Abstract
We have examined the effects of commonly used chemotherapeutic agents on human colon cancer cell lines in which the p53 pathway has been specifically disrupted by targeted homologous recombination. We found that p53 had profound effects on drug responses, and these effects varied dramatically depending on the drug. The p53-deficient cells were sensitized to the effects of DNA-damaging agents as a result of the failure to induce expression of the cyclin-dependent kinase inhibitor p21. In contrast, p53 disruption rendered cells strikingly resistant to the effects of the antimetabolite 5-fluorouracil (5-FU), the mainstay of adjuvant therapy for colorectal cancer. The effects on 5-FU sensitivity were observed both in vitro and in vivo, were independent of p21, and appeared to be the result of perturbations in RNA, rather than DNA, metabolism. These results have significant implications for future efforts to maximize therapeutic efficacy in patients with defined genetic alterations.
Introduction
The p53 gene is inactivated in the majority of human cancers, resulting in profound effects on cell birth and death processes (1, 2). Much effort has therefore gone into determining the effects of p53 inactivation on the response of cancer cells to therapeutic agents. The results have been conflicting, with some studies indicating enhanced sensitivity and others indicating increased resistance to the same compounds (see references in ref. 3).
Clinical studies have repeatedly shown that some patients respond to chemotherapeutics or radiation, whereas others, with the same histologic tumor type, do not (4). Because genetic alterations are in large part responsible for the generation and biologic properties of tumors, it is reasonable to expect that the specific alterations in tumors determine their responses to therapeutic agents. Many studies have examined the role of p53 in therapeutic responses, but the results have varied considerably. While some clinical studies have been encouraging with respect to the ability to predict responses based on p53 genotype (5–7), such studies are often confounded by tumor variability and technical difficulties in reliably assessing p53 inactivation in naturally occurring tumors (8). Preclinical studies have been pivotal in documenting effects of p53 on therapeutic responses, but also have had limitations. Studies with mouse cells, for example, have provided unequivocal evidence of drug resistance after p53 inactivation (9, 10), but the extrapolation of these results to humans is far from straightforward. The systematic screening of panels of human tumor-derived cell lines for sensitivity to therapeutic agents has revealed associations between p53 status and drug sensitivity (11), but the role of p53-unrelated genetic and epigenetic differences among diverse cell lines has made the interpretation of such results difficult. Similarly, the use of human papilloma virus–derived E6 to inactivate p53 (10) has complicated the interpretation of experiments on drug sensitivity, because E6 has major effects on cells other than just those mediated by p53 inhibition (12).
The role of p53 in the responses to therapeutics in human cells has yet to be demonstrated in an unambiguous fashion. Recent technological advances have allowed the successful targeting of individual genes in human somatic cell lines and the identification and isolation of the desired homologous recombinants (13). Here, we describe the testing of drug sensitivity in a set of isogeneic lines in which the p53 gene (14), or the gene encoding its downstream mediator p21 (15), was disrupted through homologous recombination. Because the only difference among these lines is the absence or presence of a single gene, the interpretation of results is particularly straightforward and is uncomplicated by the overexpression of exogenous genetic elements. These results suggest that p53 has a profound influence on the responses to therapeutic agents, but that the response varies considerably depending on the drug.
Methods
Cell culture and staining.
Cells were grown as monolayers in 12-well plates in McCoy’s 5A media supplemented with 10% FCS and penicillin/streptomycin. At indicated times, cells were collected by incubation with trypsin/EDTA, centrifuged, and fixed in a solution containing 3.7% formaldehyde, 0.5% Nonidet P40, and 10 μg/mL HOECHST 33258 in PBS. Stained nuclei were viewed using fluorescence microscopy and scored. A minimum of 300 cells were counted for each determination.
FACS® analysis.
Approximately 10,000 stained cells were analyzed by flow cytometry, performed as described (14).
Western analysis.
Equal numbers of cells were lysed in Laemmli sample buffer and subjected to electrophoresis and protein immunoblotting. Filters were probed with antibodies against p53 (DO-1) and p21 (EA-10). Reactive proteins were viewed using enhanced chemiluminescence (Pierce Chemical Co., Rockford, Illinois, USA).
Colony formation assay.
Cells were treated with 375 μM 5-FU for various periods of time. Cells were then washed with HBSS and collected in trypsin/EDTA. Between 1.5 × 103 and 3.8 × 104 cells were plated in drug-free medium in T-25 flasks, and then incubated for 12 days in the absence of drug. Colonies were stained with crystal violet and counted.
