Abstract
Objective
Serous borderline tumor (SBT) is a unique histopathologic entity of the ovary, believed to be intermediate between benign cystadenoma and invasive low-grade serous carcinoma. While somatic mutations in the KRAS or BRAF, and rarely ERBB2, genes have been well characterized in SBTs, other genetic alterations have not been described. Toward a more comprehensive understanding of the molecular genetic architecture of SBTs, we undertook whole exome sequencing of this tumor type.
Methods
Following pathologic review and laser capture microdissection to enrich for tumor cells, whole exomes were prepared from DNA of two independent SBTs and subjected to massively parallel DNA sequencing.
Results
Both tumors contained an activating mutation of the BRAF gene. A total of 15 additional somatic mutations were identified, nine in one tumor and six in the other. Eleven were missense mutations and four were nonsense or deletion mutations. Fourteen of the 16 genes found to be mutated in this study have been reported to be mutated in other cancers. Furthermore, 12 of these genes are mutated in ovarian cancers. The FBXW7 and KIAA1462 genes are noteworthy candidates for a pathogenic role in serous borderline tumorigenesis.
Conclusions
These findings suggest that a very small number of somatic genetic mutations are characteristic of SBTs of the ovary, thus supporting their classification as a relatively genetically stable tumor type. The mutant genes described herein represent novel candidates for the pathogenesis of ovarian SBT.
Introduction
The classification of “borderline” epithelial ovarian neoplasms was originally introduced to describe tumors that are noninvasive but that occasionally seem to behave in a malignant fashion [1]. Approximately 2% of all ovarian tumors of serous histology are borderline, as compared to 78% that are benign tumors and 20% that are invasive carcinomas [2]. There appears to exist a pathological range of serous borderline tumors (SBTs), with those at the lower end of the spectrum behaving in a benign fashion, and referred to as “atypical proliferative serous tumors” (APSTs) and those at the upper end of the spectrum behaving more like low grade invasive carcinomas and referred to as “micropapillary serous tumors” (MPSTs) [ref. 1]. The current consensus is that the terms “borderline” and “atypical proliferative” are synonymous, and that “low malignant potential” not be used to describe bordeline tumors [3].
An emerging theory suggests classification of ovarian neoplasms into two types, wherein borderline tumors represent an intermediate pathologic lesion between benign cystadenomas and low-grade carcinomas in the “type I” category [1,4,5]. In contrast, “type II” tumors consist of high grade serous and other histologic type carcinomas, with no well accepted precursor lesion. This model of ovarian cancer pathogenesis is supported by traditional morphologic observations but also by molecular genetic analyses of various ovarian tumor types [5,6]. Type II serous ovarian carcinomas are notable for the ubiquitous nature of TP53 mutations [7,8], low but statistically recurrent somatic mutations in nine additional genes, and an average of 61 additional rare somatic mutations per tumor [8]. Notably, a recent study involving whole exome analysis of low-grade serous carcinomas of the ovary identified an average of only 10 somatic mutations per tumor in seven cases [9]. Thus, the genetic mutational landscape of type II serous tumors appears dramatically distinct from that of type I tumors.
Since the initial observation that SBTs frequently harbor KRAS mutations [10], subsequent studies have confirmed this observation and further demonstrated that KRAS and BRAF mutations are common in SBTs and low-grade serous carcinomas [11–14]. Mutation of KRAS and BRAF are mutually exclusive for a given tumor, and one or the other is present in approximately one-half to two-thirds of SBTs and low-grade serous carcinomas [6], although a more recent report suggests that the prevalence of these mutations in advanced-stage, low-grade serous carcinomas may be substantially lower [15]. Finally, a 12-bp insertion mutation in ERBB2, which ultimately results in KRAS activation, has also been described in a small proportion of SBTs that lack mutations in KRAS or BRAF [16,17]. Otherwise, the molecular genetic architecture of SBTs of the ovary remains unknown. The purpose of this study was to perform whole exome sequencing of SBTs to identify additional genetic alterations that may contribute to the initiation and/or progression of type I serous ovarian neoplasms.
