manufacturer's protocol. Two mice for each genotype and time point were analyzed.
Results
Development of an E2f1S29A/S29A knock-in mouse model
The E2f1 knock-in targeting construct contained point mutations that changed codon 29 from a serine to alanine and also created an AviII restriction site (Figure 1a). The Genetically Engineered Mouse Facility at MD Anderson Cancer Center generated embryonic stem (ES) cell clones using this construct and, after Southern blot analysis of ES cell genomic DNA (Figure 1b), created chimeric mice from correctly targeted ES clones. The Neo cassette was then excised by crossing F1 generation heterozygous mice with FLPeR mice (Figure 1a). Sequencing of E2f1 exon 1 confirmed the presence of the S29A mutation and no other mutation. PCR across the remaining Frt site in the knock-in allele was subsequently used to distinguish wild type, heterozygous and homozygous S29A mutant mice (Figure 1c). As expected, E2f1S29A/S29A mice were obtained with the expected Mendelian frequency and appeared developmentally normal, similar to E2f1-/- mice (3, 4).
It has been reported that young adult E2f1-/- mice have an enlarged thymus, increased thymic cellularity and a higher fraction of mature, single positive (CD4+ and CD8+) thymocytes (3). E2f1S29A/S29A mice lack these phenotypes, demonstrating that the S29A mutation is insufficient to cause a defect in thymocyte homeostasis as is observed in E2f1-/- mice (Supplementary Figure 1). In addition, older E2f1S29A/S29A mice do not appear to have phenotypes associated with older E2f1-/- mice, such as testicular atrophy or exocrine gland hyperplasia (4).
Effects of the S29A mutation on E2F1 protein stabilization, expression of target genes, and induction of apoptosis in response to DNA damage
Various forms of DNA damage lead to E2F1 protein stabilization in a process involving E2F1 phosphorylation at S31/S29 by ATM or ATR (12, 13, 27, 28). Consistent with those studies, E2F1 protein levels are induced at 1h and peak at 8h in wild type primary keratinocytes exposed to UV radiation. In contrast, E2F1 protein induction in response to UV radiation was impaired in E2F1S29A/S29A primary keratinocytes (Figure 2a). Treatment with doxorubicin, which causes DNA double-strand breaks, did increase E2F1 protein levels in S29A knock-in MEFs but to a lesser extent than in wild type cells (Figure 2b). This finding is consistent with previous studies demonstrating that E2F1 can be stabilized in response to DNA double-strand breaks independent of ATM/ATR phosphorylation through a process involving E2F1 acetylation (15).
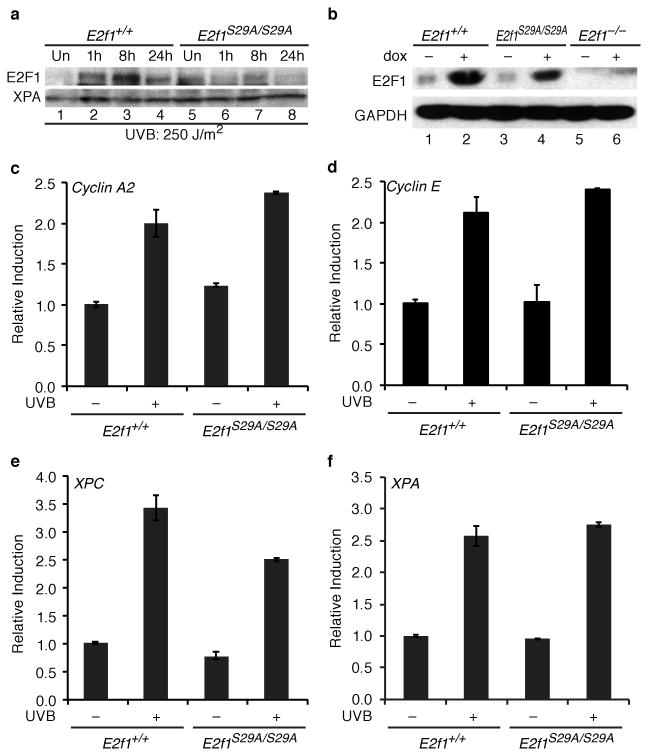
Figure 2. E2F1 stabilization and target expression in response to DNA damage in E2f1S29A/S29A cells.
