Significance
Genes must be annotated with their correct functions if genome data are to support hypothesis building and metabolic engineering. PlantSEED was developed to streamline the process of annotating plant genome sequences, to construct metabolic models based on genome annotations automatically, and to use models to test the annotation of these sequences, allowing the detection of gaps and errors in gene annotations and the prediction of new functions. PlantSEED is designed to grow in an iterative manner by including new plant genome sequences, new annotations harvested from the literature, and improved biochemical data, all of which are integrated in a consistent manner into the PlantSEED genomes and metabolic models.
Keywords: systems biology, computational biochemistry, plant metabolism, plant genomics
Abstract
The increasing number of sequenced plant genomes is placing new demands on the methods applied to analyze, annotate, and model these genomes. Today’s annotation pipelines result in inconsistent gene assignments that complicate comparative analyses and prevent efficient construction of metabolic models. To overcome these problems, we have developed the PlantSEED, an integrated, metabolism-centric database to support subsystems-based annotation and metabolic model reconstruction for plant genomes. PlantSEED combines SEED subsystems technology, first developed for microbial genomes, with refined protein families and biochemical data to assign fully consistent functional annotations to orthologous genes, particularly those encoding primary metabolic pathways. Seamless integration with its parent, the prokaryotic SEED database, makes PlantSEED a unique environment for cross-kingdom comparative analysis of plant and bacterial genomes. The consistent annotations imposed by PlantSEED permit rapid reconstruction and modeling of primary metabolism for all plant genomes in the database. This feature opens the unique possibility of model-based assessment of the completeness and accuracy of gene annotation and thus allows computational identification of genes and pathways that are restricted to certain genomes or need better curation. We demonstrate the PlantSEED system by producing consistent annotations for 10 reference genomes. We also produce a functioning metabolic model for each genome, gapfilling to identify missing annotations and proposing gene candidates for missing annotations. Models are built around an extended biomass composition representing the most comprehensive published to date. To our knowledge, our models are the first to be published for seven of the genomes analyzed.
Next-generation sequencing technology is revolutionizing genomics and transcriptomics (1–3). Some 30 plant genomes are already available, and hundreds more soon will be (4). Currently these genomes are being collected and annotated by various resources including Phytozome, Plant Metabolic Networks (PMN) (5, 6), Gramene/EnsemblPlants, PlantGDB, National Center for Biotechnology Information (NCBI) Plants (7), Plaza (8), and MetNet Online (9). These resources use various homology-based functional annotation algorithms to make new predictions and to propagate annotation between genes and species. This process often leads to overannotation in which many paralogous genes are incorrectly associated with the same reaction. This problem is particularly acute for several types of proteins: transporters, multidomain proteins, and large enzyme families such as methyltransferases and aminotransferases that act on similar but distinct substrates. The propagation of inconsistent and incorrect annotations among genomes, especially given the size and mosaic nature of plant genomes (10), degrades the value of annotations for modeling and other downstream analyses. Standardization of plant genome annotation is thus essential. In addition to considerations of consistency and standardization, there also is the issue of scalability of human curation and manual annotation effort in the midst of an exponentially growing body of plant genome sequences. The paradigm of annotating individual genes and genomes must give way to a paradigm of annotating gene families across all genomes, and the consequences of functional assignments must be considered across all impacted genomes rather than for a single genome of particular focus.
Similar problems emerge as we move downstream from genome annotation to the reconstruction of genome-scale metabolic models. Metabolic models are valuable tools for the annotation, engineering, and analysis of any organism (11–13). Models are capable of predicting host–microbe interaction (14), gene essentiality (15), growth phenotypes, and gene-expression profiles (16). More recently, models have proven to be a valuable resource for the annotation process itself by identifying and filling gaps in metabolic pathway annotations (17, 18) and supporting the reconciliation of genome annotations with observed phenotypes (19–21). At this time, numerous metabolic models exist for a small number of plant species, namely Arabidopsis (22–25), maize (Zea mays) (24, 26), and rice (Oryza sativa) (27). These models vary substantially in their representation of biochemical data, their biomass compositions, their genome annotations, and the algorithms used in their reconstruction and refinement (11). Some of these models have unique strengths in representing specific phenotypes (e.g., C4 metabolism), but their many differences make their application in comparative studies challenging.
