Significance
How cardiac and skeletal muscle function is maintained and regulated is important in terms of understanding normal muscle physiology and muscle-based disease (myopathy). The two striated muscle types are structurally and functionally divergent, suggesting alternate molecular regulation. However, our discovery that the transcription factor Prox1 controls the same fast skeletal muscle gene program in both cardiac and slow skeletal muscle is significant in assigning a common genetic mechanism to striated muscle development. These studies establish a novel model of human dilated cardiomyopathy and reveal how loss of a single factor underpins conversion of slow to fast skeletal muscle, with implications for the molecular specification of endurance (stamina) versus rapidly contractile muscle function (speed).
Abstract
Correct regulation of troponin and myosin contractile protein gene isoforms is a critical determinant of cardiac and skeletal striated muscle development and function, with misexpression frequently associated with impaired contractility or disease. Here we reveal a novel requirement for Prospero-related homeobox factor 1 (Prox1) during mouse heart development in the direct transcriptional repression of the fast-twitch skeletal muscle genes troponin T3, troponin I2, and myosin light chain 1. A proportion of cardiac-specific Prox1 knockout mice survive beyond birth with hearts characterized by marked overexpression of fast-twitch genes and postnatal development of a fatal dilated cardiomyopathy. Through conditional knockout of Prox1 from skeletal muscle, we demonstrate a conserved requirement for Prox1 in the repression of troponin T3, troponin I2, and myosin light chain 1 between cardiac and slow-twitch skeletal muscle and establish Prox1 ablation as sufficient to cause a switch from a slow- to fast-twitch muscle phenotype. Our study identifies conserved roles for Prox1 between cardiac and skeletal muscle, specifically implicated in slow-twitch fiber-type specification, function, and cardiomyopathic disease.
During mouse embryogenesis, Prospero-related homeobox factor 1 (Prox1) regulates lymphatic specification, neuronal development, and the genesis of the liver and pancreas (1–4). Recently, Prospero-related homeobox factor 1 (Prox1) expression was shown to be enriched in murine slow-twitch skeletal muscle (5), and this is conserved in the zebrafish, where Prox1 is required for slow-twitch myofibril assembly (6). Previously we demonstrated an essential role for Prox1 in normal heart development in the mouse, with cardiac-specific Prox1 mutant embryos characterized by impaired cardiac growth, reduced fetal cardiomyocyte hypertrophy, persistent ventricular septal defects (VSDs), and myofibrillar disarray (7). Expression analyses revealed misregulation of a number of genes, including components of the sarcomere (7).
Cardiac and skeletal muscle progenitors are derived from mesoderm and differentiate into striated muscle tissues. The structure of the sarcomere is comparable between cardiac and skeletal muscle, but differs in the composition of sarcomeric protein isoforms. This includes cardiac-, slow-twitch–, and fast-twitch–specific isoforms encoded by myosin heavy chain (MyHC), myosin light chain (MyLC), and troponin complex genes, which regulate the speed and Ca2+ sensitivity of muscular contraction (8).
A number of components of the regulatory network that restrict the expression of cardiac, slow-twitch, and fast-twitch contractile protein gene isoforms to their respective muscle types in the mouse have been identified. These include hormones, such as thyroid hormone (9), as well as epigenetic modifications, micro RNAs (miRs), and a cohort of transcription factors (10, 11). Several loss-of-function studies, including targeting of Histone deacetylases 1 and 2 (HDAC 1/2) and miR-208a deletion in the mouse heart and skeletal-muscle–specific knockout of Sox6 and miR-208b/miR-499, have reported aberrant expression of fast-twitch skeletal MyHC, MyLC, and troponin complex gene isoforms (5, 12, 13). In the slow-twitch skeletal muscle of the zebrafish, these fast-twitch genes are negatively regulated by the transcription factor Prdm1 (Blimp-1), which also inhibits the expression of Sox6, a transcriptional repressor of slow-twitch contractile genes (6, 14). Although Sox6 appears to function to this end in the fast-twitch skeletal muscle of both mice and zebrafish (15), recent studies have revealed Prdm1 function is not conserved between these species (16). The transcription factor directly responsible for regulating fast-twitch contractile protein gene expression in skeletal and/or cardiac muscle remains to be determined.
Following on from our previous studies (7), microarray analyses revealed that loss of Prox1 in the embryonic heart was associated with increased fast-twitch skeletal muscle gene expression. This finding, alongside the fact that Prox1 is enriched in mouse slow-twitch skeletal muscle and functions in slow-twitch myofibrillogenesis in the zebrafish, suggested Prox1 might act to regulate contractile protein genes in both cardiac and skeletal striated muscle. Here we reveal Prox1 directly represses a program of fast-twitch skeletal muscle genes in the heart and that this function is uniquely conserved across skeletal muscle during normal development. We further demonstrate that loss of Prox1 is sufficient to cause a switch from slow- to fast-twitch fiber type and contractility in mouse skeletal muscle and reveal that cardiac-specific Prox1 knockout mice, which survive to adulthood, have significantly elevated fast-twitch skeletal gene expression in their hearts and develop severe dilated cardiomyopathy (DCM). Thus, we provide insight into the mechanisms that regulate fiber-type specification and myopathic disease.