Recent advances in technology, bioinformatics, and computational biology have led to a revolution in the mining of the cancer landscape. The application of second-generation DNA sequencing technologies, also known as next-generation sequencing, allowing for whole-genome, whole-exome, and whole-transcriptome tumor analyses, is rapidly transforming cancer genomics [18]. In the near-term, the complete molecular genetic dissection of individual tumors may be anticipated to impact not only mechanisms of cancer pathogenesis, but suggest novel approaches to diagnosis and therapeutic selection as well. To further these goals with respect to SBTs of the ovary, we sequenced the entire coding regions (exomes) of two independent tumors, SBT-s2, and SBT-s5.
Methods
Tumor specimens
The tumor and corresponding blood samples used in this study were obtained from the Fox Chase Cancer Center Biosample Repository Facility under a protocol approved by the Institutional Review Board. Two tumors, SBT-s2 and SBT-s5, were initially identified from pathology reports to meet the criteria of SBTs. Additional pathologic review confirmed the original diagnosis. Photomicrographs of H&E-stained sections of the tumors are shown in Fig. 1. Both tumors were unilateral from the left ovary, and in neither case was there evidence of additional pathology in the reproductive tract. Both tumors SBT-s2 and SBT-s5 were removed from 49-year-old patients, both approximately two years prior to this study. The tumors were flash frozen in liquid nitrogen following pathologic processing at the time of surgery, and embedded in OCT medium prior to preparation for laser capture microdissection.
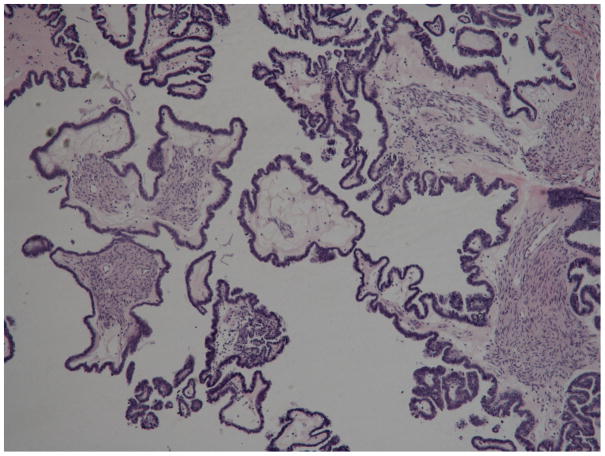
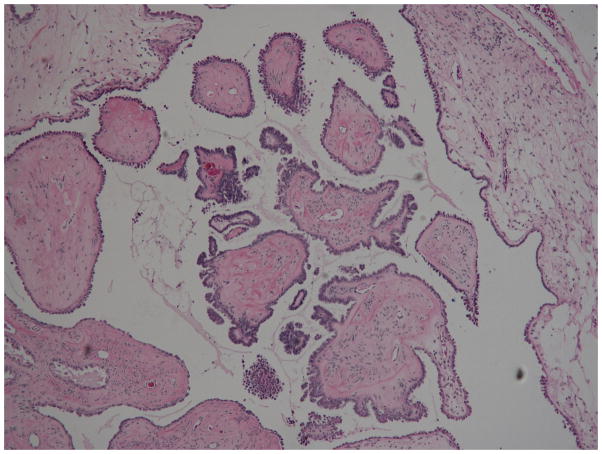
Fig. 1.
Serous borderline tumors SBT-s2 (top) and SBT-s5 (bottom). Characteristic features of SBTs include hierarchical branching of micropapillae emanating from larger, more centrally located papillae, tufting, and epithelial stratification [1].
