Abstract
Excess dietary salt intake induces the activity of the kidney arachidonate epoxygenase and markedly increases the urinary excretion of its metabolites. The epoxyeicosatrienoic acids, products of the kidney P-450 arachidonate epoxygenase, inhibit distal nephron Na+ reabsorption. Nucleic acid hybridization studies demonstrated the expression of P-450s 2C23, 2C24, and 2C11 as the predominant kidney 2C isoforms and the lack of significant dietary salt-dependent transcriptional regulation of these proteins. Recombinant P-450s 2C11, 2C23, and 2C24 catalyze arachidonate metabolism to mixtures of epoxy- and monohydroxylated acids. Whereas the arachidonate 11,12-olefin was the preferred target for epoxidation by P-450 2C23 (57% of total products), P-450s 2C11 and 2C24 epoxidized the 11,12-olefins and 14,15-olefins with nearly equal efficiency. Stereochemical comparisons demonstrated that the regiochemical and enantiofacial selectivity of P-450 2C23 matched that of the kidney microsomal epoxygenase and that excess dietary salt does not alter the regiochemical or stereochemical selectivity of the kidney arachidonate epoxygenase. Inhibition and immunoelectrophoresis experiments using antibodies raised against recombinant P-450s 2C11 and 2C23 demonstrated that P-450 2C23 is the major 2C arachidonic acid epoxygenase in the rat kidney and the renal P-450 isoform regulated by excess dietary salt intake.
Introduction
The rat kidney microsomal cytochrome P-450 (P-450) arachidonic acid (AA) epoxygenase catalyzes the asymmetric epoxidation of AA to 8(R),9(S)-EET, 11(R),12(S)-EET, and 14(S),15(R)-EET (1). Antibody inhibition experiments indicated that AA epoxidation by these microsomal fractions was catalyzed predominantly by P-450 2C gene subfamily isoforms (1–5). The potent biological activities associated with the several metabolites of the AA epoxygenase and the demonstration that P-450 participates in the in vivo metabolism of AA pools established the epoxygenase as a member of the AA cascade and suggested functional roles for this enzyme in the generation of bioactive eicosanoids (6–11).
The EETs and/or their hydration products, the vic-dihydroxyeicosatrienoic acids (DHETs), have been implicated in a variety of physiologically important processes, including the regulation of renal tubular Na+ and K+ fluxes and water permeability (6–11), and have been shown to be potent systemic vasodilators and enantioselective intrarenal vasoconstrictors (6–11). Studies with the spontaneously hypertensive rat models (6, 10), as well as marked changes in the urinary excretion of epoxygenase metabolites during pregnancy-induced hypertension (12), suggested that this pathway may be of importance to the pathophysiology of hypertension (6–12). The regulatory control of the renal epoxygenase by dietary salt intake (1), as well as the inhibitory effect of the EETs on tubular Na+ reabsorption provided further support for a role of this pathway in kidney physiology (6–11). Importantly, clotrimazole inhibition of the rat kidney epoxygenase activity leads to the development of salt-sensitive, clotrimazole-dependent hypertension (13). In previous studies it was established that excess dietary salt increased the kidney concentration of a P-450 isoform(s) recognized by polyclonal antibodies raised against human liver P-450 2C9 or rat liver P-450 2C11 (1) and that, although it is different from P-450 2C11, the salt-induced protein was a member of the P-450 2C gene subfamily. P-450 2C23, the predominant P-450 2C isoform in the rat kidney, was cloned, expressed, and shown to be an active AA epoxygenase (2, 3). However, nucleic acid hybridization studies failed to demonstrate a dietary salt-dependent transcriptional regulation of this P-450 isoform (1, 2).
Salt sensitivity is the causative agent for an important subgroup of humans with essential hypertension. However, in spite of extensive studies, the molecular and/or genetic basis of salt sensitivity remains to be defined. Whereas salt sensitivity is likely to be a multigenetic trait, a paucity in the identification of novel candidate genes has limited progress in this clinically important area of research. We report here (a) the structural and catalytic characterization of the predominant rat kidney AA epoxygenases; (b) the identification of P-450 2C23, a P-450 2C gene subfamily isoform, as a salt-sensitive renal epoxygenase; and (c) the characterization of a P-450 2C23 product, the 11(R),12(S)-EET, as the major rat kidney epoxygenase metabolite.
Methods
Animals, microsomal fractions.
Adult male Sprague Dawley rats (300–350 g; Harlan Sprague-Dawley Inc., Indianapolis, Indiana, USA) or male Dahl salt-resistant (DR) and Dahl salt-sensitive (DS) rats (280–320 g)(outbred, Brookhaven National Laboratories, Upton, New York, USA) were used for these studies. Salt loading was done either by providing 2% NaCl as drinking water for 10 days or by feeding rats a modified lab chow containing 8% NaCl (Special Mix 5001-2; Purina Mills Inc., St. Louis, Missouri, USA) for 30–40 days (Sprague-Dawley) or 12–18 days (Dahl) and allowing free access to drinking water. Kidney and liver microsomal fractions were isolated as described (14), maintained at 2–4°C, and discarded after 48 hours. P-450 and protein concentrations were determined as in ref. 14.
Enzymatic studies.
Microsomal incubations were done exactly as in ref. 14. Briefly, reaction mixtures containing 50 mM Tris-Cl buffer (pH 7.5), 150 mM KCl, 10 mM MgCl2, 8 mM sodium isocitrate, 0.5 IU of isocitrate dehydrogenase, microsomal protein (0.5–2.0 mg/mL), [1-14C]AA (2–8 μCi/μmol; 70–100 μM final concentration), and 1 mM NADPH were incubated at 35°C. At different time points the reaction products were extracted into acidified ethyl ether, resolved by reverse-phase HPLC as in refs. 14 and 15, and quantified by on-line liquid scintillation (Flow-One β Counter; Radiomatic Instruments, Tampa, Florida, USA). Initial rates of metabolism were calculated from plots of product concentrations vs. incubation times.
Purified recombinant P-450s 2C11, 2C23, or 2C24 (50–200 pmol) were sequentially mixed with dilauroylphosphatidylcholine (5–50 μL of a 1 mg/mL suspension), NADPH cytochrome P-450 reductase (to obtain P-450/NADPH P-450 reductase molar ratios between 1:1 and 1:15), and, when needed, cytochrome b5 (to obtain P-450/cytochrome b5 molar ratios between 0.5:1 and 1:5). After 10 minutes at 25°C, the mixture was diluted 10-fold with Tris-Cl buffer (pH 7.5) containing 0.15 M KCl, 10 mM MgCl2, 8 mM sodium isocitrate, and isocitrate dehydrogenase (0.5 I./mL), then was preincubated for 1 minute at 35°C before the addition of [1-14C]AA (20–50 μCi/μmol; 50–70 μM, final concentration) and NADPH (1 mM, final concentration). The AA metabolites were extracted and analyzed as above (14). For chiral analysis, EETs were purified by a combination of reverse-phase and normal-phase HPLC (14, 15), derived, and characterized by chiral-phase HPLC as in (15). Absolute configurations were assigned using published data (15).
Nucleic acid hybridizations, cDNA cloning, and characterization.