Results
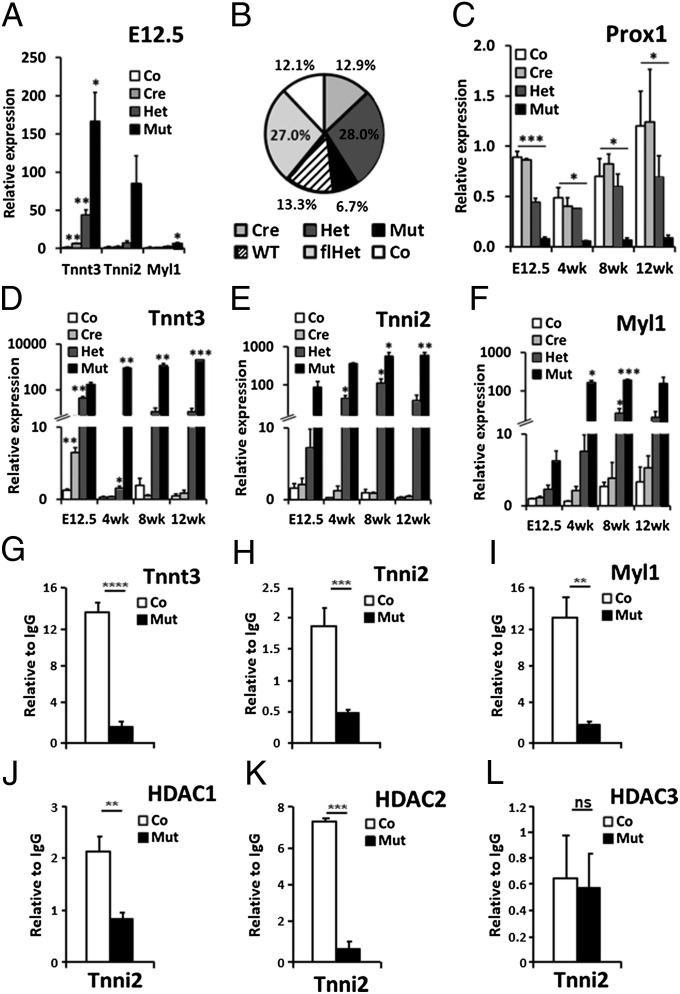
Analysis of gene expression in Prox1-deficient hearts (mutant, Nkx2.5Cre/+;Prox1fl/fl) (7) at embryonic day (E) 12.5 by Affymetrix microarray identified the fast-twitch skeletal isoforms of three contractile protein genes—troponin T3 (Tnnt3), troponin I2 (Tnni2), and myosin light chain 1 (Myl1)—as the most differentially expressed relative to litter-matched controls (Prox1fl/fl; SI Appendix, Table S1). Quantitative RT-PCR (qRT-PCR) confirmed overexpression of these genes in the embryonic mutant heart from E8.5 with levels reaching 137-, 54-, and fivefold greater than control by E12.5 for Tnnt3, Tnni2, and Myl1, respectively (Fig. 1A). This implicated Prox1 in the transcriptional repression of fast-twitch contractile protein genes during normal cardiac development.
Fig. 1.
Direct regulation of fast-twitch skeletal muscle genes by Prox1 and HDACs. (A) qRT-PCR analysis of Tnnt3, Tnni2, and Myl1 expression in control (Co, Prox1fl/fl), cre (Cre, Nkx2.5Cre/+;Prox1+/+), heterozygous (Het, Nkx2.5Cre/+;Prox1fl/+), and mutant (Mut, Nkx2.5Cre/+;Prox1fl/fl) E12.5 hearts (n = 3) to confirm microarray findings (SI Appendix, Table S1). (B) Ratio of genotypes at P10 from Nkx2.5Cre/+;Prox1fl/+ × Prox1fl/+ cross (n = 556) deviated significantly from expected Mendelian inheritance (P = 0.002). WT, Prox1+/+; flhet, Prox1fl/+. (C) Prox1 expression in control, cre, het, and mutant hearts at E12.5 (n = 3) and 4 (n = 2), 8 (n = 3), and 12 wk (n = 3) expressed relative to an E12.5 control heart. Significance determined by Student t test in comparison with control levels at each stage: *P ≤ 0.05; **P ≤ 0.01; ***P = ≤ 0.001. Analysis of control, cre, het, and mutant hearts to demonstrate increased expression of Tnnt3 (D), Tnni2 (E), and Myl1 (F) in mutant hearts at E12.5, and 4, 8, and 12 wk. Relative expression, N numbers, and significance are as in C. ChIP–qPCR quantification of Prox1 (G–I), HDAC1 (J), HDAC2 (K), and HDAC3 (L) antibody-immunoprecipitated chromatin from adult hearts of control (Prox1fl/fl; n = 6; white bars) and mutant (Nkx2.5Cre/+;Prox1fl/fl; n = 3; black bars) hearts using primers targeting responsive elements within Tnni2 (A and J–L), Tnnt3 (H), and Myl1 (I) genomic loci. ChIP–qPCR signals were standardized to the percentage of input DNA and normalized to IgG ChIP signals to determine the relative enrichment of anti-Prox1, HDAC1, HDAC2, or HDAC3 antibody over IgG immunoprecipitated chromatin. Each ChIP–qPCR was done in triplicate. Loss of binding in mutant hearts determined by Student t test comparison with control heart chromatin: ns, not significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Genotyping of offspring from Nkx2.5Cre/+;Prox1fl/fl and Prox1fl/+ crosses identified approximately half of the expected Mendelian frequency of mutants at P10 (Fig. 1B). A range of phenotypic severity, which we attributed to mosaic Cre efficiency, was observed in our analysis of Prox1-null embryos, suggesting the loss of mutant mice before P10 likely represents those with more acute myofibrillar disarray and/or persistent VSDs (7). In the Prox1 mutants that survived postnatally, Prox1 levels were significantly reduced (Fig. 1C) and lethality became fully penetrant between 7 and 14 wk. Consistent with previously published results (7), myofibrillar disarray, as demonstrated by α-actinin mislocalization (SI Appendix, Fig. S1) and valvuloseptal defects, was observed (SI Appendix, Fig. S2). However, in contrast to that described in embryonic Prox1 mutants, these defects were relatively mild and therefore compatible with survival beyond birth. Despite this milder myocardial phenotype, fast-twitch skeletal muscle gene expression was significantly elevated in the postnatal mutant hearts compared with embryonic counterparts; remarkably, the level of Tnnt3 was over 20,000-fold greater in mutant hearts relative to controls at 12 wk, whereas Tnni2 was up-regulated by over 3,500-fold (Fig. 1 D and E). Moreover, the degree of overexpression of all three fast-twitch skeletal muscle genes was found to increase with age, relative both to E12.5 mutant levels and to the respective control expression at each postnatal stage: Myl1 expression in the mutant heart at 12 wk was 25-fold higher than at E12.5 and 35-fold higher than control levels at 12 wk (Fig. 1F).
Increased expression of Tnnt3 and Myl1 has previously been associated with cardiac stress or specific cardiomyopathies (17, 18), suggesting that up-regulation of these genes in Prox1 mutants may represent a secondary defect. To determine whether Prox1 directly represses fast-twitch contractile protein gene expression in the heart, we performed chromatin immunoprecipitation (ChIP). Previous ChIP-on-chip analyses for Prox1 using E12.5 heart chromatin (7) identified binding within the first intron of Tnnt3, which was further investigated here by qPCR (SI Appendix, Fig. S3). For Tnni2 and Myl1, we focused the ChIP on previously identified responsive elements (19–21). DNA from adult-heart–derived chromatin, following Prox1 immunoprecipitation, was enriched for Tnnt3, Tnni2, and Myl1, with binding specificity confirmed using chromatin from age-matched Prox1 mutant hearts (Fig. 1 G–I). These data indicated that Prox1 localizes to putative regulatory regions in these fast-twitch skeletal contractile protein genes in vivo to directly represses their transcription in the heart.