(a) Primary keratinocytes were isolated from wild type (E2f1+/+, lanes 1-4) and E2f1S29A/S29A (lanes 5-8) mice and left untreated (lanes 1 and 5) or exposed to 250 J/m2 of UVB and harvested at the time points indicated (lanes 2-4 and 6-8). Western blot analysis was performed for E2F1 and XPA. b) MEFs were isolated from wild type (E2f1+/+, lanes 1 and 2), E2f1S29A/S29A (lanes 3 and 4) or E2f1-/- mice and left untreated (lanes 1, 3, and 5) or exposed to 1μM of doxorubicin for 24 h (lanes 2, 4, and 6). Western blot analysis was performed for E2F1 and GAPDH. (c-f) Primary keratinocytes isolated from wild type (E2f1+/+) and E2f1S29A/S29A mice were untreated or exposed to 250 J/m2 of UVB and harvested 8 h post irradiation. Real-time PCR was used to examine expression levels of (c) cyclin A2, (d) cyclin E and (e) XPC, and (f) XPA. The average of two independent samples per group is presented.
Although E2F1 protein stabilization is defective in E2f1S29A/S29A keratinocytes, expression of the E2F target genes cyclin A2 and cyclin E is similar between wild type and S29A knock-in keratinocytes before and after UV irradiation (Figure 2c and d). Expression of XPC, another E2F target gene known to be upregulated in response to UV (29), is only modestly affected by the E2F1 S29A mutation (Figure 2e). Expression of another NER gene, XPA, is also similar between wild type and E2f1S29A/S29A keratinocytes before and after UV exposure (Figure 2f). E2F1 is also implicated in the induction of the E2F target gene p73 in response to agents that cause DNA double-strand breaks (18, 22, 30). As expected, treatment of MEFs with doxorubicin strongly induced p73 gene expression but this was unaffected by the E2F1 S29A knock-in mutation (Supplemental Figure 2a).
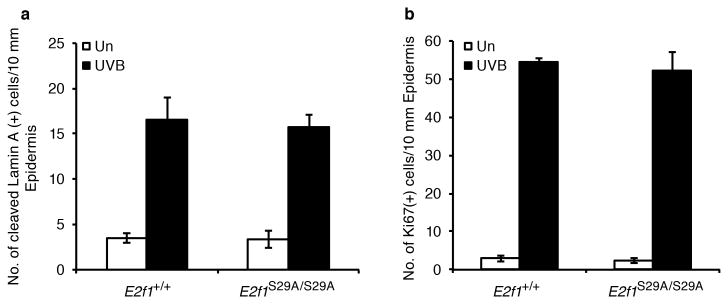
Previous studies demonstrated that E2f1-/- mice have an enhanced apoptotic response to UV radiation (19, 20). To determine the effect of the E2F1 S29A knock-in mutation on the apoptotic response to UV radiation, E2f1S29A/S29A mice and wild type sibling controls were exposed to 337 J/m2 of UVB. Skin samples were taken 24 h after exposure and immunohistochemistry was performed for cleaved lamin A as a marker of apoptosis. The numbers of epidermal cells staining positive for cleaved lamin A was similar between wild type and E2f1S29A/S29A mice before and after UVB exposure (Figure 3a). This indicates that the E2F1 S29A knock-in mutation does not recapitulate the increased sensitivity to UV-induced apoptosis observed in E2f1-/- mice. Skin samples were also stained for Ki67 as a marker of proliferation. Consistent with cyclin A2 and cyclin E expression in response to UV being unaltered by the E2F1 S29A mutation, proliferation in response to UV was also similar between wild type and E2f1S29A/S29A mice (Figure 3b).