Liver and kidney total RNAs were isolated by the guanidinium isothiocyanate/CsCl method (16). Poly(A)+ RNA was isolated using a mRNA extraction kit (Amersham Pharmacia Biotech, Piscataway, New Jersey, USA). The mRNA samples (2–5 μg each) were electrophoresed in agarose gels (1.2%) containing 0.2 M formaldehyde as described (2, 16). After capillary pressure transfer to nitrocellulose membranes, the blots were hybridized to the following 32P-labeled P-450 isoform–specific probes: (a) P-450 2C6, 48-mer 5′-TGGATGATAGAGGGAAGGGACTGGATAAAAACCTGAAGGGGATT GCAG-3′; (b) P-450 2C7, 35-mer 5′-AAGTGGATTTGTGCTATA ACTCCAGAAACTGCATG-3′; P-450 2C11, 48-mer 5′-ACTGGATCCGAAGGAAATGGGGATATGTGAGTTAAAGCTACT ATAGTA-3′; P-450 2C13, 48-mer 5′-GGATGATAGAGGGAAGGGACTGGATAAAAACCTGA AGGGGATTGCAG-3′; P-450 2C22, 47-mer 5′-AATGCCTAACCCTTGGCTGACTTTACTGAG CATTGACGAATGCATT-3′; P-450 2C23, 48-mer 5′-TCTGGTGTCAGTTGTGCTTGGGGCCT GGTTTCTAGAGCAGTTGAGGGA-3′; P-450 2C24, 48 mer 5′-GTTATAGACTGTGGGAGAC AAAGGATAGATTTGCGAGATAACTCTGG-3′. End labeling of oligonucleotides was done using [γ-32P]ATP and T4 polynucleotide kinase. Hybridizations were performed at 42°C in Hybrisol (Oncor Inc., Gaithersburg, Maryland, USA). After 3 consecutive 15-minute washes — the first in 2× SSC, 0.1% SDS (wt/vol) at 42°C; the second in 2× SSC, 0.1% SDS (wt/vol) at 25°C; and the last in 0.5× SSC, 0.1% SDS (wt/vol) at 25°C — the membranes were exposed to x-ray film.
P-450 2C11 was cloned from a Uni-Zap λgt11 male rat liver cDNA library (Stratagene, La Jolla, California, USA). The library was plated in XL1-Blue Escherichia coli as host (105 plaque-forming units per plate), and approximately 106 clones were screened using a 1-kb probe coding for the published 3′-end sequence of P-450 2C11 (17). A 1,896-bp clone, containing the entire P-450 2C11 coding sequence and 396 bp of the 3′-end untranslated sequence, was isolated, purified, rescued into pBluescript SR(+) (Stratagene), and characterized. A 1,693-bp cDNA coding for P-450 2C24 was cloned by RT-PCR amplification of total rat kidney RNA using the following PCR primers: forward, 5′-CTTCTTGAGAAGAGAAGGCTGTCATGGATC-3′; reverse, 5′-TAGACTGTGGGA GACAAAGGATAGATTTGC-3′. The resulting cDNA was cloned into the PCR 2.1 vector (Invitrogen, Carlsbad, California, USA) and characterized. The cDNA coding for P-450 2C23 was cloned and characterized as described (2). The P-450 2C11 and 2C24 cDNAs were sequenced using a Thermo Sequenase Kit (Amersham Pharmacia Biotech, Piscataway, New Jersey, USA). Between 25 and 30 oligonucleotides (20–25 mer each), spanning the length of each cDNA, were used as sequencing primers for the corresponding sense or antisense cDNA strands.
Heterologous expression and purification of recombinant P-450s.
The cDNAs coding for P-450s 2C11, 2C23, and 2C24 were expressed using a MAXBAC Baculovirus Expression System (Invitrogen), following the manufacturer’s instructions. Recombinant P-450 baculoviruses were used to infect sf9 insect cell cultures in the presence of 0.2 μM hemin. P-450 expression levels were determined in Emulgen 911 (Kao Chemical Co., Tokyo, Japan) cell lysates by CO-difference spectroscopy (18).
Recombinant P-450s 2C11, 2C23, and 2C24 were purified from insect cell lysates as described (19). Briefly, insect cell microsomes were collected by centrifugation and solubilized in 0.1 M sodium phosphate buffer (pH 7.4) containing 20% (vol/vol) glycerol, 0.1 mM EDTA, 0.1 mM DTT, and 1% (wt/vol) sodium cholate. After 1 hour at 4ºC, insoluble proteins were removed by centrifugation (1 hour, 140,000 g), and the solubilized enzymes were purified by octyl-Sepharose 4B (Pharmacia Biotech AB, Uppsala, Sweden), hydroxylapatite (Bio-Rad Laboratories Inc., Richmond, California, USA), and Bio-Gel A DEAE (Bio-Rad Laboratories Inc.) column chromatography, exactly as in ref. 19. Residual Emulgen 911 was removed from purified proteins by hydroxylapatite chromatography, as in ref. 19. Before use, proteins were dialyzed vs. 100 vol of 20 mM Tris-Cl buffer (pH 7.4) containing 20% glycerol (vol/vol), 0.1% sodium cholate (wt/vol), and 0.1 M mM EDTA.
Other methods.
Polyclonal antibodies against purified recombinant P-450s 2C11 and 2C23 were raised in female New Zealand white rabbits and purified by affinity chromatography on a protein G Superose HR 10/2 column (Pharmacia Biotech AB, Uppsala, Sweden) as published (19). Cytochrome b5 and rat liver NADPH–P-450 reductase were purified to electrophoretic homogeneity (20). Discontinuous SDS-PAGE (10% wt/vol acrylamide, 100 × 60 × 1 mm slabs) were as described in refs. 1 and 19. Resolved proteins were transferred onto PVDF membranes and immunoblotted as described (21). Antigen-antibody reactions were detected using either a SuperSignal Substrate Western Blotting kit (Pierce Chemical Co., Rockford, Illinois, USA) according to the manufacturer’s instructions or alkaline phosphatase detection methods per refs. 1, 4, and 19. For NH2-terminal amino acid analyses, purified P-450s were submitted to SDS-PAGE in 200 × 120 × 1.5–mm slabs using a 7–12% (wt/vol) polyacrylamide gradient as separating gel. Proteins were electroblotted onto PVDF membranes and sequenced (21). Cycle yields were calculated by comparison with internal standards.
Results
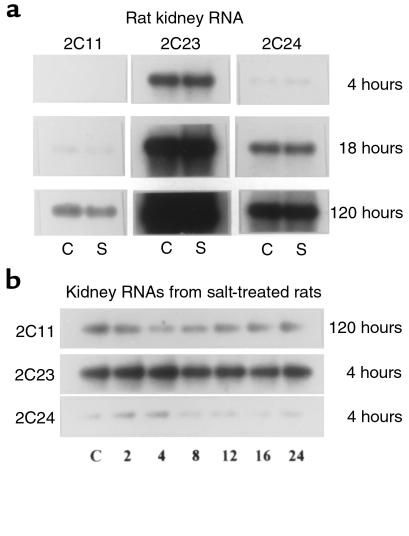
The P-450 supergene family is characterized by an increasing complexity of isoforms that, despite extensive sequence homology, display substantial degrees of catalytic heterogeneity (22–25). Under basal conditions, most of the AA epoxygenase activity present in rat or rabbit liver and kidney microsomes is catalyzed by P-450 2C gene subfamily isoforms (1–5). In rats the P-450 2C gene subfamily is composed of 8 members (23), 2 of which are sex-specific (P-450s 2C11 and 2C12, ref. 23) and a third, P-450 2C22, that is expressed only in cultured hepatocytes (26). To compare the isoform heterogeneity, the relative abundance of transcripts coding for P-450 2C isoforms, and their transcriptional regulation by dietary salt intake, mRNAs isolated from the livers and kidneys of control and salt-treated rats (2% NaCl as their drinking water for 2 weeks) were electrophoresed and hybridized to 2C isoform–specific oligonucleotides (35–48 mer each, coding for the 3′-end untranslated sequences of each cDNA; ref. 23). As reported (22, 23, 27, 28), the mRNAs coding for P-450s 2C6, 2C7, 2C11, 2C13, 2C23, and 2C24 are all expressed in the male rat liver, with P-450 2C11 as the most abundant 2C isoform (27, 28) followed, in decreasing order of relative abundance, by P-450s 2C6, 2C13, 2C23, 2C7, 2C24, and nearly undetectable levels of P-450 2C22 (data not shown). On the other hand, the limited 2C isoform heterogeneity of the rat kidney is illustrated by the expression, at levels detectable by nucleic acid hybridization techniques, of only P-450s 2C23, 2C24, and 2C11 (Figure 1) (2). Based on these studies, we concluded that P-450 2C23 was the predominant 2C isoform expressed in the rat kidney (1–3), followed by P-450 2C24 and by noticeably lower levels of P-450 2C11, and excess dietary salt did not significantly alter the transcriptional regulation of liver (not shown) or kidney P-450 2C genes (Figure 1).