Xenograft tumors.
Tumors were established in athymic nude mice by subcutaneous injection of 5 × 106 cells suspended in Hank’s Balanced Salt Solution (HBSS) into both rear legs. Irradiation of xenograft tumors (n = 7–13 animals in each group) was performed as described (16). 5-FU (Pharmacia Inc., Kalamazoo, Michigan, USA) in HBSS was administered by injection into the tail vein (n = 5 for each group). Tumor growth rate was determined by measuring 3 orthogonal diameters of each tumor every 3 days. Tumor volume was estimated as π/6[D1D2D3].
Results
P53- and p21-deficient cells undergo apoptosis after treatment with DNA-damaging agents.
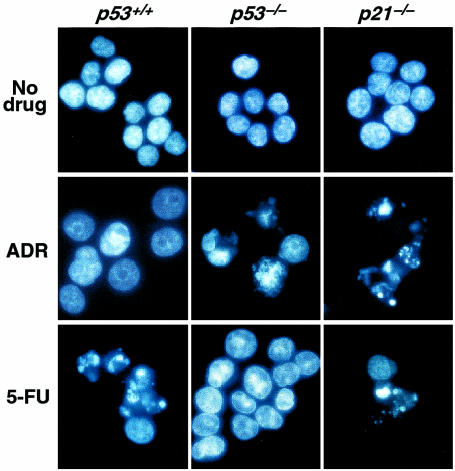
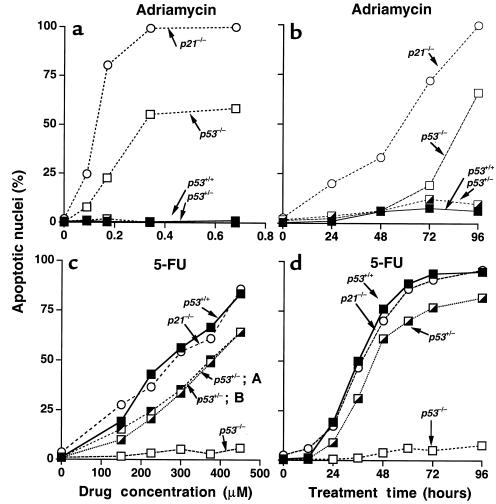
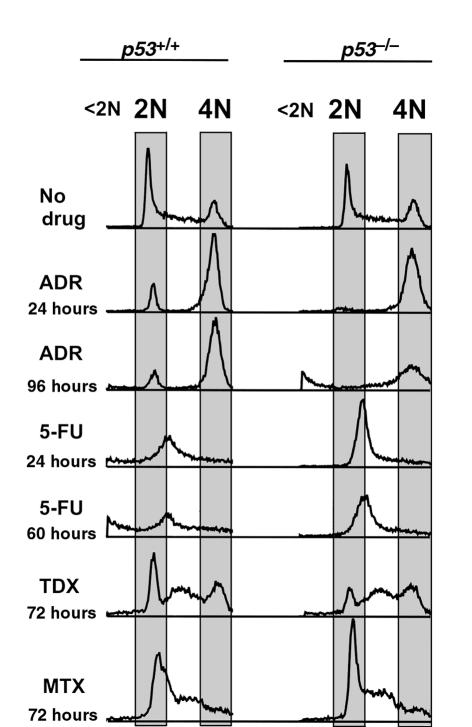
When p53-deficient cells were incubated with the DNA-damaging drug adriamycin (doxorubicin), a large proportion of cells showed nuclear changes consistent with apoptosis (Figure 1; Figure 2, a and b). Flow cytometry revealed that adriamycin-treated p53-deficient cells did not arrest in G1, but accumulated in a single peak with 4N DNA content (Figure 3). After 96 hours of treatment, a substantial proportion of nuclei had a sub-G1 DNA content characteristic of apoptosis. Similar results were seen upon treatment with ionizing radiation, which also induces DNA strand breaks (not shown). Loss of the p21 gene has been shown to result in the failure of the checkpoints that control entry into the S and M phases of the cell cycle after DNA damage (14–19). Because p53 induces p21 expression, it was reasonable to assume that the apoptotic responses of p53-deficient cells to adriamycin were mediated by the loss of p21 induction. Indeed, isogenic colorectal cancer cells with p21 disrupted by homologous recombination behaved very similarly to cells with p53 disruption (Figure 1; Figure 2, a and b). The onset of cell death after adriamycin treatment in p53-deficient cultures was somewhat delayed and reduced in extent compared with that in p21-deficient cells (Figure 2, a and b). We attribute this difference to the small amount of p53-independent p21 induction following DNA damage that occurs after adriamycin treatment of p53-deficient cells (not shown), similar to that observed after γ-irradiation (14). Aberrant progression of cells through the S and M phases of the cell cycle therefore appears to be the lethal event triggered by DNA-damaging agents in the absence of p21, whether that absence is caused by deletion of the p21 gene or its upstream inducer, p53.