Analogous to the Prox1 mutants, Tnnt3, Tnni2, and Myl1 are overexpressed in hearts lacking both HDAC1 and HDAC2 (12). HDACs limit the accessibility of regulatory regions by transcriptional activators through deacetylating histone proteins. The binding of both HDAC1 and HDAC2 to an intronic regulatory element (IRE) within Tnni2 has previously been published, implicating HDAC-mediated repression of Tnni2 expression in the heart (12). We previously revealed that an additional member of the class I HDAC family, HDAC3, interacts directly with Prox1 in the heart to negatively regulate the cardiac-specific transcription factor Nkx2.5 and modulate cardiac conduction (22). Therefore, we sought to investigate the relationship between Prox1 and HDAC-mediated repression of Tnni2. ChIP analyses confirmed binding of both HDAC1 and HDAC2 and further revealed HDAC3 localization to the Tnni2 IRE in wild-type adult hearts. Critically, however, we found that chromatin from Prox1 mutant hearts following HDAC1/2 immunoprecipitation failed to show equivalent enrichment for the Tnni2 IRE (Fig. 1 J–L). A similar trend was seen for HDAC2 and HDAC3 localization at regulatory elements within Tnnt3 and Myl1 and for the loss of HDAC2 enrichment in Prox1 mutant hearts (SI Appendix, Fig. S4). This suggests that Prox1 is involved in the normal recruitment of class I HDACs and that together they may form a regulatory complex required for the inhibition of fast-twitch skeletal muscle gene transcription in the heart.
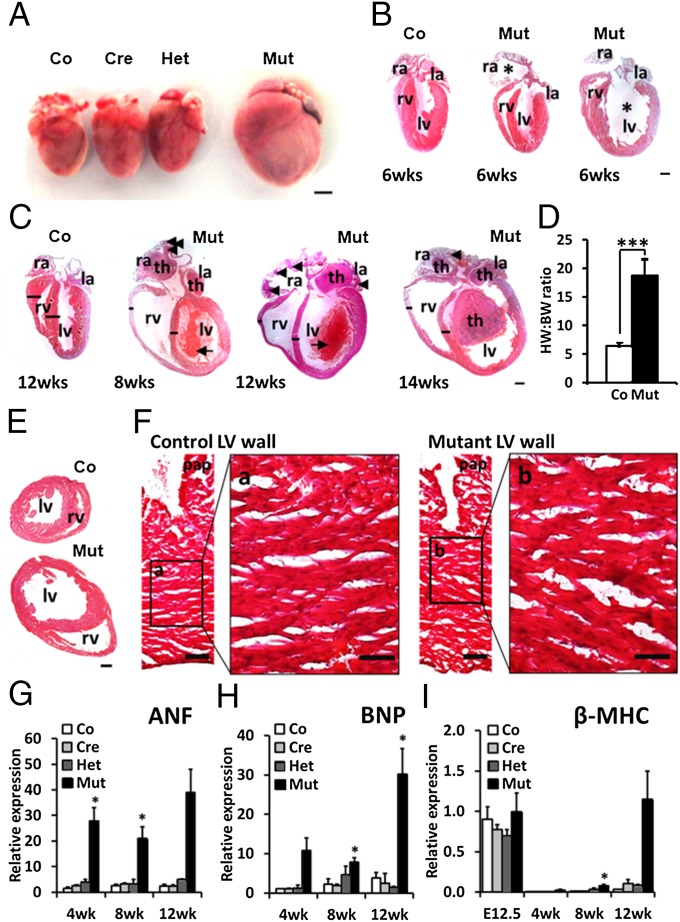
Heart-specific Prox1 mutants that survived to adulthood aberrantly expressed very high levels of fast-twitch skeletal contractile protein genes (Fig. 1 D–F), and all died between 7 and 14 wk postnatally. Before 6 wk, mutant hearts were grossly normal, but investigation of myocardial structure (SI Appendix, Fig. S2) and cytoarchitecture (SI Appendix, Fig. S1) revealed abnormalities similar to those in embryonic Prox1 mutants (7) and there was no evidence of aortic stenosis (SI Appendix, Fig. S5). At later stages, however, mutant hearts underwent remodeling and presented with marked cardiomegaly compared with litter-matched controls (Fig. 2A). Chamber dilation was evident in both the atria and ventricles (Fig. 2B), with the severity of dilation increasing with age to ultimately affect all cardiac chambers. Between 8 and 14 wk, all hearts from Prox1 mutants were characterized by cardiac enlargement, chamber dilation, and ventricular wall thinning (Fig. 2C)—changes that when taken together are consistent with DCM.
Fig. 2.
Adult cardiac-specific Prox1 mutants develop DCM without marked hypertrophy. (A) Gross morphology of 12 wk control (Co, Prox1fl/fl), cre (Cre, Nkx2.5Cre/+;Prox1+/+), heterozygous (Het, Nkx2.5Cre/+;Prox1fl/+), and mutant (Mut, Nkx2.5Cre/+;Prox1fl/fl) indicates specificity of dilation to the Prox1 mutant heart. (Scale bar, 500 μm.) (B) Examples of long-axis sections of control and mutant mice at 6 wk to show initiation of chamber dilation, marked by an asterisk. (Scale bar, 1 mm.) (C) Examples of long-axis sections of control and mutant hearts at 8, 12, or 14 wk, with dilation apparent in all cardiac chambers and associated with thinning of the right ventricular wall and interventricular septum (black lines). Location of large intra-atrial and intraventricular thrombi (th), smaller intra-atrial thrombi (arrowheads), and developing intraventricular thrombi (small arrows) illustrated on mutant sections. Right atria, ra; left atria, la; right ventricular lumen, rv; left ventricular lumen, lv. (Scale bar, 1 mm.) (D) Heart weight (HW, mg) to body weight (BW, g) ratio of control (Co, n = 18) and mutant (Mut, n = 4) mice at 12 wk: ***P = ≤ 0.001. (E) Transverse sections of 8 wk control (Co) and mutant (Mut) hearts. (Scale bar, 1 mm.) (F) Histological staining of comparable regions of left ventricular wall and papillary muscle (pap) of control and mutant hearts in E. (Scale bar, 100 μm.) High power insets of control (a) and mutant (b) ventricular myocardium reveal equivalent cardiomyocyte size between genotypes. (Scale bar, 50 μm.) Cardiac stress marker expression determined by qPCR analysis of ANF (G), BNP (H), and β-MHC (I) in control, cre, het, and mutant hearts at E12.5 (n = 3) and 4 (n = 2), 8 (n = 3), and 12 wk (n = 2). Levels expressed relative to control individual at 4 wk (G and H) or E12.5 (I), with significance determined by Student t test in comparison with control levels at each stage: *P ≤ 0.05; **P ≤ 0.01.