Figure 3. The S29A mutation does not affect the apoptotic or proliferative responses to UV.
Young adult wild type and E2f1S29A/S29A mice were shaved and 24 h later mock treated or exposed to 337 J/m2 of UVB. 24 h post irradiation, skin samples were taken, fixed with formalin, and slides processed for immunohistochemistry. (a) An antibody specific for cleaved lamin A was used to identify apoptotic cells and the average number of positive keratinocytes per 10 mm of epidermis was determined from three independent mice per group. (b) The average number of epidermal keratinocytes staining positive for Ki67, a marker of proliferation, was determined from three independent mice per group.
The S29A mutation impairs recruitment of E2F1 to sites of DNA damage
Phosphorylation of human E2F1 at serine 31 was shown to recruit E2F1 to sites of DNA double-strand breaks through a phospho-specific interaction with the BRCT domain-containing protein TopBP1 (21). To examine E2F1 recruitment to DNA double-strand breaks we employed a system using an inducible form of the I-PpoI endonuclease followed chromatin immunoprecipitation (ChIP) (31, 32). Like phosphorylated ATM (pATM), which is used as a positive control, wild type E2F1 was recruited to sequences flanking an I-PpoI site on mouse chromosome 10 following I-PpoI activation (Supplemental Figure 2B). In contrast, enrichment of E2F1 at sequences flanking the I-PpoI cut site was abolished in cells from E2f1S29A/S29A mice. E2F1 also accumulates at sites of UV-induced DNA damage and this also requires phosphorylation of human E2F1 at serine 31 (24). To examine the association of E2F1 and other proteins with UV damaged DNA, ChIP was performed followed by an enzyme-linked immunosorbant assay (ELISA) that quantitatively measures CPD photoproduct in DNA. Antibody to E2F1 efficiently immunoprecipitated CPD-containing DNA from wild type MEFs treated with UV but not from UV-treated E2f1S29A/S29A MEFs (Figure 4a, Supplemental Figure 3a). Taken together, these results demonstrate that blocking E2F1 phosphorylation by ATM/ATR impairs association of E2F1 with both DNA double-strand breaks and UV-induced DNA damage.
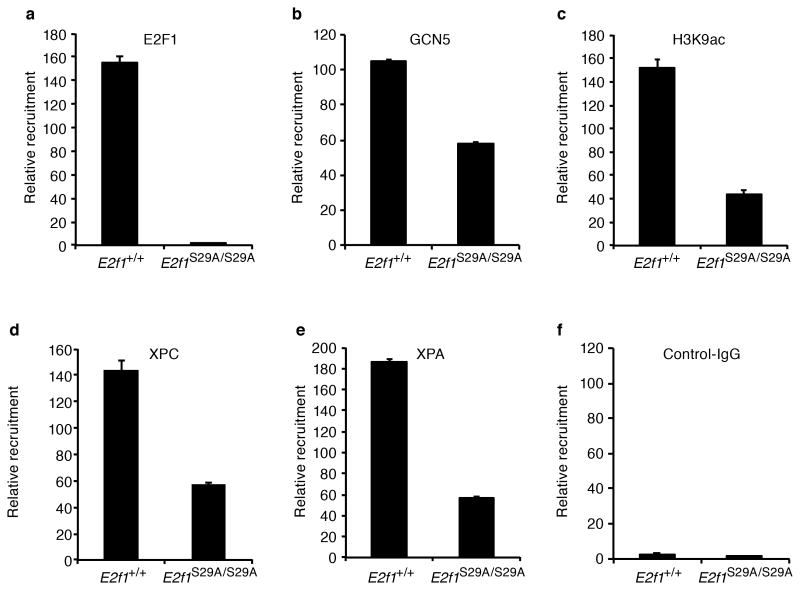
Figure 4. Association of E2F1, GCN5, H3K9ac, and NER factors with UV-induced DNA damage is impaired in E2f1S29A/S29A cells.