Figure 1.
Nucleic acid hybridization analysis of male rat kidney RNAs. Samples of poly(A)+ RNA (5 μg each) isolated from the kidneys of control rats (C) and rats fed a 2% NaCl solution as drinking water for either 10 days (S) (a) or for the indicated times (2, 4, 8, 12, 14, or 24 hours) (b) were electrophoresed and blot-hybridized to P-450 isoform–specific oligonucleotide probes coding for portions of the 3′-end untranslated sequences of P-450s 2C11, 2C23, and 2C24. After exposure to x-ray film, the blots were stripped by boiling and probed with a β-tubulin to confirm equal loading. To optimize the analysis of mRNA abundance, the P-450 blots were exposed to the x-ray film for 4, 18, or 120 hours. See Methods for further details.
To probe for early time-dependent and dietary salt–dependent changes in the transcription of kidney P-450s 2C11, 2C23, or 2C24, kidney poly(A)+ RNAs were isolated from control rats and from rats exposed to a 2% NaCl solution for the indicated time periods, then were analyzed using gene-specific probes. Within the first 24 hours of salt loading, the levels of P-450 2C23 mRNA transcripts remained more or less constant, whereas P-450s 2C11 and 2C24 appeared to be slightly downregulated (Figure 1). Nucleic acid hybridization data also showed that whereas excess dietary salt caused small decreases in the kidney concentration of P-450 2C11 transcripts (1, 2), the levels of mRNAs coding for P-450s 2C6, 2C7, 2C13, 2C22, 2C23, or 2C24 were not significantly altered by excess salt (1, 2, 13). Based on these and previous studies (1, 2, 13), it was concluded that the salt-dependent upregulation of the renal AA epoxygenase occurred in the absence of measurable changes in the transcriptional activities of known P-450 2C gene subfamily isoforms. Furthermore, using similar approaches, we demonstrated that the salt loading had no effect in the transcriptional regulation of P-450s: 1A1, 1A2, 2B1, 2B2, 2A1, 2A2, 2D1, 2D2, 2D3, 2D4, 2E1, 2E2, 2G1, 2J1, 3A4, 4A1, 4A2, and 4A3. Transcriptional and/or translational events are known to play important and, in some cases, isoform-specific roles in the regulation of many P-450 proteins (22). Additionally, the transcription-independent regulation of P-450 isoforms has been demonstrated (22, 28, 29). The absence of demonstrable salt-dependent transcriptional upregulation of a particular P-450 isoform made the characterization of the salt-responsive epoxygenase(s) at the nucleic acid level impractical. As an alternative, we chose to clone and express the cDNAs coding for kidney 2C P-450s and to use the recombinant proteins for enzymatic characterization and the generation of immunospecific probes.
Cloning of cDNA, characterization, and heterologous expression.
Among the rat kidney P-450 2C isoforms, P-450s 2C11 and 2C23 have been shown to catalyze AA epoxidation (2–4). However, low rates of catalysis and limited regioselectivity and stereoselectivity suggested the presence of contaminant P-450 proteins in the purified preparations of liver P-450 2C11 (4). To generate recombinant P-450s 2C11 and 2C24 suitable for unequivocal catalytic characterization we screened a rat liver cDNA library using either a 1-kb fragment coding for the 3′-end sequence of P-450 2C11 (17) and an oligonucleotide complementary to a portion of the 5′-end of P-450 2C11 (25-mer, 5′-CTGCCATGGATCCAGTCCTAGTCCT-3′) (17) or a 200-bp cDNA probe coding for a portion of the 3′-untranslated end of P-450 2C24 (starting at nucleotide 1,600 from the ATG initiation) (30). Approximately 40 duplicated positive clones were isolated, one of which, containing an insert with a 5′-end sequence identical to that reported for P-450 2C11 (17), was selected for further characterization. Sequence analysis showed that this clone contained an insert of 1,896 nucleotides with a 99.8% sequence identity to the published rat liver P-450 2C11 (17, 23). An open reading frame coding for a polypeptide of 500 amino acids (mol wt = 57,159) was flanked by initiation and termination codons (ATG and TAA, respectively) between nucleotides 2 and 1,500, respectively. Compared with the published cDNA (17), our clone was 141 nucleotides longer, contained a 3′-end poly(A) tail, and showed the following base substitutions: G→A, C→T, and C→A at nucleotides 1,006, 1,594, and 1,822 of the published sequence (17, 23), respectively. With the exception of a conserved histidine→arginine substitution at residue 329, the sequence of the 2C11 translated protein was identical to that published previously (17), including the invariant cysteine at position 435. It is of interest that a similar strategy, i.e., screening of liver or kidney cDNA libraries with gene-specific oligonucleotide probes, failed to yield a single clone containing a full-length P-450 2C24 coding insert. The abnormal processing of P-450 2C24 transcripts, including the existence of circular mRNA transcripts, has been reported (31).
A cDNA coding for rat kidney P-450 2C24 was obtained by RT-PCR of total rat kidney RNA and cloned into the PCR 2.1 vector. The P-450 2C24 cDNA was 1,693 bp long and contained the open reading frame coding for P-450 2C24 (ATG and TAA initiation and termination codons were at bp 5 and 1,499 of the published sequence, respectively) (30). With the exception of a single base substitution (G→C at bp 1,162, from the initiation ATG), this cDNA was identical to the published P-450 2C24 cDNA (30). The cloned cDNA coded for a protein of 490 amino acids (55,997 Da), with a conserved heme-binding peptide (FSTGKRMCVG) and a distal cysteine heme iron ligand at position 435. The molecular properties of the P-450 2C23 cDNA clone are published (2, 3). The cDNA and protein sequence homology analysis showed that whereas P-450 2C11 and 2C24 share a 76% sequence identity, their similarities to P-450 2C23 were more limited (61.6 and 60.5% sequence identity for P-450s 2C11 and 2C23, respectively; ref. 23). The sequence differences between P-450 2C23 and either 2C11 or 2C24 are more or less randomly distributed along the entire polypeptides, suggesting that the 2C23 gene diverged from the other 2C genes at an early evolutionary stage.
Heterologous expression and protein purification.