Although the Prox1 mutant DCM phenotype was associated with an increased heart weight to body weight ratio (Fig. 2D), there was no obvious concentric cardiomyocyte hypertrophy (Fig. 2 E and F). Instead we observed associated myocardial thinning at late stages, suggesting that if a degree of compensatory hypertrophic growth was activated in the Prox1 mutant heart, it was insufficient to offset the rate of dilation. Postnatal mutants were further associated with the development of large intra-atrial (7/8) and intraventricular (4/8) thrombi at 12 wk (Fig. 2C), indicative of hemostasis associated with poor systolic function (23). To confirm the extent of impaired cardiac function, mutant hearts were analyzed by magnetic resonance imaging (MRI). This revealed dilated and poorly contracting ventricles at 6 wk of age (SI Appendix, Fig. S6 A and B), with mean left ventricular ejection fraction values less than 30%, well below the 45% threshold diagnostic criterion for DCM (24) (Fig. 3B). Subsequent administration of a gadolinium contrast agent revealed thrombi at 8 wk of age (SI Appendix, Fig. S6 A and B), which, alongside risk of thromboembolism, is commonly associated with nonischemic DCM (24).
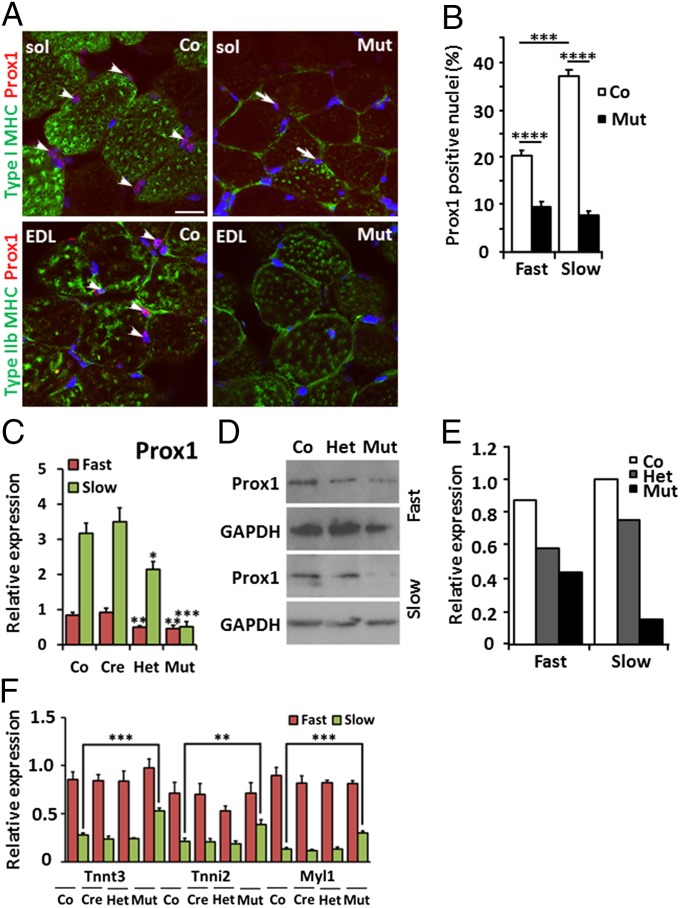
Fig. 3.
Prox1 expression is enriched in slow-twitch skeletal muscle and is significantly reduced following skeletal-muscle–specific ablation. (A) Immunostaining of sections from wild-type and mutant slow-twitch soleus (sol) and fast-twitch EDL muscles. Prox1 is expressed in the nucleus (red) in slow- and fast-twitch skeletal muscle, identified by presence of type I and type IIb fibers, respectively (green). Nuclear Prox1 staining in wild-type muscles is highlighted by white arrowheads; residual Prox1 expression in mutant soleus muscle is highlighted by white arrows and is indicative of incomplete Myf5-Cre–induced knockdown. (Scale bar, 20 μm.) (B) Analysis of the proportion of Prox1-expressing nuclei indicates significant enrichment of Prox1 in slow-twitch muscle compared with fast-twitch muscle and significant knockdown of Prox1 in fast- and slow-twitch muscle following Myf5-Cre ablation (n = 3). (C) Quantitative RT-PCR for Prox1 transcripts in control (Co, Prox1fl/fl), cre (Cre, Myf5Cre/+;Prox1+/+), het (Het, Myf5Cre/+;Prox1fl/+), and mutant (Mut, Myf5Cre/+;Prox1fl/fl) animals reveal enrichment of Prox1 in slow-twitch muscle (green) and significantly reduced expression in fast- and slow-twitch mutant muscles compared with control. (D) Western blot analysis and (E) quantification of Prox1 expression in control (Co, Prox1fl/fl), het (Het, Myf5Cre/+;Prox1fl/+), and mutant (Mut, Myf5Cre/+;Prox1fl/fl) animals show enrichment of Prox1 in slow-twitch muscle and reduced expression in slow- and fast-twitch mutant muscles. (F) Expression of Tnnt3, Tnni2, and Myl1 in fast-twitch (red) and slow-twitch (green) muscles of control (Co, Prox1fl/fl), cre (Cre, Myf5Cre/+;Prox1+/+), het (Het, Myf5Cre/+;Prox1fl/+), and mutant (Mut, Myf5Cre/+;Prox1fl/fl) animals at 4 wk. Levels are expressed relative to control fast-twitch muscle (red) (n > 5). Significance was determined by Student t test in comparison with control fast- or slow-twitch muscle levels, where appropriate: *P ≤ 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We also examined a number of cardiac stress markers in mutant hearts from embryonic stages to 12 wk postnatally. This revealed increased levels of atrial natriuretic factor (ANF) and brain natriuretic peptide (BNP), with up-regulation apparent at all postnatal stages peaking at 12 wk (Fig. 2 G and H). In addition, we observed reactivation of β-myosin heavy chain (MHC) expression (Fig. 2I) accompanied by decreased α-MHC transcription (SI Appendix, Fig. S7). In contrast to ANF and BNP, however, β-MHC up-regulation was not observed at 4 wk and was instead significantly reactivated only at 8 and 12 wk, stages associated with dilation remodeling and increased mortality risk. This suggests that an altered α-/β-MHC ratio, known to detrimentally affect adult cardiac function in both mice and humans (25), was aligned most closely with phenotype progression in Prox1 adult mutant mice. Expression profiling of β-MHC additionally revealed correct postnatal down-regulation and cardiac stress-induced reactivation of β-MHC in the Prox1 mutant heart (Fig. 2I), indicating that Prox1 is dispensable for β-MHC regulation. Together these data identify DCM in the postnatal Prox1 mutant mice, characterized by myocardial thinning, cardiac stress marker activation, and systolic dysfunction, and suggest that it is most likely caused by the aberrant expression of fast-twitch skeletal contractile proteins in combination with myofibrillar disruption within the cardiac muscle.