Wild type (E2f1+/+) and E2f1S29A/S29A MEFs were exposed to 250 J/m2 of UVB and ChIP was performed 30 min post-irradiation with the indicated antibodies: (a) E2F1, (b) GCN5, (c) H3K9ac, (d) XPC, (e) XPA, and (f) control IgG. An ELISA was used to measure the amount of CPD in immunoprecipitated DNA. The relative enrichment of CPD in genomic DNA was calculated from two independent samples per group subjected to ELISA in triplicate.
E2f1S29A/S29A mice have reduced NER efficiency and increased sensitivity to UV-induced carcinogenesis
Our previous studies demonstrated that E2F1 promotes the repair of UV-induced DNA damage by recruiting the GCN5 histone acetyltransferase to damaged sites to induce H3K9 acetylation and increase accessibility to the DNA repair machinery (25). Consistent with those findings, the E2F1 S29A mutation impaired association of GCN5 and acetylated H3K9 with chromatin containing UV damaged DNA (Figure 4b and c). Likewise, association of the DNA repair factors XPC and XPA with CPD-containing chromatin was also diminished by the E2F1 S29A mutation (Figure 4). The amount of total protein immunoprecipitated with these antibodies was similar between genotypes before and after UV treatment (Supplemental Figure 3a). The E2F1 S29A mutation did not affect the association between E2F1 and GCN5 either before or after UV exposure (Supplemental Figure 3b). Taken together, these findings suggest that E2F1 S29 is involved in recruiting GCN5 to chromatin containing CPD photoproducts and that this is important for H3K9 acetylation and efficient recruitment of NER factors to damaged DNA.
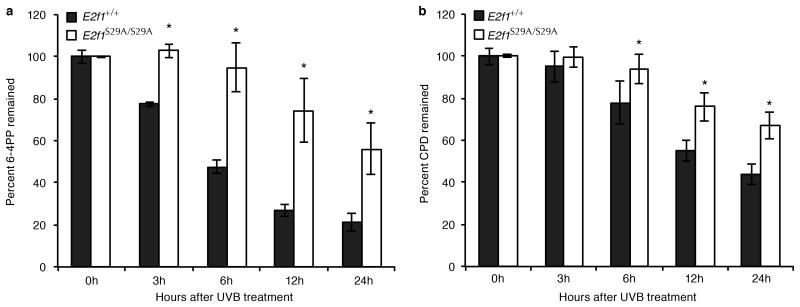
Mice and cells lacking E2F1 repair UV-damaged DNA less efficiently than wild type mice and cells (19, 24, 29). To determine whether the S29A mutation would also reduce NER efficiency, the dorsal skins of E2f1S29A/S29A and wild type control mice were shaved and exposed to a single dose of UVB at 1,000 J/m2. Genomic DNA was isolated from UV-exposed epidermis at various time points post-irradiation and an ELISA was used to measure the amounts of pyrimidine-pyrimidone (6-4) photoproducts (6-4PP) and CPD. The levels of 6-4PP remaining in UV-treated epidermal DNA was significantly higher in E2f1S29A/S29A mice compared to wild type mice between 3 and 24h post UV (Figure 5a). The CPD is repaired less efficiently than 6-4PP but a significant difference between E2f1S29A/S29A and wild type mice in CPD removal could be observed at later time points (Figure 5b). These results demonstrate that like E2f1-/- mice, E2f1S29A/S29A mice have reduced capacity to repair UV-induced DNA damage (19).
Figure 5. E2f1S29A/S29A mice display decreased NER efficiency.
(a) The dorsal skin of E2f1S29A/S29A and wild type sibling control mice were shaved and exposed once to 1,000 J/m2 of UVB. Genomic DNA was isolated from the exposed epidermis at the indicated time points and the amounts of 6-4PP (a) and CPD (b) were measured by ELISA using anti-6-4PP and anti-CPD antibodies, respectively. Two mice per genotype for each time point were used and each sample was subjected to ELISA in duplicate. *indicates statistical significance using 2 sample test for equality of proportions with continuity correction.