Among the heterologous expression systems for P-450 cDNA expression, we chose the baculovirus/insect cell system because (a) its use for the expression of cloned P-450 cDNAs was well documented (32); (b) all reagents, including the vectors, viruses, and cells are commercially available; (c) it requires minimal manipulations of the cloned cDNA, different than bacterial expression systems; (d) the primary structure of the recombinant protein is preserved (32); and (e) yields of 50–100 nmol of P-450 protein per liter of cell culture have been reported (32). Using this expression system, we achieved high-yield expression of P-450s 2C11 and 2C23 (70–120 nmol of hemoprotein per liter of culture, respectively). On the other hand, under similar conditions, the yields of recombinant P-450 2C24 were substantially lower (≤10 nmol of P-450 2C24 per liter of culture). Recombinant proteins were purified as described in Methods and, before use, nonionic detergents such as Emulgen 911 were removed by chromatography on hydroxylapatite (19). Pilot experiments demonstrated that low concentrations of nonionic detergents and DTT inhibited the metabolism of AA by the purified enzymes. Purified recombinant P-450s 2C11, 2C23, and 2C24 (approximately 12, 14, and 5 nmol of P-450/mg of protein, respectively) were used for molecular and enzymatic characterizations. The SDS-PAGE properties of purified P-450s 2C11 and 2C23 (i.e., the presence of a single protein band) suggested that these preparations contained significant amounts of the corresponding apoproteins (19, 32). On the other hand, the SDS-PAGE analysis of purified P-450 2C24 showed several additional protein contaminants. However, low expression levels and poor purification efficiency precluded further attempts to improve its purity.
Spectral, structural, and enzymatic characterization.
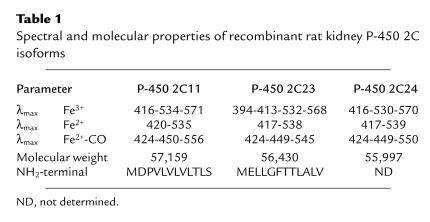
Discrepancies in the reported sequences made the translation of the P-450 2C23 protein sequence equivocal (2, 3, 33, 34). To determine the primary sequence of P-450s 2C11 and 2C23, we microsequenced the NH2-terminal of both recombinant proteins. The protein NH2-terminal sequence of recombinant P-450 2C23 is in agreement with recently published data (13, 14), but is different from the translation data of Cook et al. (33) (Table 1). On the other hand, the NH2-terminal amino acid sequence of recombinant P-450 2C11 matches literature data (17) (Table 1). The NH2-termini of P-450s 2C11 and 2C23 show the predominance of hydrophobic and low-polarity amino acids, typical of microsomal P-450s (Table 1) (23). P-450 2C23 differs from other 2C isoforms in that it contains a few extra residues, and a glutamic acid next to the NH2-terminal methionine (Table 1) (23). The absolute spectra of P-450s 2C11 and 2C24 indicate that they were isolated as predominantly low spin proteins (Fe3+ Soret band, λmax at 413–416 nm) (Table 1). Conversely, P-450 2C23 was recovered as mixtures of low- and high-spin proteins (Fe3+ Soret band, λmax at 394 and 413 nm) (Table 1). In their oxidized state (Fe3+), all 3 hemoproteins show typical P-450 α, β, and γ absorption bands (Table 1). As with most P-450s, reduction of the heme iron (Fe2+) resulted in a smaller Soret band and the replacement of the α and β bands by a single broad absorption band at around 550–560 nm (Table 1). The CO complexes of reduced P-450s (Fe2+-CO) showed typical hyperchromic shifts of their reduced Soret γ bands to 449–450 nm, the development of small shoulders at approximately 424 nm, and the presence of a single broad band centered between 545 and 556 nm (Table 1).
Table 1.
Spectral and molecular properties of recombinant rat kidney P-450 2C isoforms
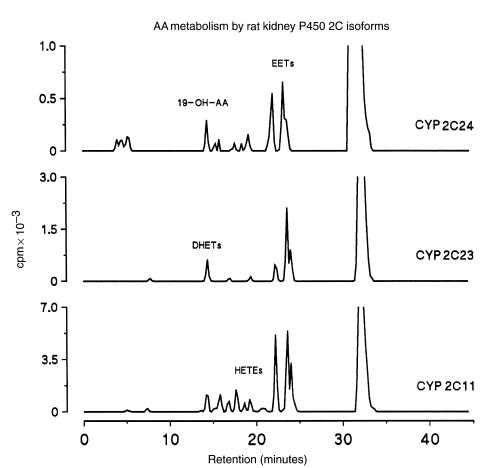
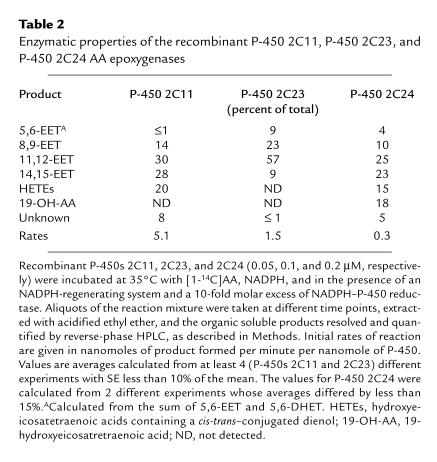
In previous studies, microsomal fractions isolated from COS-1 cells expressing recombinant P-450 2C23 were shown to catalyze the regio- and stereoselective epoxidation of AA (2). More recently, a rat kidney P-450 2C23 was cloned, expressed, and shown to catalyze AA epoxidation although EET chirality was not determined (3, 34). As discussed, the cDNA coding for P-450 2C11 was cloned several years ago (17); however, its role in the renal metabolism and/or bioactivation of AA was not addressed. Also, P-450 2C24, the second most abundant kidney 2C isoform, has not been characterized enzymatically. For enzymatic analysis recombinant P-450s 2C11 and 2C23 were reconstituted in the presence of 1,2-dilauroyl-phosphatidylcholine (50–100 μg/mL), 0.15 M KCl, and variable concentrations of NADPH–cytochrome P-450 reductase. Conversely, optimal catalytic turnover with P-450 2C24 was obtained in the absence of KCl and dilauroylphosphatidylcholine. Recombinant P-450s 2C11, 2C23, and 2C24 actively catalyzed the NADPH-dependent metabolism of AA to regioisomeric EETs (retention time [Rt] = 24–28 minutes; Figure 2) and small quantities of DHETs (Rt = 14–18 minutes; Figure 2), derived from EET chemical hydration (14). Whereas epoxidation accounted for nearly 98% of the reaction products generated from AA by P-450 2C23 (Table 2), P-450s 2C11 and 2C24 generated, in addition to EETs, small quantities of metabolites with reverse- and normal-phase HPLC retention times similar to mixtures of 12-hydroxyeicosatetraenic acid (12-HETE) and 15-HETE (20% of total, Rt ∼17–20 minutes; Figure 2 and Table 2) (14). Similar product profiles were reported for a P-450 2C11 protein purified from rat liver (4). As has been shown with P-450 2E1 (35), P-450 2C24 catalyzed both AA epoxidation and ω-1 hydroxylation and generated, in addition to EETs, small amounts of 19-OH-AA (Table 2). Finally, while the epoxygenase activity of P-450 2C23 showed a preference for oxygenation at the AA 11,12-olefin (Table 2), P-450s 2C11 and 2C24 displayed significantly lower overall regioselectivity and generated nearly equimolar mixtures of 11,12-EET and 14,15-EET (Table 2), in addition to nonepoxygenase products (Table 2).
Figure 2.