We next sought to determine whether Prox1 had an analogous role in the regulation of fast-twitch skeletal muscle genes in the skeletal muscle lineage. Skeletal-muscle–specific loss of Prox1 function was achieved using a Myf5Cre/+ mouse strain that targets the developing myotome from approximately E8.0 (26). Genotyping offspring from Myf5Cre/+;Prox1fl/fl crosses at P10 revealed mutants were born at the expected Mendelian ratio (n = 338), indicating that Prox1 expression in skeletal muscle is dispensable for survival. Consequently, phenotype characterization was focused on exemplar postnatal fast-twitch muscle types, the gastrocnemius and extensor digitorum longus (EDL), and the slow-twitch soleus muscle of the hind limb (27).
Prox1 was shown to be expressed in the nuclei of fast-twitch and slow-twitch skeletal muscle cells (Fig. 3A). An assessment of the proportion of nuclei expressing Prox1 via qRT-PCR and Western blot analyses revealed enrichment in wild-type slow-twitch skeletal muscle (Fig. 3 A–E). In the conditional Prox1 mutants (Myf5Cre/+;Prox1fl/fl), the level of Prox1 expression at 4 wk was significantly reduced in both fast- and slow-twitch skeletal muscle (Fig. 3 A–E), as well as in the slow-twitch muscle of heterozygote littermates (het; Myf5Cre/+;Prox1fl/+; Fig. 3 C-–E), relative to control mice (cre, Myf5Cre/+; control, Prox1fl/fl).
In contrast to mutant hearts, loss of Prox1 in skeletal muscles did not result in myofibril disarray (SI Appendix, Fig. S8). However, gene expression analysis of gastrocnemius and soleus muscles revealed that the fast-twitch skeletal contractile protein genes up-regulated in the Prox1 mutant heart—Tnnt3, Tnni2, and Myl1—were also significantly overexpressed in the slow-twitch skeletal muscle of Prox1 mutants by ∼twofold (Fig. 3F). This suggests that Prox1 represses an equivalent fast-twitch skeletal muscle gene program in developmentally segregated slow-twitch skeletal and cardiac muscle.
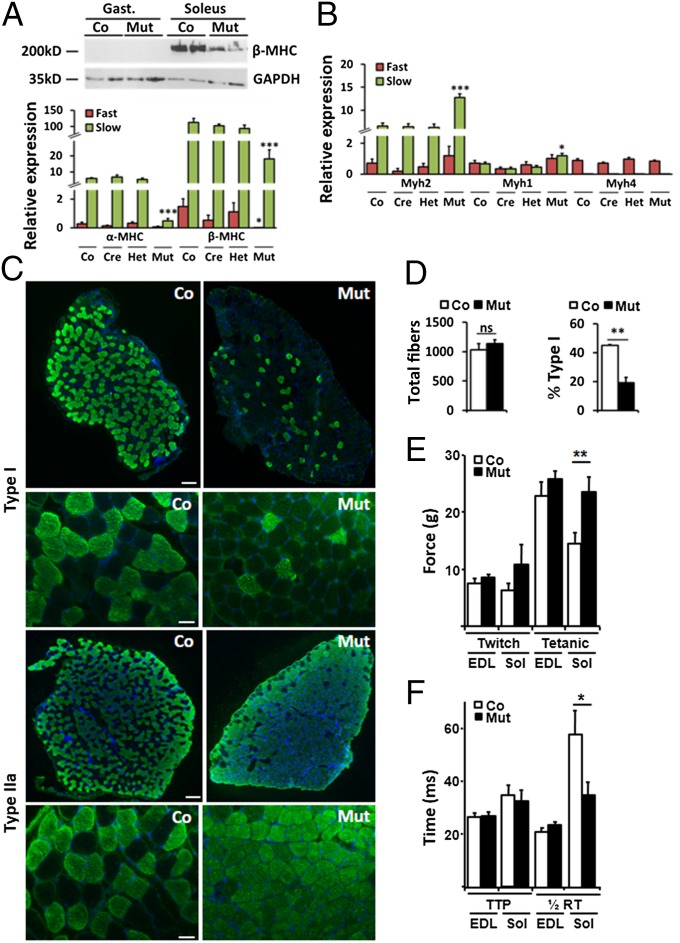
The mouse soleus muscle (slow twitch) is normally composed of type I (β-MHC) and type IIa (Myh2) fibers, whereas the EDL (fast twitch) is composed of type IIx (Myh1) and type IIb (Myh4) fibers (27, 28). In contrast to mutant hearts, loss of Prox1 in fast- and slow-twitch skeletal muscle resulted in a significant reduction of β-MHC expression, most notable at the protein level in the soleus muscle, supported by an approximately sixfold reduction in β-MHC mRNA in both fast- and slow-twitch muscle (Fig. 4A).
Fig. 4.