Laser capture microdissection and DNA isolation
Tissue sections of 7 μm thickness were prepared from embedded tumors with a cryostat and adhered to uncharged Superfrost microscope slides (Fisher Scientific, Pittsburgh, PA). The first, last, and every tenth tissue section from each tumor specimen (n = 30 for SBT-s2 and n = 75 for SBT-s5) was stained with H&E and subjected to pathologic review to confirm the clinical diagnosis and homogeneity of the tissue specimen. For laser capture microdissection, slides were stained with H&E immediately prior to loading into an Arcturus Veritas instrument (Life Technologies, Carlsbad, CA). Tumor cells were selected and laser captured onto CapSure Macro LCM Caps (MDS Analytical Technologies, Sunnyvale, CA), which were then placed onto sterile nuclease-free 0.5 mL PCR tubes (Eppendorf) and stored at −80°C. Genomic DNA was isolated from the cells using the QIAamp DNA Micro kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol for isolation of genomic DNA from laser microdissected tissue, with one exception. After the addition of lysis buffer and proteinase K, tubes were incubated at 56°C for 16 hr before proceeding with the protocol. Purified DNA was eluted in 25 μl of TE buffer, quantitated with a Nanodrop ND 1000 spectrophotometer (Fisher Scientific), and stored at 4°C.
Library construction and exon capture
Samples of DNA were adjusted to a concentration of 20 μg/ml with 1x TE buffer, and 100 μl of each sample were placed in a Covaris microTUBE 6x16 mm round bottom glass tube, AFA fiber, and pre-slit snap-cap system (Covaris, Woburn, MA). Fragmentation of DNA to 300-bp was accomplished by sonication with a Covaris S2 sonicator using the following parameters: duty cycle 10%; intensity 4; cycles per burst 200; and time 120 sec. Sheared DNA was purified and concentrated using the Agencourt AMPure XP system (Beckman Coulter, Beverly, MA) and eluted into nuclease-free water according to the manufacturer’s instructions.
Libraries were prepared using reagents from the NEBNext DNA Sample Prep Master Mix Set 1 (New England Biolabs, Ipswich, MA) and custom designed adaptors and primers (Integrated DNA Technologies, Coralville, IA) for paired-end library construction as described [18]. Damaged ends of the fragmented DNA were repaired and a single A base was added to the 3′-ends using the End Repair and dA-Tailing Modules, respectively, according to the manufacturer’s protocols (New England Biolabs). Before performing adaptor ligation, 5′- and 3′-adaptors were annealed to a final concentration of 15 μM using 1x annealing buffer (100 mM Tris-HCl, pH 8.0, 1 mM NaCl) in a thermal cycler under the following conditions: 95°C for 3 min, decrease 1°C and hold for 3 min successively, until 4°C hold. Adaptors were ligated to the dA-tailed DNA fragments using the Quick Ligation Module protocol (New England Biolabs) and 3 μM annealed adaptors. Purification was performed between each enzymatic step using the Agencourt AMPure XP system (Beckman Coulter), and eluted into nuclease-free water according to the manufacturer’s instructions. Spectrophotometric readings were taken as needed using the Nanodrop ND-1000. The adaptor-ligated library was PCR amplified for 6–8 cycles with 1x Phusion High-Fidelity Master Mix (New England Biolabs) and 200 nM paired-end primers in a 200 μl final volume, aliquoted into 50 μl reaction volumes. Products from PCR were purified and combined using the Agencourt AMPure XP system and quantitated with the Nanodrop ND-1000.
Both tumor and germline DNA libraries were enriched for exomic sequence using the SureSelect Target Enrichment System (Agilent) according to the manufacturer’s instructions, with the following exceptions: the volume of adaptor-ligated library was increased from 3.4 μl per reaction to 13.8 μl, the volume of all hybridization buffers and blocking agents was doubled, and no additional water was added. The Agencourt AMPure XP system was used for purification of the exon-enriched capture library and final concentrations were measured with a High Sensitivity DNA Chip (Agilent) on an Agilent 2100 Bioanalyzer.