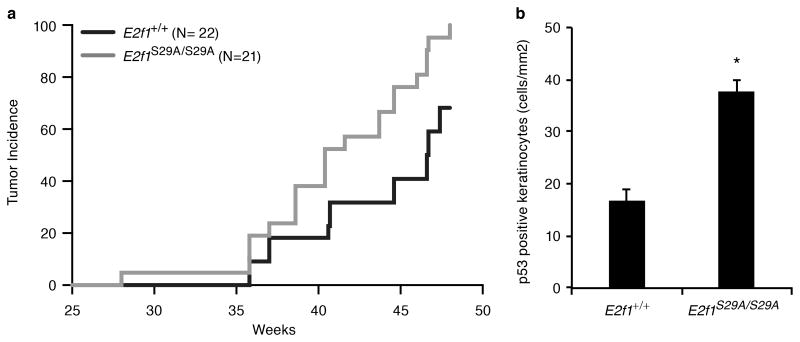
Humans and mice impaired for NER are highly susceptible to skin cancer (33). To determine the effect of the S29A mutation on UV carcinogenesis, E2f1S29A/S29A and wild type sibling control mice were exposed to 337 J/m2 of UVB three times per week for 48 weeks. E2f1S29A/S29A mice were significantly more susceptible to UV-induced skin cancer with a 95% skin tumor incidence at the end of the study compared to 68% incidence for wild type sibling mice (Figure 6a). Consistent with this increased sensitivity to UV carcinogenesis, E2f1S29A/S29A mice had more than twice as many p53-positive keratinocytes in their epidermis after 5 weeks of UV treatment compared to similarly treated wild type mice (Figure 6b). UV exposure is known to induce clones of p53-positive keratinocytes in human and mouse skin that represent precancerous lesions with p53 mutations (34, 35). These findings suggest that the impaired NER due to the E2F1 S29A mutation leads to increased p53 mutations and skin tumor development in response to UV radiation.
Figure 6. E2f1S29A/S29A mice have increased sensitivity to UV-induced skin carcinogenesis.
(a) The dorsal skin of 4-5 weeks old E2f1S29A/S29A (N=21) and wild type sibling control (N=22) mice were shaved and exposed to 337 J/m2 of UVB three times per week for 48 weeks. A statistically significant difference in skin tumor incidence over time was observed using the log-rank Mantel-Cox test (p = 0.0012). (b) Young adult wild type (E2f1+/+) and E2f1S29A/S29A mice were exposed to 337 J/m2 UVB three times per week as above for 5 weeks. 24 h after the last treatment skin samples were taken and immunohistochemically stained for p53. The average number of positive cells per mm2 of epidermis is presented from 3 mice per genotype. Bars represent standard deviation and * indicates statistical significance using two sample independent t test (p = 0.0026).
Discussion
ATM/ATR-mediated phosphorylation of E2F1 is thought to be a major mechanism by which E2F1 transcriptional activity is regulated in response to DNA damage (7, 10, 12). However, we find that mutation of this conserved phosphorylation site in murine E2F1 has little effect on the regulation of classical E2F target genes in response to UV radiation or doxorubicin. Consistent with those findings, the apoptotic and proliferative responses of E2F1S29A/S29A mice to UV was indistinguishable from wild type mice. It is possible that other E2F family members can compensate for the impaired stabilization of the E2F1 S29A mutant protein in response to DNA damage. Indeed, several other E2F family members, including E2F3, are also known to respond to DNA damage signals (36-38).
In addition to regulating E2F1 protein stability, phosphorylation of E2F1 at S31/S29 also regulates its subcellular localization and recruitment to sites of DNA damage (7, 21, 24). Accumulation of E2F1 at DNA double-strand breaks involves a phospho-specific interaction between E2F1 phosphorylated at S31/S29 and the sixth BRCT domain of TopBP1 (21). This same mechanism is likely involved in the recruitment of E2F1 to sites of UV-induced DNA damage (24). Interestingly, the DNA-binding domain of E2F1 is dispensable for E2F1 foci formation at double-strand breaks and the co-localization of E2F1 with DNA photoproducts (21, 24). The association of E2F1 with damaged DNA correlates with an enhanced recruitment of DNA repair factors and increased repair efficiency (23, 24). This transcription-independent function for E2F1 at sites of DNA damage appears to be unique among the E2F family since the S31/S29 phosphorylation site is not found in other E2F proteins.