Functional annotation, comparison, and modeling of plant genomes therefore must evolve from a customized, artisanal process to a uniform, industrial one, allowing the ongoing curation and propagation of consistent annotations among all existing and incoming plant genome sequences. A key step to assuring consistency is to impose a uniform functional ontology on precisely defined protein families, so that the known functions of each family can be propagated faithfully among plant species. Furthermore, metabolic functions, in turn, must be linked to a set of stoichiometrically consistent reactions that occur in each organism, to allow construction of in silico metabolic models to test the accuracy and coverage of annotations of metabolic genes. Misannotated or unannotated genes cause these models to fail; then the cause of this failure can be identified and used to improve the gene annotations. To enable consistent annotations and the integration of metabolic models in the annotation process, we built the PlantSEED, a metabolism-centric resource for annotating the genomes of a core set of plant species and constructing models of plant metabolism. Although PlantSEED was designed to incorporate several resources, notably that of AraCyc, every gene–reaction association was reviewed manually for supporting evidence, and every biochemical reaction was balanced and modified for stoichiometric consistency. This highly conservative approach to annotation is a defining characteristic of PlantSEED and enables PlantSEED to serve as a reliable resource for comparative genomics and biochemistry.
Results
Overview of PlantSEED.
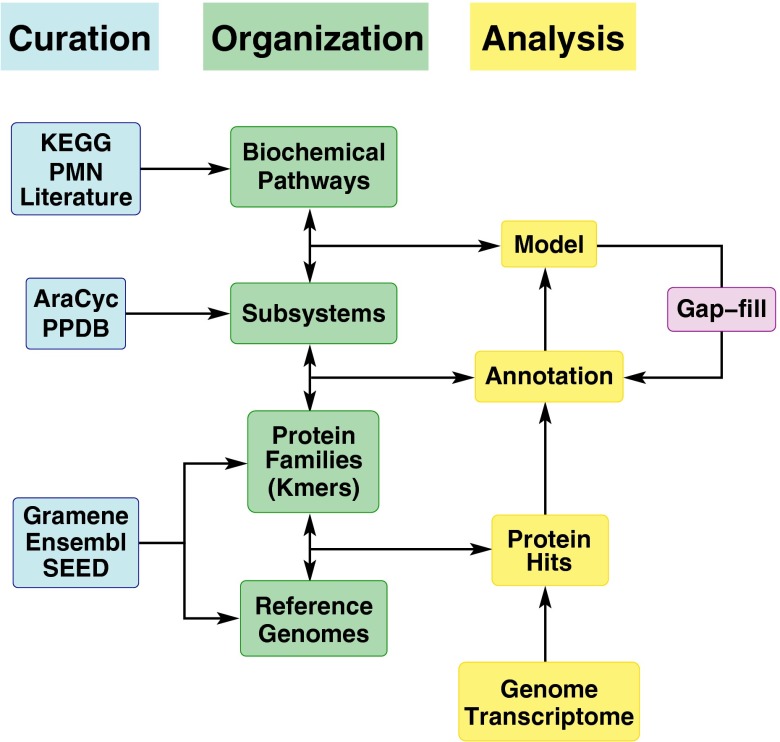
PlantSEED has four core components: biochemical pathways, subsystems, genomes, and protein families (Fig. 1). The biochemical pathway database in the PlantSEED integrates biochemical data found in numerous resources (6, 28–30), with an emphasis on AraCyc (6), which served as the primary source for our initial set of pathways and gene–reaction associations. The reactions in our biochemical pathways are mapped to biological functions contained in 97 plant-specific subsystems created to support genome annotation and curation in the PlantSEED. Subsystems are curated groups of related biological functions mapped to corresponding genes in a database of reference genomes (31). The PlantSEED subsystems were constructed starting from the excellent annotations and pathway data found in 209 AraCyc pathways (6). As each new subsystem was constructed, we extended our subsystems-based annotations beyond Arabidopsis to maize and eight other core plant genomes. We used protein families computed by Ensembl Compara (32) to propagate annotations among all genomes, but we conservatively trimmed these families to minimize the overannotation of paralogs, which often presents a challenge in plant genomes. This combined use of subsystems and protein families is patterned on SEED annotation services (33), particularly RAST (34) and ModelSEED (28, 35). Because PlantSEED is fully integrated with its parent, the larger prokaryotic SEED, it empowers users with unique cross-kingdom genome comparison tools, including the capacity to view prokaryotic homologs of plant genes and the genomic context of these homologs, as illustrated later.