Switch to fast-twitch gene expression, fiber type, and maximal contractile strength in Prox1-deficient soleus muscle. (A) Decreased β-MHC protein in mutant soleus muscle revealed by Western blot, with reduced expression of α-MHC and β-MHC in fast- and slow-twitch mutant muscles determined by qRT-PCR. (B) Quantification of fast-twitch MyHC gene isoforms in fast- and slow-twitch muscle of control (Co, Prox1fl/fl), cre (Myf5Cre/+), het (Myf5Cre/+;Prox1fl/+), and mutant (Myf5Cre/+;Prox1fl/fl) mice (n > 5 at 4 wk). (C) Immunostaining to reveal fewer type I and more type IIa (green) muscle fibers in transverse sections through soleus muscles of 8 wk mutant animals compared with controls. (Scale bar, 200 μm and 50 μm.) (D) Quantification of total fiber number in soleus muscle section of control and mutant animals (n = 3) and significant reduction in the percentage of type I fibers in mutant versus control soleus (n = 3). (E) Maximal contractile strength of Prox1 mutant slow-twitch soleus muscle is equivalent to fast-twitch skeletal muscle: average twitch and tetanic force for fast-twitch EDL and slow-twitch soleus muscle of control (Co, Prox1fl/fl) and mutant (Mut, Myf5Cre/+;Prox1fl/fl) animals at 8 wk (n = 6). (F) Average time taken to reach maximum force (time-to-peak, TTP) and return to half maximum force (½ relaxation time, RT), where ½ RT in Prox1 mutant slow-twitch soleus muscle is equivalent to control fast-twitch muscle. Levels are expressed relative to control fast-twitch muscle. Significance is determined by Student t test in comparison with control fast- or slow-twitch muscle levels, where appropriate: *P ≤ 0.05; **P < 0.01; ***P < 0.001.
Further examination of the MyHC expression in mutant slow-twitch soleus muscle revealed decreased α-MHC levels (Fig. 4A), which is ordinarily enriched in slow-twitch muscle (5, 29). In contrast, levels of the fast-twitch MyHC genes Myh2 and Myh1 were significantly up-regulated, although Myh4—the fastest MyHC gene—expression remained equivalent between all genotypes (Fig. 4B). These data collectively suggested a molecular switch in phenotype from slow- to fast-twitch muscle, which was subsequently confirmed by immunofluorescent staining of muscle sections from 8-wk-old mutant animals. In slow-twitch soleus muscle, there was an increase in Myh2- and Myh1-expressing type IIa (Fig. 4C) and type IIx fibers (SI Appendix, Fig. S9A), respectively, and a decrease in β-MHC–expressing type I fibers (Fig. 4C) but comparable fiber-type composition between Prox1 mutant and control fast-twitch EDL muscles (SI Appendix, Fig. S9B). Quantification of type I fibers in the soleus muscle revealed a decrease from ∼45% in control as comparable with previous analyses (27) to ∼19% in the mutant, with no decrease in total fiber number (Fig. 4D). Further confirmation of the fiber-type switch in mutant soleus muscles was demonstrated by silver staining of high-resolution polyacrylamide glycerol gels and complementary Western blot analyses (SI Appendix, Fig. S10), which highlighted a decrease in type I and an increase in type IIa. Staining with wheat germ agglutinin–Alexa594 revealed that myofiber size was reduced in Prox1 mutant slow-twitch soleus muscle and increased in Prox1 mutant fast-twitch EDL muscle (SI Appendix, Fig. S11). Succinate dehydrogenase staining demonstrated there were no changes in the overall oxidative capacity of mutant soleus or EDL muscles (SI Appendix, Fig. S12). Functional studies subsequently revealed increased maximal (tetanic) contractile strength and decreased half relaxation time exclusively in the mutant soleus, both of which are characteristic features of fast-twitch EDL muscles (Fig. 4 E and F). Collectively these data implicate loss of Prox1 from slow-twitch skeletal muscle as sufficient not only to alter the molecular signature and fiber-type composition from slow to fast twitch but also to induce functionality more akin to fast skeletal muscle.
Discussion
The restricted expression of contractile protein isoforms contributes to the functional distinction between cardiac and skeletal striated muscle types, with the encoding genes often misregulated in myopathic disease. In this study, loss of Prox1 from either cardiac or skeletal muscle lineages was sufficient to de-repress the fast-twitch skeletal genes Tnnt3, Tnni2, and Myl1, identifying a conserved role for Prox1 in their regulation across striated muscle. Confirmation of direct transcriptional repression was obtained by ChIP analyses, which revealed Prox1 protein localization to putative regulatory regions within all three fast-twitch genes in the adult heart and concomitant decrease in an association of class I HDACs to these regulatory regions in the absence of Prox1. This suggests that HDACs and Prox1 may form a repressive regulatory complex to inhibit fast-twitch skeletal genes in cardiac and skeletal muscle.
Despite the role for Prox1 in the regulation of fast-twitch contractile genes during development, Prox1 knockdown in developing skeletal muscle was not associated with embryonic lethality. This contrasts with conditional deletion of Prox1 from the heart, which resulted in loss of half of the mutants by P10, with lethality fully penetrant by ∼14 wk of age. This difference in phenotypic severity reflects the relative importance of the heart for embryonic survival, compared with slow-twitch skeletal muscle, and the increased sensitivity of cardiac function to altered contractile protein gene expression (30, 31). In addition to a unifying role across striated muscle, Prox1 also appears to have independent functions in the two muscle subtypes, such that Prox1 has no effect on MyHC gene regulation in the heart but its loss in skeletal muscle causes a significant reduction in α-MHC and β-MHC expression. This was a surprising finding given that prox1 has no effect on β-MHC in the skeletal muscle of the zebrafish (6). Importantly, the decrease in β-MHC expression likely results in the switch from slow- to fast-twitch fiber type in the soleus muscle, with type I fibers replaced by type IIa and type IIx fibers. The latter are as fatigue resistant as type I (28) and already make up a significant proportion of wild-type soleus muscle (27), but nevertheless, the change in fiber type was accompanied by altered mutant soleus muscle function, which manifested as elevated tetanic strength and a steady state of contraction characteristic of fast-twitch muscle. This is a significant finding at the level of loss of a single transcription factor: Prox1 appears to be pivotal for the molecular programming that specifies slow-twitch skeletal muscle fiber type and consequently the maintenance of a physiological balance between endurance versus rapid skeletal muscle function.