A comparative HPLC analysis of AA metabolism by recombinant kidney P-450s 2C11, 2C23, and 2C24. The AA monooxygenase activities of recombinant P-450s 2C11, 2C23, and 2C24 were reconstituted in the presence of NADPH–P-450 reductase and NADPH, exactly as indicated in Methods and in Table 2. After 10 minutes at 35°C, the reaction products were extracted into acidified ethyl ether and analyzed by reverse-phase HPLC. Shown are the radiochromatograms derived from reaction mixtures containing 0.1 μM P-450, 1 μM NADPH–P-450 reductase, 70 μM [1-14C]AA, and 1 mM NADPH. Approximate HPLC retention times for selected groups of P-450 eicosanoids are indicated in the chromatograms.
Table 2.
Enzymatic properties of the recombinant P-450 2C11, P-450 2C23, and P-450 2C24 AA epoxygenases
During measurements of AA turnover numbers for P-450s 2C11 and 2C23, it was noted that the reaction rates increased linearly when the NADPH–cytochrome P-450 reductase/P-450 molar ratios were varied between 1 and 10 (not shown). For example, for P-450s 2C11 and 2C23, maximal reaction rates (5.1 ± 0.5 and 1.5 ± 0.1 nmol of product/min per nanomole of P-450 for P-450s 2C11 and 2C23, respectively) (not shown) were obtained at NADPH–cytochrome P-450 reductase/P-450 molar ratios of approximately 10. Importantly, variations in the P-450/NADPH–cytochrome P-450 reductase ratios did not alter the enzymes’ regioselectivity and/or stereoselectivity of AA oxidation. Whereas these flavoprotein-dependent rate effects are not unique to P-450s 2C11 and 2C23 (19), they suggest P-450 isoform–specific differences in flavoprotein-hemoprotein coupling and, therefore, electron flow from NADPH to the heme iron. In control experiments it was determined that AA metabolism by P-450s 2C11, 2C23, or 2C24 required P-450 and NADPH–cytochrome P-450 reductase and that the addition of cytochrome b5 had little or no effect on reaction rates or product regiochemistry (19). The limited availability and purity of recombinant P-450 2C24, due to its low expression yields, precluded the titration of optimal P-450 2C24/reductase ratios. In analogy with P-450s 2C11 and 2C23, all P-450 2C24 reconstitution studies were performed using a NADPH–cytochrome P-450 reductase/P-450 molar ratio of 10. Under optimal conditions, the partially purified P-450 2C24 preparation metabolized AA at a rate that was substantially lower than that of P-450s 2C11 and 2C23 (0.3 ± 0.01 nmol of product per minute per nanomole of P-450 at 35°C (Table 2). This low turnover could be either an intrinsic property of P-450 2C24 or the result of (a) poor coupling between the exogenously added NADPH–cytochrome P-450 reductase and the P-450 heme iron; (b) the inhibition of electron transport and/or enzyme activity by contaminant proteins; (c) the presence of catalytically unproductive AA binding sites in the contaminant proteins; or (d) vestiges of Emulgen 911 used during solubilization and purification. However, hydroxylapatite chromatography efficiently removed Emulgen 911 from purified P-450s 2C23, 2C11, 2C8, and 2C10 (2, 19).
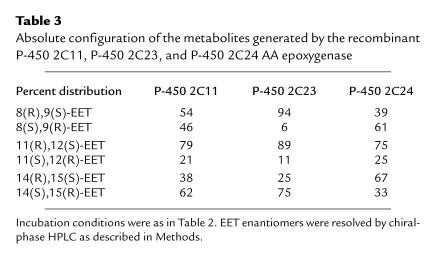
Comparisons of regioselectivity and enantioselectivity of AA metabolism, as well as antibody inhibition profiles, have been used to study the contribution of individual P-450 isoforms to the overall metabolism of AA (1, 4, 19). Chiral analysis of the metabolites (15) showed that AA epoxidation by recombinant P-450 2C23 is highly asymmetric and generates 8(R),9(S)-EET, 11(R),12(S)-EET, and 14(S),15(R)-EET with optical purities of 94, 89, and 75%, respectively (Table 3). The similarity of these values to those obtained using microsomal forms of P-450 2C23 (1, 2) show that stereoselectivity is an intrinsic property of the protein catalyst, not determined and/or affected by the microsomal membrane environment or protein purity. Compared with P-450 2C23, overall AA epoxidation by P-450 2C11 was less stereoselective (Table 3), and whereas 11(R),12(S)-EET was also the predominant 11,12-EET enantiomer formed by P-450 2C11, 14,15-EET and 8,9-EET were recovered with low enantiomeric purity or were nearly racemic (Table 3). Finally, the stereoselectivity of the P-450 2C24 AA epoxygenase was unique in that, compared with P-4502 2C11 and 2C23, the 2C24 protein catalyzes asymmetric oxygen insertion to the opposite enantiotopic faces of the 8,9-olefins and 14,15-olefins (the si,re and re,si faces of the 8,9-olefins and 14,15-olefins, respectively; Table 3). The fact that P-450 2C23 oxygenates AA with a high degree of molecular asymmetry (Table 3), but limited regioselectivity, can be understood if the protein active site molecular coordinates responsible for heme–fatty acid spatial orientation are remarkably rigid in restricting olefin rotation along the AA bis-allylic methylene bridges, but do allow for limited lateral displacement of the AA molecule (36, 37). Because with recombinant proteins the regioselectivity and stereoselectivity of oxygenation is under the control of a single protein catalyst, these results further demonstrate that the regiochemical and stereochemical control of the AA epoxygenase is P-450 isoform–specific (1, 2, 4, 5) and a single protein catalyst can catalyze the asymmetric epoxidation of the AA molecular template at more than one olefin (2–5, 19).
Table 3.
Absolute configuration of the metabolites generated by the recombinant P-450 2C11, P-450 2C23, and P-450 2C24 AA epoxygenase
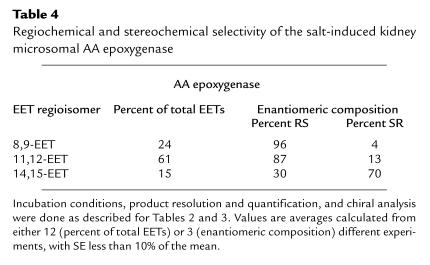
A central issue in the studies of the kidney AA epoxygenases is its role in AA bioactivation and renal physiology and its regulatory control by dietary salt–induced changes in renal function (6–11). The AA epoxygenase in microsomes from salt-loaded animals generates 11,12-EET as its major product, followed by smaller amounts of 8, 9-EET and 14,15-EET (61, 24, and 15% of total EETs, respectively) (Table 4). A small and variable amount of the labile 5,6-EET was also recovered (≤5% of total). These values are close to those reported for the AA epoxygenase in control kidney microsomes (1, 2) and to those obtained with the recombinant P-450 2C23 (Table 2). An inspection of the data in Tables 2, 3, and 4 indicates that (a) the regiofacial and enantiofacial selectivity of the recombinant P-450 2C23 epoxygenase matches that of the epoxygenase in salt-treated kidney microsomes; (b) contrary to the high degree of asymmetry displayed by the microsomal enzymes (Table 4), P-450 2C11 epoxidizes the AA 8,9-olefin to nearly racemic products (Table 3); and (c) the recombinant P-450 2C24 epoxidizes the AA 8,9-olefins and 14,15-olefins with an enantiofacial selectivity opposed to that of the microsomal enzymes (Tables 3 and 4). Based on these considerations, we concluded that, as with control rats (1, 2), P-450 2C23 is the predominant microsomal AA epoxygenase isoform in the kidneys of salt-treated animals and that salt-dependent changes in renal microsomal epoxidation occurred without changes in the regioselectivity and/or stereoselectivity of the microsomal enzymes.