Fig. 1.
Overview of PlantSEED, which consists of reference genomes, protein families, subsystems, and biochemical pathways. Reference genomes for the 10 species listed in Table 1 were installed from Gramene. Protein families computed by Ensembl Compara for these 10 genomes were installed along with Kmers (unique oligopeptide sequences representing the families) computed by SEED. Gene–reaction associations were curated primarily using AraCyc metabolic pathways to form a set of PlantSEED subsystems. Finally, a biochemical pathway database was formed by integrating KEGG, several BioCyc databases from PMN and Gramene, and published metabolic models. Gapfilling results from use of the reference genomes are reviewed and used in PlantSEED’s subsystems and resulting annotation, thereby improving PlantSEED’s output dynamically.
PlantSEED Biochemistry and Pathways.
The PlantSEED biochemistry database represents a large-scale integration of biochemical data found in ModelSEED (28), Kyoto Encyclopedia of Genes and Genomics (KEGG) (29), MetaCyc (30), PMN (5, 6), AraCyc (6), MaizeCyc, and numerous published metabolic models for plants and microbes (Table S1). The database includes a nonredundant set of all reactions found in all source repositories, including 27,470 distinct compounds and 31,528 distinct reactions (Dataset S1). All reactions and compounds in the data were standardized to a pH of 7.0, and, when possible, the group contribution method (36) was applied to estimate Gibbs free energies for compounds and reactions. Gibbs free energy estimates were used in combination with a set of heuristic rules (19) to predict thermodynamic reversibility of all database reactions. Finally, a quality-control analysis was conducted on all database reactions to ensure that they are mass and charge balanced and involve only defined reactants. Some 10,292 reactions in the database failed this quality-control analysis. Although these reactions were retained in the database, they were excluded from PlantSEED metabolic models, because flux balance models are intolerant of these types of errors in biochemistry.
Organizing Biochemical Pathways and Protein Families into Subsystems.
The subsystems in PlantSEED are, in effect, tables whose columns are a set of proteins that together constitute a metabolic pathway (or other biological process) and whose rows are genomes; the table is populated with the genes encoding each protein in each genome (31). In subsystems-based annotation, annotators curate and assign functions not to individual genes but to entire columns of the subsystem table. Curating functional annotations across all genomes in this way offers decisive advantages: (i) annotations are applied consistently, regardless of the number of genomes; (ii) annotation curation is performed by experts in the subsystem, not by experts in a particular genome; (iii) comparative genomics is exploited to identify patterns in subsystem representation in various genomes, resolving these patterns into recognizable variants that also may be consistently applied; and (iv) subsystems serve to organize and enforce the development of a controlled ontology for functional annotation, ensuring that the same protein is annotated with the same function in different organisms.
PlantSEED currently consists of 97 subsystems covering 209 pathways of plant primary and secondary metabolism. These foundational subsystems, which together encompass 1,384 metabolic reactions, were constructed from AraCyc pathways (6) and from curated functions and reactions in seven B-vitamin biosynthetic pathways (37). In addition, membrane energetic functions were encoded in 14 original subsystems that capture in detail individual membrane complexes and soluble components of respiratory and photosynthetic electron transport chains. Note that in PlantSEED these complexes are not grouped into linear “respiration” or “photosynthesis” pathways, as is usually done. Instead they are organized as mitochondrial or plastidial electron transport to emphasize that both mitochondria and chloroplasts possess independent electron transfer chains and generate metabolic energy via chemiosmotic ATP synthesis (see SI Text for details). This approach is an important step toward accurate, comprehensive encoding of this crucial area of plant function because it allows the various components to be strung together in a more flexible and realistic way in metabolic models and genome annotations. As curation continues, more subsystems will be constructed and incorporated into PlantSEED, thus dynamically extending its coverage of plant metabolism.