DNA Sequencing
Sequencing of exon-enriched libraries was performed on Illumina’s Genome Analyzer IIx as paired-end 76-bp reads at a 12 pM final concentration (one sample per lane), following the manufacturer’s instructions and using the standard sequencing primers. Image analysis and base calling were performed using the standard Genome Analyzer pipeline software, Sequencing Control Studio (SCS) and Real-Time Analysis (RTA), respectively.
Data Analysis
To remove duplicate reads and recalibrate base quality, sequence reads were mapped to the hg18 reference genome (http://www.ncbi.nlm.nih.gov/) using the publically available BWA tool [19], SAMtools [20], Picard (http://picard.sourceforge.net/command-line-overview.shtml) and GATK [21]. Single nucleotide variants and indel variants were identified using the Unified Genotyper caller of the GATK package from 44 samples, including samples from different projects. Mutations were annotated with SeattleSeq Annotation (http://gvs.gs.washington.edu/SeattleSeqAnnotation/). An SQL database was created from the annotated dataset. Somatic mutations in tumor samples were identified by comparison with sequence of the associated normal control sample, dbSNP130, 1000Genomes, and other unrelated samples in an in-house created database. In order to call somatic mutations, at least five-fold total read coverage of the area of interest was required, as well as a requirement for mutant reads to represent less than 2% of total reads in the normal control sample. In addition, manual examination was conducted with TViewer of SAMtools to identify high confidence mutations from the raw sequence data.
Mutation validation
Potential somatic mutations identified by whole exome sequencing were resequenced for validation purposes. For a given mutation, oligonucleotide primers were designed to flank the sequence variant by approximately 30-bp in both the 5′- and 3′- directions. Amplification by PCR of both somatic and corresponding germline DNA products was performed using standard methods, and products were subjected to single-pass Sanger DNA sequencing by Beckman Genomics (Beverly, MA).
Results and Discussion
The purpose of this study was to determine the degree to which ovarian SBTs of the ovary are affected by somatic genetic mutations generally, and to identify mutations potentially pathogenic for SBTs as well. To date, only three genes have been described to play such a role in ovarian SBTs, the oncogenes BRAF, KRAS, and to a lesser extent, ERBB2. It is likely that additional genes, including tumor suppressors, play a role in this tumor type; the mutually exclusive alterations observed in BRAF and KRAS are found in only a proportion of SBTs and low-grade serous ovarian carcinomas. For those low-grade carcinomas, however, mutational activation of the mitogen-activated protein kinase pathway suggests a potential target-based therapeutic approach [22]. Further elucidation of the genetic architecture of SBTs would be expected to yield significant insight into the pathogenesis of type I ovarian cancers, with those of serous histology hypothesized to develop through a morphologic continuum of benign cystadenoma, SBT, and low-grade carcinoma. The Cancer Genome Atlas (TCGA) initiative was focused exclusively on type II high-grade serous carcinomas of the ovary, but there are undoubtedly distinct genetic profiles that define type I vs. type II serous tumors, in light of their unique biological behaviors and our current knowledge of their molecular genetic features.
Not unexpected was the finding that both tumors harbored activating mutations of BRAF (Table 1), and TP53 mutations were absent from both, consistent with their pathologic diagnosis as SBTs. Interestingly, the BRAF mutation in tumor SBT-s2 (D594G) is distinct from the common activating mutation in BRAF (V600E), although it occurs in the kinase domain, has been observed in 39 independent tumor specimens listed in the Wellcome Trust Sanger Institute COSMIC (catalogue of somatic mutations in cancer) database [23], and is functionally classified as a “low-activity BRAF mutation” [24]. Other than BRAF, a total of only 15 additional unique somatic mutations were identified in the two tumors. Nine additional somatic mutations were present in SBT-s2, seven of which were missense and two of which were deletion mutations. In SBT-s5, six additional somatic mutations were present, four of which were missense and two of which were deletion mutations. Confirmation of mutations was validated using traditional Sanger technology as described in the Methods section. The mutations identified in this study were compared to those in the COSMIC database [23], and notably, 14 of the 16 genes found to be mutated in this study have been reported to be mutated in other cancers. Furthermore, 12 of these genes are mutated in ovarian cancers, the majority of which are described by the TCGA project.