Figure 1.
Nuclear morphology of cells treated with anticancer agents. Wild-type (p53+/+), p53–/–, and p21–/– cells, as indicated, were treated with 0.34 μM adriamycin (ADR) for 72 hours or with 375 μM 5-FU for 60 hours, stained with HOECHST 33258, and photographed at ×40.
Figure 2.
Induction of apoptosis by drug treatment. (a) HOECHST 33258–stained cells analyzed by fluorescence microscopy after 96 hours of treatment with adriamycin. (b) Time course of cell death after treatment with 0.34 μM adriamycin. (c) Cell death after 60 hours of treatment with 5-FU. Two different heterozygous cell lines (A and B) were assayed. (d) Time course of cell death after treatment with 375 μM 5-FU. Cells marked +/– had 1 allele of the indicated gene disrupted and were the parents of the cells with both alleles disrupted (–/–).
Figure 3.
Cell cycle distribution of drug-treated cells with wild-type (p53+/+) and disrupted (p53–/–) p53 genes. HOECHST 33258–stained nuclei were assayed by flow cytometry after treatment with 0.34 μM adriamycin (ADR), 375 μM 5-FU, 10 μM Tomudex (TDX), or 25 μM methotrexate (MTX) for the indicated time periods. Positions of peaks corresponding to diploid (2N), tetraploid (4N), and subdiploid (< 2N) DNA contents are shown. The scale on the horizontal axis shown is linear.
Cells with targeted p53 deletion are resistant to apoptosis induced by 5-FU.
Marked differences in sensitivity were observed between cell lines with and without intact p53 genes after treatment with 5-FU. Cells with wild-type p53 were quite sensitive to 5-FU, and a large proportion underwent apoptosis (Figure 1; Figure 2, c and d). This effect was completely p53 dependent, because apoptosis was not observed in p53-deficient cells (Figure 1; Figure 2, c and d). Interestingly, cells with 1 allele of p53 disrupted displayed a sensitivity between that of parental cells and cells with both p53 alleles disrupted (Figure 2c), suggesting tight control of 5-FU sensitivity by p53. Cells with targeted deletions of the p21 gene were as sensitive to 5-FU as wild-type cells (Figure 1; Figure 2, c and d). This result demonstrates that p21 does not play a role in the ability of p53 to modulate the response to 5-FU, and stands in marked contrast to the results obtained with adriamycin (Figure 1; Figure 2, a and b). The time course of cell death was rapid, with the majority of wild-type cells becoming apoptotic by 48 hours (Figure 2d). Flow cytometry of 5-FU–treated cells revealed that all cells, regardless of p53 genotype, accumulated in a single peak that spanned the G1/S phase boundary (Figure 3). This cell cycle distribution was stable in the p53-deficient cells, but not in the p53 wild-type cells, most of which displayed a sub-G1 DNA content by 60 hours (Figure 3).