Subsystems in themselves do not ensure consistent, accurate, and scalable annotations. It also is necessary to create and maintain a curated set of high-quality orthologous protein families. Fast heuristics such as BLAST detect gene homology, but homology may indicate the presence of out-paralogs as well as orthologs (38). Thus, many large enzyme families, such as aminotransferases (39), contain members that differ in function. Furthermore, plants have many paralogs resulting from whole-genome duplication events (10, 40, 41). To deal with these challenges, we use the Ensembl Compara protein families, which depend on phylogenetic trees to extract orthologous relationships (32, 42). A family of orthologs, not individual genes, then becomes the fundamental unit of annotation, ensuring that annotations are scalable and applied consistently across genomes. We used 700 such families for PlantSEED; however, we found that many of the orthologous relationships within each family were tenuous. Therefore we weeded out dubious orthologous pairs using two criteria. (i) Because many protein families contain multiple clusters of nonoverlapping orthologous relationships between Arabidopsis thaliana and Arabidopsis lyrata, we were able to divide each family into orthologous groups centered on these Arabidopsis clusters. (ii) Because many of the tenuous relationships, compared with their supposed Arabidopsis orthologs, had low sequence identity as reported by Ensembl Compara, we accepted a non-Arabidopsis protein within an orthologous family only if the sequence identity exceeded 80%. From the original 700 protein families containing a total of 19,746 genes, we derived 1,303 protein families containing 13,709 genes with which we launched PlantSEED. We then performed an initial assignment of these families to our subsystems based on gene assignments in AraCyc. At this stage, we began a subsystems-based curation of these protein families and functional assignments.
Subsystems-Based Annotation and Metabolic Reconstruction of Core Plant Genomes.
To create a core of reference genomes that captures much of the diversity among flowering plants, we applied our subsystems-based annotation approach to annotate five eudicot and five monocot genomes (Table 1). Because our initial subsystems curation focused on metabolism, nearly all genes in subsystems are associated with a metabolic reaction. The curation process refined gene annotations by (i) narrowing reaction specificity and eliminating paralogs to improve the resolution of the mapping between metabolic reactions and their genes; (ii) correcting inconsistent or erroneous annotations based on literature evidence; and (iii) refining generic reactions, improving the use of transporters to activate compartmentalized reactions, and rebalancing reactions. During this process, our 97 new plant subsystems were populated with all 10 plant genomes in a manner that is consistent with our plant protein families. In total, these subsystems include 19,566 consistently annotated genes. All subsystems and genomes are accessible via the PlantSEED website: http://plantseed.theseed.org.
Table 1.
Annotation and model data for 10 reference PlantSEED genomes
| Species | Subsystem genes* | Annotated reactions† | Restored reactions‡ |
| A. thaliana | 2,084 (27,416) | 1,080 (236) | 0 (739) |
| A. lyrata | 2,095 (32,667) | 1,078 (236) | 0 (628) |
| Glycine max | 3,334 (54,174) | 928 (215) | 33 (590) |
| P. trichocarpa | 2,331 (41,377) | 946 (220) | 23 (597) |
| Vitus vinifera | 1,778 (29,927) | 945 (220) | 30 (609) |
| Brachypodium distachyon | 1,339 (26,552) | 857 (196) | 55 (593) |
| Oryza glaberrima | 1,445 (33,164) | 854 (204) | 53 (600) |
| O. sativa | 1,320 (68,682) | 861 (198) | 54 (600) |
| Sorghum bicolor | 1,475 (36,338) | 874 (210) | 51 (602) |
| Z. mays | 1,712 (63,009) | 861 (206) | 53 (607) |
Total numbers of protein-encoding genes are stated in parentheses.
Numbers of reactions in compartments are stated in parentheses.
Total numbers of gapfilled reactions are stated in parentheses.
We also used our annotations and their mappings to our biochemistry database to construct draft genome-scale metabolic models from the 10 core genomes (Table 1) using an approach based on ModelSEED (28). Such models consist of (i) a catalog of all the biochemical and transport reactions in an organism’s metabolic pathways; (ii) a mapping of those reactions to the proteins that catalyze each reaction; and (iii) a biomass composition (“biomass reaction”) that specifies the relative quantity of each small molecule that is needed to make 1 g dry weight of biomass. We expanded the set of biomass components to more than double the average of 37 components used in previously published plant models (23, 24, 26). Thus, our models use 79 components, including many cofactors (SI Text and Dataset S2). The literature indicates that each of these components is synthesized by all land plants. Our core models thus are able to make qualitative predictions regarding central pathway utilization; further tissue-specific curation of the biomass composition will be required for quantitative predictions of pathway flux.
Finally, to model eukaryotic metabolism reliably, the localization of reactions in different organelles and other subcellular compartments is important. We extended our approach beyond that of ModelSEED to capture the presence of nine subcellular compartments. We incorporated experimental evidence listed for the localization of enzymes from AraCyc, from the Plant Proteomics Database (PPDB) (43), and from an analysis of maize B-vitamin pathways (38). In the case of the PPDB, we used only curated localization data. This evidence is reduced to a set of nine keywords, each representing a subcellular compartment, which are added to the gene annotations (SI Text and Table S2). We used this compartment-specific annotation to localize reactions when building our metabolic models.