Table 1.
Gene mutations identified in serous borderline tumors by whole exome sequencing.
| Gene | Genbank | Mutation | AA Change | |
|---|---|---|---|---|
| Tumor SBT-s2 | BRAF | NM_004333.4 | c.1781A>G | D594G |
| TRIO | NM_007118.2 | c.8881C>T | P2961S | |
| FOXRED2 | NM_001102371.1 | c.1114G>A | M328I | |
| HRAS | NM_001130442.1 | c.182A>G | Q61R | |
| DST | NM_015548.4 | c.964A>G | I322V | |
| UBR2 | NM_001184801.1 | c.994C>T | R332C | |
| HAUS6 | NM_017645.3 | c.1108G>T | V370F | |
| AR | NM_000044.2 | c.1013C>A | T338K | |
| C2orf16 | NM_032266.3 | c.5223_5246del24 | H1742_R1749del | |
| PEAR1 | NM_001080471.1 | c.2512_2526del15 | P839_F843del | |
| Tumor SBT-s5 | BRAF | NM_004333.4 | c.1799T>A | V600E |
| NCCRP1 | NM_0010011414.1 | c.781C>T | R261W | |
| OXGR1 | NM_080818.3 | c.766C>T | P256S | |
| FBXW7 | NM_001013415.1 | c.1288C>T | Q430X | |
| STX19 | NM_001001850.2 | c.835C>A | P279T | |
| SLC24A5 | NM_205850.2 | c.832C>T | L278F | |
| KIAA1462 | NM_020848.2 | c.1620_1623del4 | Q542fsX35 |
Several genes are of particular interest and potential relevance to SBT pathogenesis. The FBXW7 gene was found to harbor a deleterious nonsense mutation in tumor SBT-s5, and is a well characterized tumor suppressor gene [25]. The protein encoded by FBXW7 (F-box and WD40 domain protein 7) is a member of the F-box family of proteins, and participates in the ubiquitin-mediated proteolysis of several oncoproteins including cyclin E1, c-Myc, c-Jun, and Notch. The FBXW7 gene is mutated in a wide variety of human cancers including ovarian [23]. Interestingly, however, the only two ovarian cancers from the TCGA project that are listed in the COSMIC database as having FBXW7 mutations are characteristic of low-grade ovarian carcinomas; tumor TCGA-24-1565 has a BRAF mutation and tumor TCGA-25-1316 has a KRAS mutation, and neither has a mutation in TP53 [23]. The presence of mutant FBXW7 in SBT-s5 suggests that this mutation may represent an early event in the pathogenic progression of SBT to low-grade serous carcinoma. Furthermore, these observations raise the intriguing possibility that loss of FBXW7 function in low-grade serous tumorigenesis would lead to increased Notch signaling (as described above), a phenomenon associated with platinum resistance [26,27] and characteristic of low-grade serous tumors. Targeting Notch may therefore represent a novel therapeutic opportunity for low-grade serous ovarian tumors.
The KIAA1462 gene, which contains a deleterious 4-bp deletion in tumor SBT-s5, is widely expressed and evolutionarily conserved. The function of the 1,359-amino-acid protein encoded by KIAA1462 is largely unknown, however, with no recognizable functional domains and little homology to other protein families. Three additional TCGA ovarian tumors with KIAA1462 mutations, two of which are also potentially deleterious, are listed in the COSMIC database [23], implying a tumor suppressor function for this protein. Two of these tumors (one of which is TCGA-24-1565) are wild-type for TP53, again suggesting that inactivation of KIAA1462 may play a role in the pathogenesis of type I ovarian tumors.