The stimulation of NER by E2F1 involves the E2F1-dependent recruitment of GCN5 to damaged DNA, increased H3K9 acetylation, and relaxation of chromatin structure (11, 25). As expected, the S29A knock-in mutation prevented E2F1 association with UV-damaged DNA and decreased the association of GCN5, H3K9ac, and NER factors (e.g. XPA and XPC) with CPD-containing DNA. This correlates with E2f1S29A/S29A mice having reduced repair capacity and increased sensitivity to UV-induced skin carcinogenesis. While E2F1 can regulate the expression of some genes involved in NER (29, 39) we find little evidence that E2F1 deficiency or the S29A mutation significantly affects the expression of NER factors before or after UV exposure (24). Instead, the ability of E2F1 to alter chromatin structure at sites of damage and to enhance access to the repair machinery may be the major mechanism by which E2F1 suppresses UV carcinogenesis. Whether a similar mechanism is involved in E2F1-mediated tumor suppression in other contexts is unclear but this can now be addressed using the E2F1S29A/S29A mouse model.
Supplementary Material
Acknowledgments
We thank Jennifer Smith, Philip Khan, Lakshmi Paniker, Carla Carter and Pamela Blau for technical assistance, Becky Brooks for preparation of the manuscript, Christine Brown for graphics, Jianjun Shen and Luis Coletta in the Science Park Molecular Biology Core, Donna Kusewitt, Nancy Otto and coworkers in the Science Park Histology and Pathology Core, Ellen Richie and Pam Whitney in the Science Park Flow Cytometry Core, Jan Parker-Thornburg and coworkers in the MD Anderson Genetically Engineered Mouse Facility, and Lezlee Coghlan, Dale Weiss and coworkers for animal care. We also thank Dr. Michael Kastan for providing the inducible I-PpoI retroviral construct and Dr. Mark Bedford for assistance with the knock-in construct.
Grant Support: This research was supported by grants from the NIH (CA079648 to DGJ and MD Anderson Cancer Center Support Grant CA016672).
Footnotes
Conflict of Interest: The authors have no conflict of interest to disclose.
References
- 1.DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006 Nov;6(7):739–48. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 2.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009 Nov;9(11):785–97. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Field SJ, Tsai FY, Kuo F, Zubiaga AM, Kaelin WG, Jr, Livingston DM, et al. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–61. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 4.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson NJ. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–48. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 5.Rounbehler RJ, Rogers PM, Conti CJ, Johnson DG. Inactivation of E2f1 enhances tumorigenesis in a Myc transgenic model. Cancer Res. 2002 Jun 1;62(11):3276–81. [PubMed] [Google Scholar]
- 6.Yamasaki L, Bronson R, Williams BO, Dyson NJ, Harlow E, Jacks T. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1(+/-) mice. Nat Genet. 1998;18:360–4. doi: 10.1038/ng0498-360. [DOI] [PubMed] [Google Scholar]
- 7.Biswas AK, Johnson DG. Transcriptional and nontranscriptional functions of E2F1 in response to DNA damage. Cancer Res. 2012 Jan 1;72(1):13–7. doi: 10.1158/0008-5472.CAN-11-2196. Research Support, N.I.H., Extramural Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson DG, Degregori J. Putting the Oncogenic and Tumor Suppressive Activities of E2F into Context. Curr Mol Med. 2006 Nov;6(7):731–8. doi: 10.2174/1566524010606070731. [DOI] [PubMed] [Google Scholar]