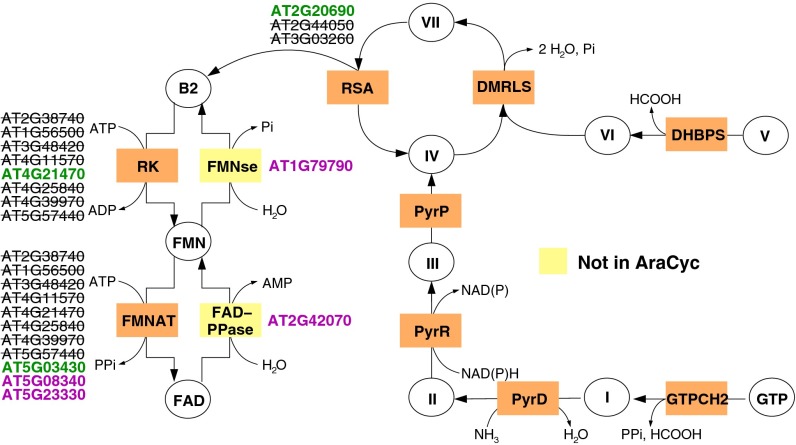
Although PlantSEED is based first and foremost on curation of AraCyc biochemistry and annotations, the final PlantSEED content differs substantially from AraCyc. In AraCyc, annotations are propagated primarily based on sequence homology, with little accounting for phylogenetic context. Conversely, PlantSEED annotation is propagated using Ensembl Compara protein families that were divided and trimmed using the strict criteria described above. This conservative approach is demonstrated in the FAD biosynthesis pathway (Table S3 and Fig. 2) (37), in which PlantSEED eliminated 17 of the gene–reaction associations proposed in the AraCyc v. 10.0 database release. Besides reducing the overannotation of paralogs in Arabidopsis, the use of the tailored PlantSEED protein families also restricted the overannotation of functions such as “FMN adenylyltransferase (EC 2.7.7.2)” in the other nine reference plant species included in PlantSEED. PlantSEED also includes additional reactions and gene–reaction associations garnered from other curated resources (e.g., PMN and Gramene) and the literature. In the FAD biosynthesis pathway, this process includes two additional reactions and four additional genes (Fig. 2). (Note that many of the differences between AraCyc and PlantSEED in FAD synthesis were corrected in the AraCyc v11.5 database release, based on the PlantSEED annotations, highlighting both the accuracy of our annotation corrections and the excellent ongoing curation of the AraCyc database.) Furthermore, we perform a global comparison of the gene–reaction associations for Arabidopsis, Populus trichocarpa, and maize in PlantSEED with those in the BioCyc databases AraCyc, PoplarCyc, and MaizeCyc (SI Text and Dataset S3). Relative to PlantSEED, an average of 4.5, 15.8, and 33.6 extra gene–reaction associations per pathway were present in AraCyc, PoplarCyc, and MaizeCyc, respectively. Overall, although PlantSEED may be less comprehensive than AraCyc, its data are substantially more precise.
Fig. 2.
Comparison of the reactions and genes available from AraCyc v.10.0 and PlantSEED for FAD biosynthesis. For the creation of the PlantSEED subsystem named “Riboflavin, FMN and FAD biosynthesis in plants,” 17 gene–reaction associations found in AraCyc were excised (black type with strike-through), and four additional gene–reaction associations curated from the literature were added (purple). Green indicates agreement between AraCyc and PlantSEED. Furthermore, two additional reactions were added to the pathway based on experimental support (yellow boxes). For the other reactions in the pathway, AraCyc contained the correct gene–reaction associations, and these were incorporated into the subsystem. The full list of genes is found in Table S3. The abbreviations and Roman numerals used to represent the reactions and metabolites, respectively, are detailed in the associated diagram found online at http://plantseed.theseed.org. The current version of AraCyc (11.5) includes the updated curation shown here.
Model-Driven Annotation of Core Angiosperm Genomes.