Of the additional genes with somatic missense mutations in tumor SBT-s5 (other than BRAF), NCCRP1 (non-specific cytotoxic cell receptor protein 1 homolog) is also mutated in a TCGA ovarian cancer with mutant TP53, as is OXGR1 (oxoglutarate receptor 1), which is mutated in a melanoma sample as well. The STX19 (syntaxin 19) and SLC24A5 (solute carrier family 24, member 5) genes have no previously documented mutations in the COSMIC database. The functional significance of these missense variants remains uncertain.
With respect to tumor BST-s2, there was the unusual finding of an activating BRAF mutation together with an activating mutation in codon 61 of the HRAS gene (Table 1). Mutations in HRAS are common in skin and thyroid tumors but have not been described in ovarian cancers, and as noted before, mutations in BRAF and KRAS are mutually exclusive in SBTs and low grade ovarian cancers. Of the remaining eight genes mutated in this tumor, six have been reported to be mutated in ovarian cancers.
The PEAR1 gene (platelet endothelial aggregation receptor 1) sustained a 15-bp deletion and has been reported as mutated in two ovarian cancers from the TCGA project [8], one of which is a splice-site mutation. Remarkably, this same tumor (TCGA-24-1565) contains mutations in PEAR1, KIAA1462, FBXW7, and BRAF, four of the genes reported as mutated in one or another of the two tumors in this study. Sequence analysis of PEAR1 predicts a type-1 membrane protein, 15 extracellular epidermal growth factor-like repeats, and multiple cytoplasmic tyrosines. Analysis of the tissue distribution of PEAR1 showed that it was most highly expressed in platelets and endothelial cells, but other tissues such as the ovary as well [28].
The functional relevance of the additional seven genes that exhibit missense or in-frame deletion mutations in BST-s2 is unclear. The TRIO gene (triple functional domain) is mutated in many cancer types and three missense mutations are reported in ovarian cancers from the TCGA project, one of which does not contain a TP53 mutation [23]. Somatic missense mutations of the DST gene (dystonin) gene are reported in five high grade serous carcinomas in the TCGA database [23]. Six additional mutations are described in melanomas. A 24-bp deletion was observed in C2orf16, an uncharacterized open reading frame, which has also been mutated in a colorectal cancer [23]. The UBR2 gene (ubiquitin-protein ligase E3 component N-recognin 2) is mutated in two ovarian cancers from the TCGA database, as well as a pancreatic cancer [23]. The HAUS6 gene is also mutated in two ovarian cancers from the TCGA database [23], one of which is wild-type for TP53. The AR gene (androgen receptor) is mutated in one TCGA sample, a high grade serous carcinoma, and one glioma sample [23]. Finally, mutation of the FOXRED2 gene (FAD-dependent oxidoreductase domain containing 2) has not been reported for ovarian carcinoma, but one mutation each has been described in a kidny and lung tumor [23].
In summary, we present the first full exome analysis of SBTs of the ovary. With respect to chromosomal instability, SBTs are relatively genetically stable, in terms of both DNA ploidy [29] and copy number alterations [30], consistent with the low number of genetic mutations identified in this study. Only 10 somatic genetic mutations were identified in tumor SBT-s2, and seven in tumor SBT-s5. These data are very similar to those described in the whole exome analysis of low grade serous carcinomas [9]. As might be expected, low grade serous invasive carcinomas have a DNA content and level of copy number alterations that more closely resembles SBTs than high grade serous tumors, but which are intermediate between the two [29,30]. In contrast, high grade serous carcinomas are generally aneuploid with a high level of copy number alterations [8,30] and data from the TCGA ovarian study indicate that these tumors typically sustain 50–70 somatic mutations, with TP53 representing a clear driver mutation in this tumor type [8].