Table 4.
Regiochemical and stereochemical selectivity of the salt-induced kidney microsomal AA epoxygenase
Immunological analysis of the rat kidney microsomal 2C P-450 proteins.
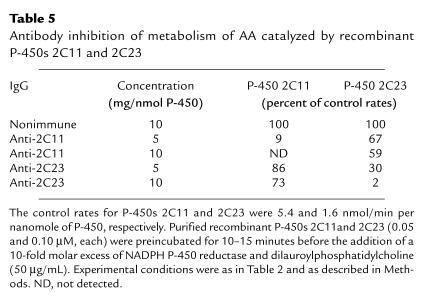
To further delineate the contributions of renal P-450 2C isoforms to the epoxygenase activity of salt-treated kidneys, we raised antibodies against recombinant P-450s 2C11 and 2C23. SDS-PAGE immunoelectrophoresis using recombinant P-450 2C11, 2C23, and 2C24 showed that the anti-2C11 antibody does not react with P-450 2C23 and recognizes P-450 2C24 only at high concentrations (Figure 3). Similarly, cross-reactivity between anti–P-450 2C23 and P-450 2C11 was observed only at high concentrations of P-450 2C11 (Figure 3). The identity of the anti-2C23 immunoreactive, protein with a Mr of approximately 77,000 present in purified P-450 2C11 remains unknown (Figure 3). In one experiment we analyzed the inhibitory potency and isoform selectivity of the purified anti-2C11 and anti-2C23 antibodies by incubating them with the recombinant proteins (at a ratio of 5 or 10 mg of IgG per nanomole of P-450) in the presence of NADPH–P-450 reductase, AA, and NADPH. As shown in Table 5, the anti-2C11 antibody was highly effective in blocking AA epoxidation by P-450 2C11. As seen in Figure 3, limited interactions between P-450 2C11 and anti-2C23 IgG were evidenced by the weak inhibition of the P-450 2C11 epoxygenase by the purified anti-2C23 IgG (Table 5). Complete inhibition (98%) of the P-450 2C23 epoxygenase was achieved at a concentration of 10 mg of anti-2C23 IgG per nanomole of P-450 2C23 (Table 5). On the other hand, under similar conditions, the anti-2C23 antibody reduced P-450 2C11 activity only partially, even at the highest concentration used (Table 5). These studies demonstrated that the degree of cross-reactivity between the anti-2C11 and anti-2C23 IgGs and the kidney epoxygenase was limited, and that, consequently, these antibodies could be useful for the analysis of the kidney P-450 isoform–specific microsomal metabolism and dietary salt regulation.
Figure 3.
Immunoelectrophoresis of recombinant kidney P-450s 2C11, 2C23, and 2C24 using purified anti–P-450 2C11 IgG (a) and anti–P-450 2C23 IgG (b). Samples of recombinant P-450 2C11, P-450 2C23, and P-450 2C24 (2.5 and 10 μg, each) were submitted to PAGE-SDS, electroblotted into PVDF membranes, which, after blocking in 5% milk, were incubated with a solution of either the anti-2C11 or anti-2C23 IgG (2 μg/mL in PBS) for 2 hours. They were washed extensively with a 0.05% solution of Tween-20 in PBS, incubated for 10 hours with an anti-rabbit IgG conjugated to horseradish peroxidase, and washed extensively before viewing, as described in Methods.
Table 5.
Antibody inhibition of metabolism of AA catalyzed by recombinant P-450s 2C11 and 2C23
To study the effects of excess dietary salt on the levels of P-450s 2C11 and 2C23, kidney microsomes were prepared from control rats and from rats that drank 2% NaCl (wt/vol) for 10 days, then were submitted to SDS-PAGE immunoelectrophoresis, and probed with the purified anti–P-450 2C11 and anti–P-450 2C23 antibodies. Salt-loading selectively increased the levels of an anti-2C23 immunoreactive protein with the electrophoretic mobility of P-450 2C23 and had only minimal effects on the levels of anti 2C11 immunoreactive material (Figure 4). To confirm these results, the anti-2C11 and anti-2C23 antibodies were incubated with kidney microsomes from control and salt-treated rats, and their effects on AA product profiles were analyzed by reverse-phase HPLC. As shown by the radiochromatograms in Figure 5, the epoxygenase activity of salt-treated microsomes was completely inhibited in the presence of the anti-2C23 antibodies; on the other hand, the nonimmune and the anti–P-450 2C11 IgGs had little or no effect on AA metabolism. Furthermore, the anti-2C23 antibody selectively blocked AA epoxidation (Rt ∼22–26 minutes; Figure 5) and had no effect on the AA ω/ω-1 hydroxylase reaction (Rt ∼14–16 minutes; Figure 5). Similar antibody inhibition patterns were observed when using microsomes from the kidneys of control rats (not shown), thus confirming that the salt-dependent changes in epoxygenase activity result mostly from increases in P-450 2C23 protein concentrations and that these changes are not accompanied by major P-450 2C isoform redistributions.
Figure 4.
Immunoelectrophoresis of microsomal fractions isolated from the kidneys of control (C) and salt-treated (S) rats. Kidney microsomal fractions isolated from either control rats or rats administered a 2% solution of NaCl as drinking water, were submitted to immunoelectrophoresis as described in Methods and the legend to Figure 3. After electroblotting into PVDF membranes, antigen-antibody reactions were viewed using the alkaline phosphatase–nitro blue tetrazolium reaction. For immunoblotting with the anti-2C11 or anti-2C23 IgGs, the gels were loaded with either 10 or 40 μg of microsomal protein per well, respectively.
Figure 5.
Antibody inhibition profiles for the arachidonate monooxygenase activities of microsomal fractions isolated from the kidneys of salt-loaded animals. Microsomal fractions were isolated from the kidneys of adult male rats fed a 2% solution of NaCl as drinking water for 10 days; suspended in a buffer system (1 mg/mL of protein) containing an NADPH-regenerating system; and incubated for 15 minutes at room temperature with either nonimmune rabbit IgG or the anti-2C11 or anti-2C23 IgGs (at a ratio of 5 or 10 mg of IgG/nmol of microsomal P-450). The reaction mixtures were then incubated for 15 minutes at 35°C in the presence of [1-14C]AA and NADPH (100 μM and 1 mM final concentrations, respectively). Shown are the radiochromatograms of the organic soluble products derived from reaction mixtures containing a total of 1 mg of microsomal protein. See Methods for further details.
Based on endothelial cell synthesis and release (38), EET-mediated relaxation of preconstricted arteries (39) and G protein–mediated EET opening of vascular smooth cell Ca2+-activated K+ channels (40, 41), the EETs — 11,12-EET in particular — were identified as endothelium-derived hyperpolarizing factors (EDHF) (40–42). These studies, with their potential for providing a molecular understanding of EET vasoactivity, are contributing to the integration of EET ion transport and vasoactive properties into a coherent mechanistic description amenable to experimental analysis. The demonstration of P-450s 2C11, 2C23, and 2C24, the major kidney 2C isoforms, as active regioselective and enantioselective AA epoxygenase and of P-450 2C23 as a dietary salt-sensitive epoxygenase provides biochemical, enzymatic, and molecular bases to current studies of the physiological and/or pathophysiological roles played by this enzyme system. These results should facilitate the analysis of biochemical, genetic, and functional correlations of protein-dependent EET biosynthesis, segmental localization, and regulatory control by functionally meaningful experimental manipulations. Indeed, the molecular characterization of the P-450 isoforms responsible for organ- and/or tissue-specific AA bioactivation is an important prerequisite for studies of the mechanisms and site of action of P-450–derived eicosanoids, the development of experimental models of gene/isoform-specific functional phenotypes, and the future development of protein-specific inhibitors.