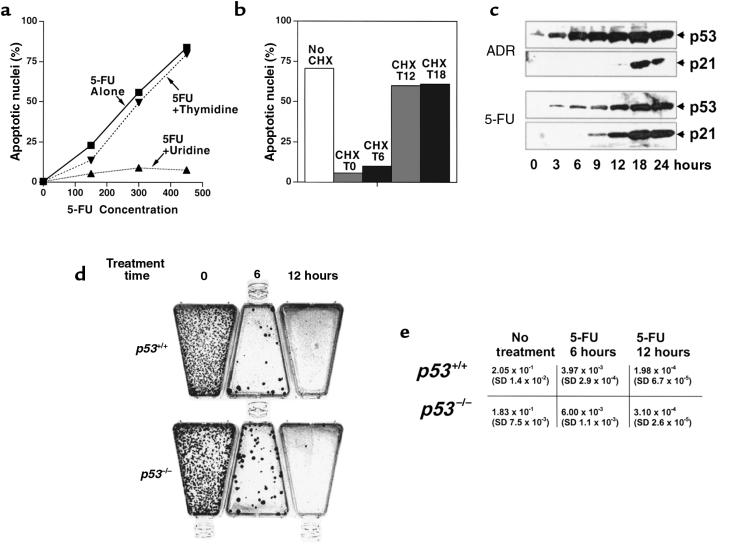
The mechanism of action underlying the therapeutic effect of 5-FU is unclear. The drug is known to be a suicide inhibitor of the enzyme thymidylate synthase (TS), which catalyzes the methylation of deoxyuridylate to thymidylate, a DNA precursor (20). Additionally, 5-FU has been shown to be misincorporated into both DNA and RNA, with consequent effects on the structure and function of these nucleic acids (20, 21). To determine whether the effects of 5-FU were the result of its effects on DNA synthesis or structure, excess thymidine was added at the time of 5-FU addition to cells. Thymidine had no effect on the apoptosis observed in these cultures (Figure 4a). In contrast, inclusion of excess uridine almost completely blocked induction of apoptosis by 5-FU. These results suggest that the impairment of thymidylate generation through inhibition of TS is not the crucial factor for the p53-mediated 5-FU sensitivity in this system. Assays of additional chemotherapeutic agents supported this idea. When incubated with the highly specific and potent TS inhibitor Tomudex (Raltitrexed; kind gift of S. Averbuch, Zeneca Pharmaceuticals, Wilmington, Delaware, USA), both p53-proficient and p53-deficient cells exhibited an S-phase block, but no differential responses between cell lines with and without intact p53 genes were observed (Figure 3). Treatment with methotrexate also induced identical responses in all cells, with an increase in the S-phase fraction consistent with inhibition of the folate-regeneration cycle required for TS function (Figure 3). High doses of either Tomudex or methotrexate caused cell death in all cell lines tested, regardless of p21 or p53 genotype (data not shown).
Figure 4.
Characteristics of 5-FU–induced apoptosis. (a) p53 wild-type cells incubated with the indicated concentrations of 5-FU alone (filled squares), or in combination with 400 μM thymidine (inverted filled triangles) or 400 μM uridine (filled triangles). (b) Cells were incubated with 5-FU in the absence (No CHX) or continuous presence (CHX) of 10 μg/mL cycloheximide. Alternatively, cycloheximide was added 6, 12, or 18 hours after addition of 5-FU (CHX T6, T12, and T18, respectively). All cells were harvested 60 hours after the addition of 5-FU. (c) Western blot of p53 and p21 proteins in whole-cell lysates after adriamycin (ADR) or 5-FU treatment. (d and e) Colony formation assay. After treatment with 375 μM 5-FU for 6 or 12 hours, cells were replated in drug-free medium and stained; colonies were counted 12 days later. Representative flasks are shown in d.
It has been shown that p53 is posttranslationally stabilized after cell stress, particularly DNA damage. Immunoblotting of cell lysates revealed that both adriamycin and 5-FU caused increases in p53 protein levels over a similar time course (Figure 4c). The stabilization of p53 was associated with increased levels of p21 (Figure 4c), a protein known to be transcriptionally regulated by p53 (1, 2). To determine whether continued protein synthesis was required for the observed p53-dependent apoptosis, the protein synthesis inhibitor cycloheximide was added to 5-FU–treated cultures. When administered up to 6 hours after 5-FU, cycloheximide was able to block cell death with intact p53 genes (Figure 4b). This result demonstrates that the p53-dependent cell death following 5-FU treatment is apoptotic in the classic sense, requiring active biosynthesis for cellular suicide (22).
Whereas 5-FU treatment led to the dramatic induction of death in cells with intact p53, a relatively small proportion of cells apparently survived and gave rise to colonies upon replating (Figure 4d). Interestingly, this proportion of clonogenic survivors did not differ significantly between cells with and without wild-type p53, indicating that clonal loss of p53 was probably not the cause of this outgrowth. A similar disparity between in vitro cell death and colony formation was observed previously in p21-deficient cells treated with ionizing radiation (16).
Treatment of xenograft tumors.