Plant genome annotations always include gaps in metabolic pathways representing reactions that are poorly understood or for which no gene has been identified. Numerous methods have been proposed to fill gaps in incomplete pathways arising from missing annotations. One of the first methods, called “pathway hole filling,” involves adding an entire pathway if a gene can be associated with one or more steps of the pathway (44), but this method can lead to the unwarranted addition of pathways (SI Text).
In PlantSEED, we use the metabolic models constructed for each genome to conduct a model-driven form of annotation refinement (17, 18). In this approach, metabolic models are used to ensure that (i) all pathways needed to make every component of biomass are present and complete and (ii) every reaction associated with an annotated gene is connected to the rest of the metabolic network. Through a process called “gapfilling,” models rapidly identify when these criteria are not satisfied and propose reactions, annotations, and genes that may be associated with missing pathway steps. This type of gapfilling is offered in other annotation and modeling environments, including the Constraints-Based Reconstruction and Analysis (COBRA) toolbox (45) and Pathway Tools (46), although PlantSEED is distinct in offering model-driven gapfilling of plant genomes based on a refined and curated database of plant biochemistry.
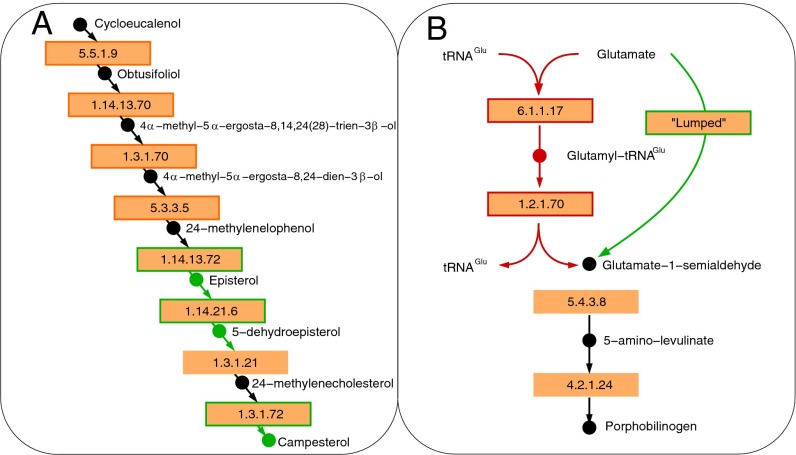
To fulfill the criteria, PlantSEED applies two stages of gapfilling. First comes the biomass-centric process called “essential gapfilling,” which ensures that each model can produce biomass during heterotrophic growth. Fig. 3A highlights a successful example of essential gapfilling in campesterol biosynthesis. Essential gapfilling added an average of 233 reactions to the 10 PlantSEED models, with an average of 21 gene candidates found for gapfilled reactions (Table 1, Fig. S1, and Dataset S4). Essential gapfilling focuses only on pathways involved in the biosynthesis of biomass, so many pathways may remain incomplete and disconnected from the metabolic network. This issue is addressed by a second process called “pathway gapfilling,” which ensures that each reaction associated with annotated genes will be functional in the model. Fig. 3B highlights a successful example of pathway gapfilling in tetrapyrrole biosynthesis. Pathway gapfilling added an average of 344 reactions to the 10 PlantSEED models, with an average of 35 gene candidates found for gapfilled reactions (Table 1, Fig. S1, and Dataset S4). All the reactions in each of the models can be found in Dataset S5. In addition, 20 transport reactions were added, activating 58 localized enzymatic reactions (Dataset S4). Transporter gapfilling and annotations are discussed in more detail in SI Text.
Fig. 3.
Examples of essential and pathway gapfilling results performed on the A. thaliana metabolic reconstruction generated by PlantSEED. (A) Essential gapfilling identified and filled the necessary reactions for the biosynthesis pathway of campesterol (all reactions shown are gap-filling reactions). This sterol is a biomass component, but its biosynthesis is not yet covered in PlantSEED subsystems. The missing reactions were added from the “Plant sterol biosynthesis” pathway in AraCyc (PWY-2541; boxes outlined in red) and the “Steroid biosynthesis” pathway in KEGG (map00100; boxes outlined in green). (B) Pathway gapfilling identified and filled a gap in the biosynthesis pathway of porphobilinogen. The tRNAGlu and glutamyl-tRNAGlu compounds in the original AraCyc representation of this pathway (boxes outlined in red) were excluded from the model because of their lack of molecular formulas, leaving the porphobilinogen pathway incomplete. The process focused on activating reaction–gene associations further downstream, such as AT1G50170 annotated as “uroporphyrinogen-III methyltransferase (EC 2.1.1.107)” (47) and identified an alternative version of this pathway in which the undefined intermediates were lumped out (box outlined in green), completing the pathway and enabling biosynthesis of many heme-like compounds.