Although it is challenging to predict the functional relevance of a specific genetic mutation to SBTs without the appropriate functional studies, the mutations found in FBXW7 and KIAA1462 render them compelling candidate genes. Both sustain clear loss of function mutations, one is a well established tumor suppressor gene (FBXW7), and mutations in both are observed in ovarian tumors that are likely to be low grade serous carcinomas. Further studies on these and other genes described in this report will be necessary to determine the relevance of these results to the pathogenesis of SBTs and their role in the type I ovarian cancer tumorigenesis pathway.
Acknowledgments
This work was supported by a grant from the Sandy Rollman Ovarian Cancer Foundation.
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Seidman JD, Cho KR, Ronnett BM, Kurman R. Surface epithelial tumors of the ovary. In: Kurman RJ, Hedrick Ellenson L, Ronnett BM, editors. Blaustein’s pathology of the female genital tract. 6. New York: Springer; 2011. pp. 679–784. [Google Scholar]
- 2.Fleming GF, Ronnett BM, Seidman J, Zaino RJ, Rubin SC. Principles and practice of gynecologic oncology. 5. Philadelphia: Lippincott Williams & Wilkins; 2009. Epithelial ovarian cancer; pp. 763–835. [Google Scholar]
- 3.Seidman JD, Soslow RA, Vang R, Berman JJ, Stoler MH, Sherman ME, et al. Borderline ovarian tumors: diverse contemporary viewpoints on terminology and diagnostic criteria with illustrative images. Hum Pathol. 2004;35:918–33. doi: 10.1016/j.humpath.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Kurman RJ, Seidman JD, Shih IM. Serous borderline tumors of the ovary. Histopathology. 2005;47:310–5. doi: 10.1111/j.1365-2559.2005.02186.x. [DOI] [PubMed] [Google Scholar]
- 5.Kurman RJ, Shih I-M. The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory. Am J Surg Pathol. 2010;34:433–43. doi: 10.1097/PAS.0b013e3181cf3d79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho KR, Shih I-M. Ovarian cancer. Annu Rev Pathol. 2009;4:287–313. doi: 10.1146/annurev.pathol.4.110807.092246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmed AA, Etemadmoghadam D, Temple J, Lynch AG, Riad M, Sharma R, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 2010;221:49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Cancer Genome Atlas Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones S, Wang T-L, Kurman RJ, Nakayama K, Velculescu VE, Vogelstein B, et al. Low- grade serous carcinomas of the ovary contain very few point mutations. J Pathol. 2012;226:413–420. doi: 10.1002/path.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mok SC, Bell DA, Knapp RC, Fishbaugh PM, Welch WR, Muto MG, et al. Mutation of K- ras protooncogene in human ovarian epithelial tumors of borderline malignancy. Cancer Res. 1993;53:1489–92. [PubMed] [Google Scholar]
- 11.Singer G, Oldt R, Cohen Y, Wang BG, Sidransky D, Kurman RJ, et al. Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma. J Natl Cancer Inst. 2003;95:484–6. doi: 10.1093/jnci/95.6.484. [DOI] [PubMed] [Google Scholar]
- 12.Sieben NL, Macropoulos P, Roemen GM, Kolkman-Uljee SM, Jan Fleuren G, Houmadi R, et al. In ovarian neoplasms, BRAF, but not KRAS, mutations are restricted to low grade serous tumors. J Pathol. 2004;202:336–40. doi: 10.1002/path.1521. [DOI] [PubMed] [Google Scholar]
- 13.Mayr D, Hirschmann A, Lohrs U, Diebold J. KRAS and BRAF mutations in ovarian tumors: a comprehensive study of invasive carcinomas, borderline tumors, and extraovarian implants. Gynecol Oncol. 2006;103:883–7. doi: 10.1016/j.ygyno.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 14.Ueda M, Toji E, Noda S. Germ line and somatic mutations of BRAF V599E in ovarian carcinoma. Int J Gynecol Cancer. 2007;17:794–7. doi: 10.1111/j.1525-1438.2006.00853.x. [DOI] [PubMed] [Google Scholar]