Survived postnatal cardiac-specific Prox1 mutants subsequently developed fatal DCM and heart failure from ∼7 wk that was fully penetrant by 14 wk. Dilation was apparent in all four cardiac chambers and was associated with prolific thrombus formation, reflecting the poor contractility and aberrant hemostasis of the Prox1 mutant heart. A number of defects likely contributed to the development of DCM, including reduced compensatory hypertrophic response, supported by the myocardial thinning in the postnatal Prox1 mutant heart, myofibrillar disarray, and impaired fetal hypertrophy (7). Although several studies have reported cardiac dilation and heart failure following the loss of factors required for cardiomyocyte hypertrophy (12, 32), the most striking feature of the postnatal Prox1 mutant heart herein was the significant overexpression of the fast-twitch skeletal genes Tnnt3, Tnni2, and Myl1. Coexpression of the cardiac and fast-twitch skeletal isoforms of troponin genes results in the formation of troponin complexes that vary in their contractile responses to Ca2+ signaling and consequently impaired systolic function. Transgenic expression of Tnnt3 in the heart negatively affects heart function in line with the degree of troponin complex heterogeneity causing dilation in aged mouse hearts (33). Moreover, a splice variant of cardiac TnT that resembles Tnnt3 causes DCM in multiple animal models (34) and is expressed in failing human hearts (35). Overexpression of the fast-twitch Myl1 gene also alters the contractile function of the heart (36) and is up-regulated exclusively in patients with DCM (37). Consequently, contractile defects arising from the de-repression of fast-twitch skeletal muscle gene expression, coupled with the persistence of myocardial sarcomeric defects from the embryo (7), likely underpins the development of DCM in the Prox1 mutant mice.
The onset and manifestation of DCM in the cardiac-specific Prox1 knockout mice parallels the human condition. Cardiac dilation is first apparent in postnatal mutant hearts around puberty (6–8 wk), in line with a second peak of human DCM that occurs during early adolescence (38). There is also a subset of patients that develop DCM in later life following earlier presentation with congenital heart defects, such as incomplete septation and failed myocardial compaction (23, 39). This is comparable with the known embryonic phenotype of Prox1 mutant hearts (7) and suggests that future studies to investigate PROX1 as a candidate disease gene, causative for contractile protein misregulation, may be of significant prognostic value for DCM patients.
Materials and Methods
All animal experiments were carried out according to UK Home Office project licences compliant with the UK Animals (Scientific Procedures) Act 1986 and approved by the University College London (UCL) Biological Services Ethical Review Process. Animal husbandry at UCL Biological Services was in accordance with UK Home Office Certificate of Designation. Cardiac- and skeletal-muscle–specific loss of Prox1 mice were generated by crossing Prox1fl/fl with Nkx2.5Cre/+ and Myf5Cre/+ [B6.129S4–Myf5tm3(cre)Sor/J] strains, respectively. Hearts and soleus skeletal muscle were histologically processed for hematoxylin and eosin staining or immunofluorescence, RNA was extracted for qRT-PCR/exon arrays, and chromatin was prepared for immunoprecipitation using primer sets and antibodies as detailed in SI Appendix, SI Materials and Methods. Functional analyses were carried out by MRI on postnatal hearts at 6 and 8 wk and isometric muscle tension electron physiology on soleus and EDL slow and fast skeletal muscle, respectively. More information is available in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank A. Cook for expert assessment of the structural defects in the adult Prox1 mutant hearts. This study was supported by funding from the Child Health Research Appeal Trust, in association with Great Ormond Street Children’s Hospital, and from the British Heart Foundation (PG/09/043/27565).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406191111/-/DCSupplemental.
References
- 1.Wigle JT, et al. An essential role for Prox1 in the induction of the lymphatic endothelial cell phenotype. EMBO J. 2002;21(7):1505–1513. doi: 10.1093/emboj/21.7.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elkouris M, et al. Sox1 maintains the undifferentiated state of cortical neural progenitor cells via the suppression of Prox1-mediated cell cycle exit and neurogenesis. Stem Cells. 2011;29(1):89–98. doi: 10.1002/stem.554. [DOI] [PubMed] [Google Scholar]
- 3.Wang J, et al. Prox1 activity controls pancreas morphogenesis and participates in the production of “secondary transition” pancreatic endocrine cells. Dev Biol. 2005;286(1):182–194. doi: 10.1016/j.ydbio.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 4.Sosa-Pineda B, Wigle JT, Oliver G. Hepatocyte migration during liver development requires Prox1. Nat Genet. 2000;25(3):254–255. doi: 10.1038/76996. [DOI] [PubMed] [Google Scholar]
- 5.An CI, Dong Y, Hagiwara N. Genome-wide mapping of Sox6 binding sites in skeletal muscle reveals both direct and indirect regulation of muscle terminal differentiation by Sox6. BMC Dev Biol. 2011;11:59. doi: 10.1186/1471-213X-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy S, Wolff C, Ingham PW. The u-boot mutation identifies a Hedgehog-regulated myogenic switch for fiber-type diversification in the zebrafish embryo. Genes Dev. 2001;15(12):1563–1576. doi: 10.1101/gad.195801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Risebro CA, et al. Prox1 maintains muscle structure and growth in the developing heart. Development. 2009;136(3):495–505. doi: 10.1242/dev.030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon AM, Regnier M, Homsher E. Skeletal and cardiac muscle contractile activation: Tropomyosin “rocks and rolls”. News Physiol Sci. 2001;16:49–55. [PubMed] [Google Scholar]
- 9.Vadászová A, Zacharová G, Machácová K, Jirmanová I, Soukup T. Influence of thyroid status on the differentiation of slow and fast muscle phenotypes. Physiol Res. 2004;53(Suppl 1):S57–S61. [PubMed] [Google Scholar]
- 10.Clark KL, Yutzey KE, Benson DW. Transcription factors and congenital heart defects. Annu Rev Physiol. 2006;68:97–121. doi: 10.1146/annurev.physiol.68.040104.113828. [DOI] [PubMed] [Google Scholar]