As with most enzymes of the AA cascade, P-450s will not metabolize phospholipid-bound AA pools (6–11, 43); consequently, EET formation must be preceded by a phospholipase-catalyzed release of the fatty acid from glycerolipid stores (41). In most cases, it is the hormonal regulation of the releasing reaction that controls AA metabolism and eicosanoid biosynthesis. Whereas the hormonal pathways and/or signaling pathways linking dietary salt loading to increases in P-450 2C23 protein concentration are yet to be determined, our data show that regardless of the rate-limiting step(s), salt loading does lead the augmented EET formation, as revealed by the observed marked increases in urinary EET/DHET levels (1), and that these in vivo increases in the excretion of epoxygenase metabolites (1) are accompanied by the increases in renal P-450 2C23 concentrations documented here. Preliminary studies indicate that the microsomal epoxygenase activity reaches a maximum after 7–8 days on a high-salt diet, whereas increased levels of P-450 2C23 immunoreactive protein can be observed after 4–5 days on a high-salt diet and reach a maximum within 8–10 days.
Prohypertensive and antihypertensive roles have been attributed to the products of the kidney P-450 AA monooxygenase, and a role for the EETs and the ω-alcohol of AA in the pathophysiology of experimental salt-sensitive hypertension has been suggested (6–11). However, studies with the Dahl rat model of salt-sensitive hypertension have failed to establish direct associations between the salt-sensitive phenotype and either structural and/or regulatory alterations in a specific P-450 gene, even though P-450s 2C and 4A have been suggested as potential candidate genes (9, 13, 44, 45). Preliminary studies show that (a) under normal diets, salt-sensitive Dahl rats show substantially lower levels of renal P-450 2C23; (b) both salt-sensitive and salt-resistant rats respond to excess dietary salt by inducing the synthesis of P-450 2C23; and (c) the cDNA open reading frames coding for P-450s 2C23, 2C11, 4A1, 4A2, and 4A3 in Dahl salt-sensitive and Dahl salt-resistant rats are identical.
Finally, the recognized pharmacological and toxicological importance of microsomal P-450 provided the needed rationale for past extensive studies of the chemistry, biochemistry, and molecular biology of this important and ubiquitous catalyst. More recently, however, studies from several laboratories are providing the experimental framework needed for the analysis of functional roles played by this enzyme system in the metabolism of endogenous substrates such as vitamins, fatty acids, prostanoids, etc. Among these, AA occupies a unique and important position as the precursor for a variety of physiologically important signaling molecules and lipid mediators. The demonstration of a role for P-450 as an in vivo participant in the AA metabolic cascade has opened new opportunities for the understanding of the biological significance of these proteins, vis-a-vis their accepted toxicological and pharmacological roles.
Acknowledgments
These studies were supported by United States Public Health Service grant NIHDK 38226 and by an American Heart Association Fellowship to K. Makita. Protein sequences were performed at the Vanderbilt Medical Center Protein Sequencing Core Facility, a Vanderbilt Cancer Center Shared Resource supported by a National Cancer Institute grant (CA 68485).
References
- 1.Capdevila JH, et al. Cytochrome P-450 arachidonic acid epoxygenase: regulatory control of the renal epoxygenase by dietary salt loading. J Biol Chem. 1992;267:21720–21726. [PubMed] [Google Scholar]
- 2.Karara A, et al. Molecular cloning, expression, and enzymatic characterization of the rat kidney cytochrome P-450 arachidonic acid epoxygenase. J Biol Chem. 1993;268:13565–13570. [PubMed] [Google Scholar]
- 3.Imaoka S, et al. Identification of CYP2C23 expressed in rat kidney as an arachidonic acid epoxygenase. J Pharmacol Exp Ther. 1993;267:1012–1016. [PubMed] [Google Scholar]
- 4.Capdevila JH, et al. Cytochrome P-450 enzyme-specific control of the regio- and enantiofacial selectivity of the microsomal arachidonic acid epoxygenase. J Biol Chem. 1989;265:10865–10971. [PubMed] [Google Scholar]
- 5.Oliw EH. Oxygenation of polyunsaturated fatty acids by cytochrome P450 monooxygenases. Prog Lipid Res. 1994;33:329–354. doi: 10.1016/0163-7827(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 6.McGiff JC. Cytochrome P-450 metabolism of arachidonic acid. Annu Rev Pharmacol Toxicol. 1991;31:339–369. doi: 10.1146/annurev.pa.31.040191.002011. [DOI] [PubMed] [Google Scholar]
- 7.Harder DR, Campbell WB, Roman RJ. Role of cytochrome P-450 enzymes and metabolites of arachidonic acid in the control of vascular tone. J Vasc Res. 1995;32:79–92. doi: 10.1159/000159080. [DOI] [PubMed] [Google Scholar]
- 8.Capdevila, J.H., Zeldin, D., Makita, K., Karara, A., and Falck, J.R. 1995. Cytochrome P450 and the metabolism of arachidonic acid and oxygenated eicosanoids. In Cytochrome P450: structure, mechanism, and biochemistry. 2nd edition. P. Ortiz de Montellano, editor. Plenum Press. New York, NY. 443–471.
- 9.Makita K, Falck JR, Capdevila JH. Cytochrome P450, the arachidonic acid cascade, and hypertension: new vistas for an old enzyme system. FASEB J. 1996;10:1456–1463. doi: 10.1096/fasebj.10.13.8940291. [DOI] [PubMed] [Google Scholar]
- 10.McGiff JC, Steinberg M, Quilley J. Missing links: cytochrome P450 arachidonate products, a new class of lipid mediators. Trends Cardiovasc Med. 1996;6:4–10. doi: 10.1016/1050-1738(95)00106-9. [DOI] [PubMed] [Google Scholar]
- 11.Rahman M, Wright JT, Douglas JG. The role of the cytochrome P450-dependent metabolites of arachidonic acid in blood pressure regulation and renal function: a review. Am J Hypertens. 1997;10:356–365. doi: 10.1016/s0895-7061(96)00381-0. [DOI] [PubMed] [Google Scholar]
- 12.Catella F, Lawson JA, Fitzgerald DJ, Fitzgerald GA. Endogenous biosynthesis of arachidonic acid epoxides in humans: increased formation in pregnancy-induced hypertension. Proc Natl Acad Sci USA. 1990;87:5893–5897. doi: 10.1073/pnas.87.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makita K, et al. Experimental and/or genetically controlled alterations of the renal microsomal cytochrome P450 epoxygenase induce hypertension in rats fed a high salt diet. J Clin Invest. 1994;94:2414–2420. doi: 10.1172/JCI117608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capdevila JH, Falck JR, Dishman E, Karara A. Cytochrome P-450 arachidonate oxygenase. Methods Enzymol. 1989;187:385–394. doi: 10.1016/0076-6879(90)87045-5. [DOI] [PubMed] [Google Scholar]
- 15.Capdevila JH, Dishman E, Karara A, Falck JR. Cytochrome P450 arachidonic acid epoxygenase: stereochemical characterization of epoxyeicosatrienoic acids. Methods Enzymol. 1991;206:441–453. doi: 10.1016/0076-6879(91)06113-h. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press. Plainview, NY. 7.3–7.52.