- 15.Wong K-K, Tsang YTM, Deavers MT, Mok SC, Zu Z, Sun C, et al. BRAF mutation is rare in advanced-stage low-grade ovarian serous carcinomas. Am J Pathol. 2010;177:1611–17. doi: 10.2353/ajpath.2010.100212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakayama K, Nakayama N, Kurman RJ, Cope L, Pohl G, Samuels Y, et al. Sequence mutations and amplification of PIK3CA and AKT2 genes in purified ovarian serous neoplasms. Cancer Biol Ther. 2006;5:779–85. doi: 10.4161/cbt.5.7.2751. [DOI] [PubMed] [Google Scholar]
- 17.Wang SE, Narasanna A, Perez-Torres M, Xiang B, Wu FY, Yang S, et al. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell. 2006;10:25–38. doi: 10.1016/j.ccr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 18.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nature Rev Genet. 2010;11:685–96. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a Map Reduce framework for analyzing next-generation DNA sequencing data. Genomics. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pohl G, Ho CL, Kurman RJ, Bristow R, Wang TL, Shih IeM. Inactivation of the mitogen- activated protein kinase pathway as a potential target-based therapy in ovarian serous tumors with KRAS or BRAF mutations. Cancer Res. 2005;65:1994–2000. doi: 10.1158/0008-5472.CAN-04-3625. [DOI] [PubMed] [Google Scholar]
- 23.http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/
- 24.Smalley KS, Xiao M, Villanueva J, Nguyen TK, Flaherty KT, Letero R, et al. CRAF inhibition induces apoptosis in melanoma cells with non-V600E BRAF mutations. Oncogene. 2009;28:85–94. doi: 10.1038/onc.2008.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, et al. FBXW7/hCDCD4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–12. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 26.Rahman MT, Nakayama K, Rahman M, Katagiri H, Katagiri A, Ishibashi T, et al. Notch3 overexpression as potential therapeutic target in advanced stage chemoresistant ovarian cancer. Am J Clin Pathol. 2012;138:535–44. doi: 10.1309/AJCPKDLRQ8F3EWNS. [DOI] [PubMed] [Google Scholar]
- 27.McAuliffe SM, Morgan SL, Wyant GA, Tran LT, Muto KW, Chen YS, et al. Targeting Notch, a key pathway for ovarian cancer stem cells, sensitizes tumors to platinum therapy. Proc Natl Acad Sci USA. 2012;109:E2939–48. doi: 10.1073/pnas.1206400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nanda M, Bao M, Lin H, Clauser K, Komuves L, Quertermous T, et al. Platelet endothelial aggregation receptor 1 (PEAR1), a novel epidermal growth factor repeat-containing transmembrane receptor, participates in platelet contact-induced activation. J Biol Chem. 2005;280:24680–9. doi: 10.1074/jbc.M413411200. [DOI] [PubMed] [Google Scholar]
- 29.Pradhan M, Davidson B, Tropé CG, Danielsen HE, Abeler VM, Risberg B. Gross genomic alterations differ between serous borderline tumors and serous adenocarcinomas – an image cytometric DNA ploidy analysis of 307 cases with histogenetic implications. Virchows Arch. 2009;454:677–83. doi: 10.1007/s00428-009-0778-y. [DOI] [PubMed] [Google Scholar]
- 30.Kuo K-T, Guan B, Feng Y, Mao T-L, Chen X, Jinawath N, et al. Analysis of DNA copy number alterations in ovarian serous tumors identifies new molecular genetic changes in low-grade and high-grade carcinomas. Cancer Res. 2009;69:4036–42. doi: 10.1158/0008-5472.CAN-08-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]