To test whether the differences in 5-FU–induced apoptosis translated to differences in drug sensitivity in vivo, cells with intact or disrupted p53 genes were grafted into athymic nude mice, and tumor growth and therapeutic sensitivity were monitored once tumors were established. To control for possible dosage variation in different animals, each mouse received 2 xenografts, 1 of each genotype. Tumors grew at similar rates once they were established, independent of p53 genotype.
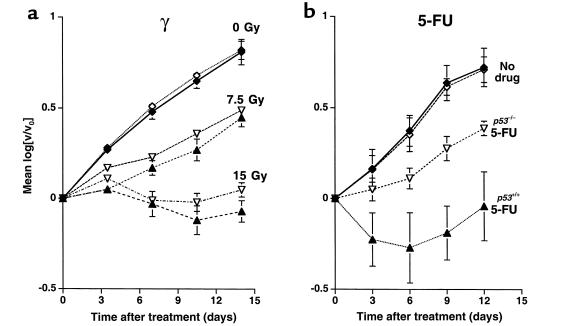
For the treatment of live animals, γ-radiation is more convenient than intravenous drug administration as a means to induce local DNA strand breakage. To examine the effects of DNA-damaging agents in vivo, we therefore chose to compare the responses of xenografts to treatment with 7.5- and 15-Gy doses of γ-radiation (Figure 5a). No significant difference in the response of p53 wild-type and p53-deficient tumors was observed (P > 0.05 for all time points, Student’s paired t test; Figure 5a). All tumors responded within 3 days of treatment and subsequently regrew at similar rates.
Figure 5.
Growth of xenograft tumors. (a) Animals with 1 tumor of each genotype were treated with either 7.5 Gy or 15 Gy doses of γ-radiation. (b) Intravenous 5-FU (12.5 mg/kg) or carrier alone (No drug) was administered on 2 consecutive days, ending on day 0. Filled triangles: tumors composed of p53++-containing cells; open triangles: tumors composed of p53–/– cells.
In contrast, there was a marked difference in the xenografts’ response to 5-FU treatment. Tumors with intact p53 genes regressed during the treatment (Figure 5b), whereas the tumors with deleted p53 genes continued to grow. There was a highly significant difference in the degree of regression in tumors with intact p53 genes compared with those with p53 deficiency (P < 0.05). After cessation of drug treatment, tumors of both genotypes grew at similar rates.
Discussion
The data presented here have several important implications for understanding and evaluating the treatment of human cancers with therapeutic agents. They confirm some previous studies that have indicated that p53 mutations confer resistance to therapeutics (23–25). However, they significantly extend these results by demonstrating that while DNA-damaging and antimetabolic drugs both function in a p53-dependent manner, the outcomes of treatment are markedly distinct. In particular, p53 disruption makes these human colorectal cancer cells more sensitive to apoptosis induced by adriamycin and radiation, but less sensitive to the apoptotic effects of 5-FU. Interestingly, our 5-FU results were in excellent accord with those on normal mouse intestinal cells. It has been shown that p53 deficiency leads to increased resistance to 5-FU in mice (26). Furthermore, the apoptosis induced in normal mouse colorectal epithelial cells by 5-FU was shown to be related to RNA metabolism rather than to DNA metabolism (26), just as in the human cells we studied (Figure 4a). The fact that similar 5-FU–related observations have been made in normal murine colorectal epithelial cells, as well as in malignant human colorectal epithelial cells, suggests that they are basic to p53 biology.
Although p53 induction has long been known to occur after DNA damage, the induction of p53 and the p53-dependent apoptosis after 5-FU treatment raises fascinating issues for further study. It has been widely believed that 5-FU works by altering DNA metabolism, thereby causing strand breaks that, in turn, activate p53-dependent apoptosis. The radically different responses of p53-deficient cells to DNA-damaging agents and 5-FU, the blockage of apoptosis by uridine but not thymidine, and previous studies (27) all indicate that this model is insufficient and that at least some major effects of the drug are likely to be mediated through defects in RNA metabolism. Many current efforts to mimic or enhance the efficacy of 5-FU employ agents that target thymidylate synthesis or DNA metabolism. Our data suggest that agents that target RNA metabolism might also be worth investigating. Further elucidation of the molecular mechanisms of 5-FU–mediated induction of p53 and the resulting apoptosis should provide insights into how this important chemotherapeutic agent functions.