- 17.Yoshioka H, et al. Structural analysis and specific expression of microsomal cytochrome P-450(M-1) mRNA in male rat livers. J Biol Chem. 1987;262:1706–1711. [PubMed] [Google Scholar]
- 18.Omura T, Sato R. The carbon monoxide binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 19.Zeldin DC, DuBois RN, Falck JR, Capdevila JH. Molecular cloning, expression, and characterization of an endogenous human cytochrome P450 arachidonic acid epoxygenase isoform. Arch Biochem Biophys. 1995;322:76–86. doi: 10.1006/abbi.1995.1438. [DOI] [PubMed] [Google Scholar]
- 20.Yasokochi Y, Masters BSS. Some properties of a detergent-solubilized NADPH-cytochrome c (cytochrome P-450) reductase purified by biospecific affinity chromatography. J Biol Chem. 1976;251:5337–5344. [PubMed] [Google Scholar]
- 21.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 22.Gonzalez FJ. The molecular biology of cytochrome P450s. Pharmacol Rev. 1988;40:243–288. [PubMed] [Google Scholar]
- 23.Nelson DR, et al. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 24.Hsu MH, Griffin KJ, Wang Y, Kemper B, Johnson EF. A single amino acid substitution confers progesterone 6 beta-hydroxylase activity to rabbit cytochrome P450 2C3. J Biol Chem. 1993;268:6939–6944. [PubMed] [Google Scholar]
- 25.Negishi M, Uno T, Darden TA, Sueyoshi T, Pedersen LG. Structural flexibility and functional versatility of mammalian P450 enzymes. FASEB J. 1996;10:683–689. doi: 10.1096/fasebj.10.7.8635685. [DOI] [PubMed] [Google Scholar]
- 26.Emi Y, Chijiwa C, Omura T. A different cytochrome P450 form is induced in primary cultures of rat hepatocytes. Proc Natl Acad Sci USA. 1990;87:9746–9750. doi: 10.1073/pnas.87.24.9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waxman DJ. Rat hepatic P450IIA and P450IIC subfamily 7 expression using catalytic, immunochemical, and molecular probes. Methods Enzymol. 1991;206:249–267. doi: 10.1016/0076-6879(91)06095-k. [DOI] [PubMed] [Google Scholar]
- 28.Legraverend C, Mode A, Welss T, Robinson I, Gustafsson JA. Hepatic steroid hydroxylating enzymes are controlled by the sexually dimorphic pattern of growth hormone secretion in normal and dwarf rats. FASEB J. 1992;6:711–718. doi: 10.1096/fasebj.6.2.1537461. [DOI] [PubMed] [Google Scholar]
- 29.Song BJ, Veech RL, Park SS, Gelboin HV, Gonzalez FJ. Induction of rat hepatic N-nitrosodimethylamine demethylase by acetone is due to protein stabilization. J Biol Chem. 1989;264:3568–3572. [PubMed] [Google Scholar]
- 30.Zaphiropoulos PG. cDNA cloning and regulation of a novel rat cytochrome P450 of the 2C gene subfamily (P450IIC24) Biochem Biophys Res Commun. 1991;180:645–651. doi: 10.1016/s0006-291x(05)81114-3. [DOI] [PubMed] [Google Scholar]
- 31.Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci USA. 1996;93:6536–6541. doi: 10.1073/pnas.93.13.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez FJ, Kimura S, Tamura S, Gelboin HV. Expression of mammalian cytochrome P450 using baculovirus. Methods Enzymol. 1991;206:93–99. doi: 10.1016/0076-6879(91)06080-m. [DOI] [PubMed] [Google Scholar]
- 33.Cook EA, et al. cDNA and deduced amino acid sequence of a novel cytochrome P-450 from female rat liver mRNA with high homology to P-450 IIC family. Nucleic Acids Res. 1990;18:7156–7160. doi: 10.1093/nar/18.23.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandrine M, Roussel F, Cresteil T. Age- and tissue-dependent expression of CYP2C23 in the rat. Biochim Biophys Acta. 1993;1172:124–130. doi: 10.1016/0167-4781(93)90278-l. [DOI] [PubMed] [Google Scholar]
- 35.Laethem RM, Balazy M, Falck JR, Laethem CL, Koop DR. Formation of 19(S)-, 19(R)-, and 18(R)-hydroxyeicosatetraenoic acids by alcohol-inducible cytochrome P450 2E1. J Biol Chem. 1993;268:12912–12918. [PubMed] [Google Scholar]
- 36.Capdevila JH, et al. The highly stereoselective oxidation of polyunsaturated fatty acids by cytochrome P450BM-3. J Biol Chem. 1996;271:22663–22671. doi: 10.1074/jbc.271.37.22663. [DOI] [PubMed] [Google Scholar]
- 37.Laethem RM, Halpert JR, Koop DR. Epoxidation of AA as an active site probe of cytochrome P450 2B isoforms. Biochim Biophys Acta. 1994;1206:42–48. doi: 10.1016/0167-4838(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 38.Rosolowsky M, Campbell WB. Synthesis of hydroxyeicosatetraenoic (HETEs) and epoxyeicosatrienoic acids (EETs) by cultured bovine coronary artery endothelial cells. Biochim Biophys Acta. 1996;1299:267–277. doi: 10.1016/0005-2760(95)00216-2. [DOI] [PubMed] [Google Scholar]
- 39.Hecker M, Bara AT, Bauersachs J, Busse R. Characterization of endothelium-derived hyperpolarizing factor as a cytochrome P450-derived arachidonic acid metabolite in mammals. J Physiol. 1994;481:407–414. doi: 10.1113/jphysiol.1994.sp020449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell WB, Gebremedhin D, Pratt P, Harder DR. Identification of epoxyeicosatrienoic acids as endothelium-derived hyperpolarizing factors. Circ Res. 1996;78:415–423. doi: 10.1161/01.res.78.3.415. [DOI] [PubMed] [Google Scholar]
- 41.Li PL, Campbell WB. Epoxyeicosatrienoic acids activate K+ channels in coronary smooth muscle through a guanine nucleotide binding protein. Circ Res. 1997;80:877–884. doi: 10.1161/01.res.80.6.877. [DOI] [PubMed] [Google Scholar]
- 42.Vanhoutte PM, Mombouli JV. Vascular endothelium: vasoactive mediators. Prog Cardiovasc Dis. 1996;39:229–238. doi: 10.1016/s0033-0620(96)80003-x. [DOI] [PubMed] [Google Scholar]
- 43.Karara A, Dishman E, Falck JR, Capdevila JH. Endogenous epoxyeicosatrienoyl-phospholipids. A novel class of cellular glycerolipids containing epoxidized arachidonate moieties. J Biol Chem. 1991;266:7561–7569. [PubMed] [Google Scholar]
- 44.Stec DE, Deng AY, Rapp JP, Roman RJ. Cytochrome P4504A genotype cosegregates with hypertension in Dahl SS/Jr rats. Hypertension. 1996;27:564–568. doi: 10.1161/01.hyp.27.3.564. [DOI] [PubMed] [Google Scholar]
- 45.Gu L, et al. Genetic mapping of two blood pressure quantitative trait loci on rat chromosome 1. J Clin Invest. 1966;97:777–788. doi: 10.1172/JCI118477. [DOI] [PMC free article] [PubMed] [Google Scholar]