- 11.Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol. 2011;12(6):349–361. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- 12.Montgomery RL, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21(14):1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Rooij E, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17(5):662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Hofsten J, et al. Prdm1- and Sox6-mediated transcriptional repression specifies muscle fibre type in the zebrafish embryo. EMBO Rep. 2008;9(7):683–689. doi: 10.1038/embor.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagiwara N, Ma B, Ly A. Slow and fast fiber isoform gene expression is systematically altered in skeletal muscle of the Sox6 mutant, p100H. Dev Dyn. 2005;234(2):301–311. doi: 10.1002/dvdy.20535. [DOI] [PubMed] [Google Scholar]
- 16.Vincent SD, Mayeuf A, Niro C, Saitou M, Buckingham M. Non conservation of function for the evolutionarily conserved prdm1 protein in the control of the slow twitch myogenic program in the mouse embryo. Mol Biol Evol. 2012;29(10):3181–3191. doi: 10.1093/molbev/mss125. [DOI] [PubMed] [Google Scholar]
- 17.Sasse S, et al. Troponin I gene expression during human cardiac development and in end-stage heart failure. Circ Res. 1993;72(5):932–938. doi: 10.1161/01.res.72.5.932. [DOI] [PubMed] [Google Scholar]
- 18.Nakao K, et al. Increased expression of atrial myosin light chain 1 in the overloaded human left ventricle: Possible expression of fetal type myocytes. Int J Cardiol. 1992;36(3):315–328. doi: 10.1016/0167-5273(92)90302-j. [DOI] [PubMed] [Google Scholar]
- 19.Niro C, et al. Six1 and Six4 gene expression is necessary to activate the fast-type muscle gene program in the mouse primary myotome. Dev Biol. 2010;338(2):168–182. doi: 10.1016/j.ydbio.2009.11.031. [DOI] [PubMed] [Google Scholar]
- 20.Delgado-Olguín P, et al. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat Genet. 2012;44(3):343–347. doi: 10.1038/ng.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin H, Yutzey KE, Konieczny SF. Muscle-specific expression of the troponin I gene requires interactions between helix-loop-helix muscle regulatory factors and ubiquitous transcription factors. Mol Cell Biol. 1991;11(1):267–280. doi: 10.1128/mcb.11.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Risebro CA, et al. Epistatic rescue of Nkx2.5 adult cardiac conduction disease phenotypes by prospero-related homeobox protein 1 and HDAC3. Circ Res. 2012;111(2):e19–e31. doi: 10.1161/CIRCRESAHA.111.260695. [DOI] [PubMed] [Google Scholar]
- 23.Luk A, Ahn E, Soor GS, Butany J. Dilated cardiomyopathy: A review. J Clin Pathol. 2009;62(3):219–225. doi: 10.1136/jcp.2008.060731. [DOI] [PubMed] [Google Scholar]
- 24.Elliott P, et al. Classification of the cardiomyopathies: A position statement from the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J. 2008;29(2):270–276. doi: 10.1093/eurheartj/ehm342. [DOI] [PubMed] [Google Scholar]
- 25.Nakao K, Minobe W, Roden R, Bristow MR, Leinwand LA. Myosin heavy chain gene expression in human heart failure. J Clin Invest. 1997;100(9):2362–2370. doi: 10.1172/JCI119776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ott MO, Bober E, Lyons G, Arnold H, Buckingham M. Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development. 1991;111(4):1097–1107. doi: 10.1242/dev.111.4.1097. [DOI] [PubMed] [Google Scholar]
- 27.Wigston DJ, English AW. Fiber-type proportions in mammalian soleus muscle during postnatal development. J Neurobiol. 1992;23(1):61–70. doi: 10.1002/neu.480230107. [DOI] [PubMed] [Google Scholar]
- 28.Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91(4):1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 29.Soukup T, Pedrosa-Domellöf F, Thornell LE. Expression of myosin heavy chain isoforms and myogenesis of intrafusal fibres in rat muscle spindles. Microsc Res Tech. 1995;30(5):390–407. doi: 10.1002/jemt.1070300506. [DOI] [PubMed] [Google Scholar]
- 30.Feng HZ, Jin JP. Coexistence of cardiac troponin T variants reduces heart efficiency. Am J Physiol Heart Circ Physiol. 2010;299(1):H97–H105. doi: 10.1152/ajpheart.01105.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei B, Jin JP. Troponin T isoforms and posttranscriptional modifications: Evolution, regulation and function. Arch Biochem Biophys. 2011;505(2):144–154. doi: 10.1016/j.abb.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Berlo JH, Maillet M, Molkentin JD. Signaling effectors underlying pathologic growth and remodeling of the heart. J Clin Invest. 2013;123(1):37–45. doi: 10.1172/JCI62839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang QQ, et al. Co-expression of skeletal and cardiac troponin T decreases mouse cardiac function. Am J Physiol Cell Physiol. 2008;294(1):C213–C222. doi: 10.1152/ajpcell.00146.2007. [DOI] [PubMed] [Google Scholar]
- 34.Biesiadecki BJ, Elder BD, Yu ZB, Jin JP. Cardiac troponin T variants produced by aberrant splicing of multiple exons in animals with high instances of dilated cardiomyopathy. J Biol Chem. 2002;277(52):50275–50285. doi: 10.1074/jbc.M206369200. [DOI] [PubMed] [Google Scholar]
- 35.Anderson PA, et al. Molecular basis of human cardiac troponin T isoforms expressed in the developing, adult, and failing heart. Circ Res. 1995;76(4):681–686. doi: 10.1161/01.res.76.4.681. [DOI] [PubMed] [Google Scholar]
- 36.Morano I, et al. Changes in essential myosin light chain isoform expression provide a molecular basis for isometric force regulation in the failing human heart. J Mol Cell Cardiol. 1997;29(4):1177–1187. doi: 10.1006/jmcc.1996.0353. [DOI] [PubMed] [Google Scholar]
- 37.Hirzel HO, Tuchschmid CR, Schneider J, Krayenbuehl HP, Schaub MC. Relationship between myosin isoenzyme composition, hemodynamics, and myocardial structure in various forms of human cardiac hypertrophy. Circ Res. 1985;57(5):729–740. doi: 10.1161/01.res.57.5.729. [DOI] [PubMed] [Google Scholar]
- 38.Andrews RE, Fenton MJ, Ridout DA, Burch M. British Congenital Cardiac Association New-onset heart failure due to heart muscle disease in childhood: A prospective study in the United kingdom and Ireland. Circulation. 2008;117(1):79–84. doi: 10.1161/CIRCULATIONAHA.106.671735. [DOI] [PubMed] [Google Scholar]
- 39.van der Linde D, et al. Birth prevalence of congenital heart disease worldwide: A systematic review and meta-analysis. J Am Coll Cardiol. 2011;58(21):2241–2247. doi: 10.1016/j.jacc.2011.08.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.