Our results also suggest that p53 plays at least 2 separate roles in the responses to therapeutic agents: it is an important component of cellular checkpoints, and it can mediate apoptosis. The response to individual drugs will be determined by which of these 2 functions is paramount. The checkpoint function of p53 is mediated by the genes it transcriptionally regulates, including p21 (14, 15, 16) and 14-3-3σ (28), and keeps cells from progressing from G1 to S or G2 to M after DNA damage. The mechanisms underlying p53-induced apoptosis are less clear, but likely involve reactive oxygen species generation (29–31) and mitochondrial dysfunction, including that caused by the induction of bax (32). After treatment with DNA-damaging agents, the loss of checkpoint function in p53 mutant cells may be most important, with consequent increased sensitivity. After treatment with 5-FU, the loss of the apoptotic function of p53 may be most relevant, with a consequent decrease in sensitivity. It is also important to note that we have assumed, but not proven, that the disruption of p53 through homologous recombination yields an inactive gene that is functionally equivalent to the mutant p53 genes that arise during human tumorigenesis. It would be premature to extrapolate the results we obtained in a single cell line to the heterogeneous tumors that arise naturally.
In experiments with both DNA-damaging agents and 5-FU, tumor responses in vivo were less marked than the cellular responses observed in vitro. Whereas treatment with 5-FU did result in preferential regression of p53-proficient tumors, treatment with γ-radiation showed no discernible effect, despite the fact that the p53 knockout cells showed clear changes in the extent of apoptosis when irradiated in culture. Additionally, no effects of p53 disruption were observed in standard colony formation assays after 5-FU treatment, although the extent of 5-FU–induced apoptosis in tissue culture and the responses of tumor xenografts to 5-FU in vivo were markedly affected by p53 disruption. These results emphasize the difficulties in extrapolating from in vitro assays to in vivo situations and suggest that apoptosis may, in some cases, be a better predictor of drug responsiveness in vivo than colony formation (33).
Because 5-FU is the major drug used for colorectal cancer therapy, our study has particular relevance to the treatment of patients with this disease. It is important to note that our results do not indicate that 5-FU will be useless in p53-mutant tumors. A more reasonable interpretation is that tumors with p53 mutations are less likely to respond to 5-FU than tumors with p53 mutations. The response to all drugs, including 5-FU, is complex and unlikely to be completely explained by any single genetic alteration. The strength of the approach used here is that one can isolate a specific genetic alteration and determine its effects on drug sensitivity. This may prove to be a paradigm for determining the role of other genetic alterations in the response to established or novel drugs.
Acknowledgments
This work was supported by the Clayton Fund, the National Foundation for Cancer Research, and National Institutes of Health grants CA-57345 and CA-62924. P.M. Hwang is supported by a Howard Hughes Postdoctoral Fellowship for Physicians. We wish to thank S. Averbuch for providing Tomudex. Under an agreement between CalBiochem and The Johns Hopkins University, K.W. Kinzler and B. Vogelstein are entitled to a share of the sales royalty for the anti-p53 and anti-p21 antibodies received by the University from CalBiochem.
References
- 1.Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 2.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 3.Blandino G, Levine AJ, Oren M. Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene. 1999;18:477–485. doi: 10.1038/sj.onc.1202314. [DOI] [PubMed] [Google Scholar]
- 4.Morrow, C.S., and Cowan, K.H. 1998. Drug resistance and its clinical circumvention. In Cancer medicine. Volume 1, 4th edition. J.R. Holland et al., editors. Lea and Febiger. Philadelphia, PA. 799–815
- 5.Benhattar J, Cerottini JP, Saraga E, Metthez G, Givel JC. p53 mutations as a possible predictor of response to chemotherapy in metastatic colorectal carcinomas. Int J Cancer. 1996;6:190–192. doi: 10.1002/(SICI)1097-0215(19960621)69:3<190::AID-IJC7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 6.Brett MC, et al. p53 protein overexpression and response to biomodulated 5-fluorouracil chemotherapy in patients with advanced colorectal cancer. Eur J Surg Oncol. 1996;l2:182–185. doi: 10.1016/s0748-7983(96)90827-6. [DOI] [PubMed] [Google Scholar]