- 9.Putzer BM, Engelmann D. E2F1 apoptosis counterattacked: evil strikes back. Trends Mol Med. 2013 Feb;19(2):89–98. doi: 10.1016/j.molmed.2012.10.009. Research Support, Non-U.S. Gov't. [DOI] [PubMed] [Google Scholar]
- 10.Stevens C, La Thangue NB. The emerging role of E2F-1 in the DNA damage response and checkpoint control. DNA Repair. 2004 Aug-Sep;3(8-9):1071–9. doi: 10.1016/j.dnarep.2004.03.034. Research Support, Non-U.S. Gov't Review. [DOI] [PubMed] [Google Scholar]
- 11.Velez-Cruz R, Johnson DG. E2F1 and p53 Transcription Factors as Accessory Factors for Nucleotide Excision Repair. Int J Mol Sci. 2012;13(10):13554–68. doi: 10.3390/ijms131013554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM- dependent phosphorylation. Genes Dev. 2001;15(14):1833–44. [PMC free article] [PubMed] [Google Scholar]
- 13.Wang B, Liu K, Lin FT, Lin WC. A role for 14-3-3 tau in E2F1 stabilization and DNA damage-induced apoptosis. J Biol Chem. 2004 Dec 24;279(52):54140–52. doi: 10.1074/jbc.M410493200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galbiati L, Mendoza-Maldonado R, Gutierrez MI, Giacca M. Regulation of E2F-1 after DNA damage by p300-mediated acetylation and ubiquitination. Cell Cycle. 2005 Jul;4(7):930–9. doi: 10.4161/cc.4.7.1784. [DOI] [PubMed] [Google Scholar]
- 15.Ianari A, Gallo R, Palma M, Alesse E, Gulino A. Specific role for PCAF acetylytransferase activity in E2F1 stabilization in response to DNA damage. J Biol Chem. 2004;279:20830–30835. doi: 10.1074/jbc.M402403200. [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. The EMBO journal. 2000 Feb 15;19(4):662–71. doi: 10.1093/emboj/19.4.662. In Vitro Research Support, Non-U.S. Gov't. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marzio G, Wagener C, Gutierrez MI, Cartwright P, Helin K, Giacca M. E2F family members are differentially regulated by reversible acetylation. J Biol Chem. 2000 Apr 14;275(15):10887–92. doi: 10.1074/jbc.275.15.10887. [DOI] [PubMed] [Google Scholar]
- 18.Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L, et al. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol. 2003 Jun;5(6):552–8. doi: 10.1038/ncb998. [DOI] [PubMed] [Google Scholar]
- 19.Berton TR, Mitchell DL, Guo R, Johnson DG. Regulation of epidermal apoptosis and DNA repair by E2F1 in response to ultraviolet B radiation. Oncogene. 2005 Apr 7;24(15):2449–60. doi: 10.1038/sj.onc.1208462. [DOI] [PubMed] [Google Scholar]
- 20.Wikonkal NM, Remenyik E, Knezevic D, Zhang W, Liu M, Zhao H, et al. Inactivating E2f1 reverts apoptosis resistance and cancer sensitivity in Trp53-deficient mice. Nat Cell Biol. 2003 Jul;5(7):655–60. doi: 10.1038/ncb1001. [DOI] [PubMed] [Google Scholar]
- 21.Liu K, Lin FT, Ruppert JM, Lin WC. Regulation of E2F1 by BRCT domain-containing protein TopBP1. Mol Cell Biol. 2003 May;23(9):3287–304. doi: 10.1128/MCB.23.9.3287-3304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu K, Luo Y, Lin FT, Lin WC. TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRb-independent and E2F1-specific control for cell survival. Genes Dev. 2004 Mar 15;18(6):673–86. doi: 10.1101/gad.1180204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Zhu F, Weaks RL, Biswas AK, Guo R, Li Y, et al. E2F1 promotes the recruitment of DNA repair factors to sites of DNA double-strand breaks. Cell Cycle. 2011 Apr 15;10(8):1287–94. doi: 10.4161/cc.10.8.15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo R, Chen J, Zhu F, Biswas AK, Berton TR, Mitchell DL, et al. E2F1 localizes to sites of UV-induced DNA damage to enhance nucleotide excision repair. J Biol Chem. 2010 Jun 18;285(25):19308–15. doi: 10.1074/jbc.M110.121939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo R, Chen J, Mitchell DL, Johnson DG. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res. 2011 Mar;39(4):1390–7. doi: 10.1093/nar/gkq983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lichti U, Anders J, Yuspa SH. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat Protoc. 2008;3(5):799–810. doi: 10.1038/nprot.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blattner C, Sparks A, Lane D. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Molecular Cellular Biololgy. 1999;19(5):3704–13. doi: 10.1128/mcb.19.5.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofferer M, Wirbelauer C, Humar B, Krek W. Increased levels of E2F-1-dependent DNA binding activity after UV- or gamma-irradiation. Nucleic Acids Res. 1999;27(2):491–5. doi: 10.1093/nar/27.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin PS, McPherson LA, Chen AY, Sage J, Ford JM. The role of the retinoblastoma/E2F1 tumor suppressor pathway in the lesion recognition step of nucleotide excision repair. DNA Repair. 2009 Jul 4;8(7):795–802. doi: 10.1016/j.dnarep.2009.03.003. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ianari A, Natale T, Calo E, Ferretti E, Alesse E, Screpanti I, et al. Proapoptotic function of the retinoblastoma tumor suppressor protein. Cancer Cell. 2009 Mar 3;15(3):184–94. doi: 10.1016/j.ccr.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007 Jun;9(6):683–90. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 32.Berkovich E, Monnat RJ, Jr, Kastan MB. Assessment of protein dynamics and DNA repair following generation of DNA double-strand breaks at defined genomic sites. Nat Protoc. 2008;3(5):915–22. doi: 10.1038/nprot.2008.54. Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't. [DOI] [PubMed] [Google Scholar]
- 33.Friedberg EC. How nucleotide excision repair protects against cancer. Nat Rev Cancer. 2001 Oct;1(1):22–33. doi: 10.1038/35094000. Review. [DOI] [PubMed] [Google Scholar]
- 34.Jonason AS, Kunala S, Price GJ, Restifo RJ, Spinelli HM, Persing JA, et al. Frequent clones of p53-mutated keratinocytes in normal human skin. Proc Natl Acad Sci U S A. 1996 Nov 26;93(24):14025–9. doi: 10.1073/pnas.93.24.14025. Research Support, Non-U.S. Gov't Research Support, U.S. Gov't, P.H.S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W, Remenyik E, Zelterman D, Brash DE, Wikonkal NM. Escaping the stem cell compartment: sustained UVB exposure allows p53-mutant keratinocytes to colonize adjacent epidermal proliferating units without incurring additional mutations. Proc Natl Acad Sci U S A. 2001 Nov 20;98(24):13948–53. doi: 10.1073/pnas.241353198. Research Support, Non-U.S. Gov't Research Support, U.S. Gov't, P.H.S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez LA, Goluszko E, Chen HZ, Leone G, Post S, Lozano G, et al. E2F3 is a mediator of DNA damage-induced apoptosis. Mol Cell Biol. 2010 Jan;30(2):524–36. doi: 10.1128/MCB.00938-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milton AH, Khaire N, Ingram L, O'Donnell AJ, La Thangue NB. 14-3-3 proteins integrate E2F activity with the DNA damage response. EMBO J. 2006 Mar 8;25(5):1046–57. doi: 10.1038/sj.emboj.7600999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zalmas LP, Zhao X, Graham AL, Fisher R, Reilly C, Coutts AS, et al. DNA-damage response control of E2F7 and E2F8. EMBO Rep. 2008 Mar;9(3):252–9. doi: 10.1038/sj.embor.7401158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prost S, Lu P, Caldwell H, Harrison D. E2F regulates DDB2: consequences for DNA repair in Rb-deficient cells. Oncogene. 2007 May 24;26(24):3572–81. doi: 10.1038/sj.onc.1210151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.