- 7.Luna-Perez P, Arriola EL, Cuadra Y, Alvarado I, Quintero A. p53 protein overexpression and response to induction chemoradiation therapy in patients with locally advanced rectal adenocarcinoma. Ann Surg Oncol. 1998;5:203–208. doi: 10.1007/BF02303772. [DOI] [PubMed] [Google Scholar]
- 8.Hall PA, Lane DP. p53 in tumour pathology: can we trust immunohistochemistry?—Revisited! J Pathol. 1994;172:1–4. doi: 10.1002/path.1711720103. [DOI] [PubMed] [Google Scholar]
- 9.Lowe SW, et al. p53 status and the efficacy of cancer therapy in vivo. Science. 1994;266:807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 10.Hawkins DS, Demers GW, Galloway DA. Inactivation of p53 enhances sensitivity to multiple chemotherapeutic agents. Cancer Res. 1996;56:892–898. [PubMed] [Google Scholar]
- 11.O’Connor PM, et al. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res. 1997;57:4285–4300. [PubMed] [Google Scholar]
- 12.Klingelhutz AJ, Foster SA, McDougall JK. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- 13.Sedivy JM, et al. Gene targeting in human cells without isogenic DNA. Science. 1999;283:9. [Google Scholar]
- 14.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 15.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 16.Waldman T, et al. Cell cycle arrest versus cell death in cancer therapy. Nat Med. 1997;3:1034–1036. doi: 10.1038/nm0997-1034. [DOI] [PubMed] [Google Scholar]
- 17.Brugarolas J, et al. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 18.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 19.Waldman T, Lengauer C, Kinzler KW, Vogelstein B. Uncoupling of S phase and mitosis induced by anticancer agents in cells lacking p21. Nature. 1996;381:713–716. doi: 10.1038/381713a0. [DOI] [PubMed] [Google Scholar]
- 20.Parker WB, Cheng YC. Metabolism and mechanism of action of 5-fluorouracil. Pharmacol Ther. 1990;48:381–395. doi: 10.1016/0163-7258(90)90056-8. [DOI] [PubMed] [Google Scholar]
- 21.Geoffroy FJ, Allegra CJ, Sinha B, Grem JL. Enhanced cytotoxicity with interleukin-1 alpha and 5-fluorouracil in HCT116 colon cancer cells. Oncol Res. 1994;6:581–591. [PubMed] [Google Scholar]
- 22.Wyllie AH. Apoptosis and carcinogenesis. Eur J Cell Biol. 1997;73:189–197. [PubMed] [Google Scholar]
- 23.Fisher DE. Apoptosis in cancer therapy: crossing the threshold. Cell. 1994;78:539–542. doi: 10.1016/0092-8674(94)90518-5. [DOI] [PubMed] [Google Scholar]
- 24.Lowe SW. Cancer therapy and p53. Curr Opin Oncol. 1995;7:547–553. doi: 10.1097/00001622-199511000-00013. [DOI] [PubMed] [Google Scholar]
- 25.Weller M. Predicting response to cancer chemotherapy: the role of p53. Cell Tissue Res. 1998;292:435–445. doi: 10.1007/s004410051072. [DOI] [PubMed] [Google Scholar]
- 26.Pritchard DM, Watson AJ, Potten CS, Jackman AL, Hickman JA. Inhibition by uridine but not thymidine of p53-dependent intestinal apoptosis initiated by 5-fluorouracil: evidence for the involvement of RNA perturbation. Proc Natl Acad Sci USA. 1997;94:1795–1799. doi: 10.1073/pnas.94.5.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cory JG, Breland JC, Carter GL. Effect of 5-fluorouracil on RNA metabolism in Novikoff hepatoma cells. Cancer Res. 1979;39:4905–4913. [PubMed] [Google Scholar]
- 28.Hermeking H, et al. 14-3-3σ is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–8. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 29.Vayssiere JL, Petit PX, Risler Y, Mignotte B. Commitment to apoptosis is associated with changes in mitochondrial biogenesis and activity in cell lines conditionally immortalized with simian virus 40. Proc Natl Acad Sci USA. 1994;91:11752–11756. doi: 10.1073/pnas.91.24.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson TM, Yu Z-X, Ferrans VJ, Lowenstein RA, Finkel T. Reactive oxygen species are downstream mediators of p53-dependent apoptosis. Proc Natl Acad Sci USA. 1996;93:11848–11852. doi: 10.1073/pnas.93.21.11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 32.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 33.Lamb JR, Friend SH. Which guesstimate is the best guesstimate? Predicting chemotherapeutic outcomes. Nat Med. 1997;3:962–963. doi: 10.1038/nm0997-962. [DOI] [PubMed] [Google Scholar]