Gapfilling has a particularly critical role in the PlantSEED annotation process. The trimmed protein families used by the PlantSEED reduce the overannotation of paralogs dramatically, but this reduction comes with an increased risk of failing to propagate correct annotations: Only 69% of the reactions in the Arabidopsis model are found in all 10 plant species (Fig. S2). The two gapfilling processes were extremely effective in compensating for this overpruning in PlantSEED, with an average of 43 previously trimmed reactions being restored to the PlantSEED models during the gapfilling process (Table 1). Among the reactions that do not propagate between Arabidopsis and other species (except for A. lyrata) several from glucosinolate biosynthesis were not restored. This outcome validates our annotation propagation process, because glucosinolates are known to be confined to the order Brassicales (48).
We recognize that any given result from the pathway gapfilling could be confounded by an incorrect original gene–reaction association. Therefore, we create a second set of PlantSEED metabolic models to include the additional pathway gapfilling results. None of the gene–reaction associations proposed by gapfilling are incorporated into PlantSEED, but they are provided separately to the research community in the hope that they will be validated (Dataset S4). We do not yet incorporate other data types (e.g., proteomics, metabolomics, and transcriptomics) to validate our results further, but others have published methods for doing so (49, 50).
The Power of PlantSEED Annotations and Tools.
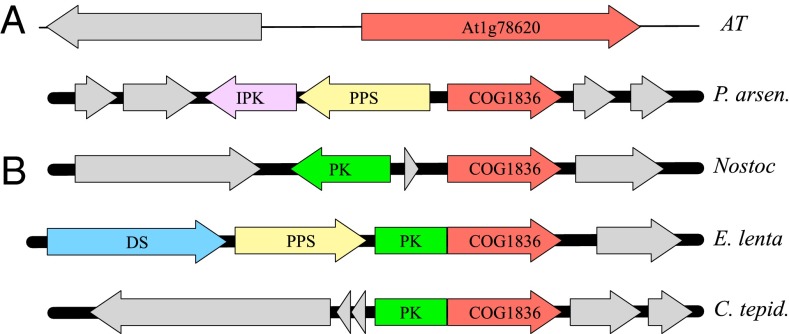
One problem that bedevils the plant annotation process is “missing annotations,” in which a function is known to exist, but the corresponding gene remains unidentified. PlantSEED offers unique support for the identification of such genes by cross-kingdom comparative genomics because it is fully integrated with the prokaryotic SEED database and uniform annotations that are consistent across all plant, bacterial, and archaeal genomes are used for orthologous genes. An example of a candidate gene discovered in this way is phytyl-phosphate kinase. Arabidopsis contains a dedicated salvage pathway for redirecting free phytol released from chlorophyll degradation during senescence into chloroplast lipid metabolism (51). Two distinct successive kinase activities associated with chloroplast envelope membranes that phosphorylate phytol to phytyl-phosphate and phytyl-diphosphate have been characterized. However, the corresponding gene (VTE5, AT5G04490) has been identified for only the first of these kinases, the CTP-dependent phytol kinase (52). The phytyl-phosphate kinase activity has not yet been associated with any sequence in any organism (i.e., the gene is “globally missing”) (53). Comparative analysis of plant and bacterial genomes in SEED and PlantSEED identified a hypothetical membrane protein, AT1G78620 (COG1836, DUF92), as a promising candidate for the missing role of phytyl-phosphate kinase, based on several types of association evidence in bacteria (SI Text and Fig. 4). The localization of AT1G78620 in the chloroplast envelope (53) and the similarity of AT1G78620 expression patterns with those of the phytol kinase VTE5 (http://csbdb.mpimp-golm.mpg.de) also are consistent with this predicted role. This discussion illustrates a method for predicting gene annotations by comparative genomics that is routinely used in microbes but is far less common in plants because of the lack of comparative genomics tools based on chromosomal localization. Here we show how the comparative genomics capabilities of PlantSEED, combined with the seamless integration of a large database of microbial genomes, enable users to apply this type of analysis rapidly to plant genomes.
Fig. 4.
Prokaryotic contextual genome evidence for the functional prediction of AT1G78620 as putative phytyl-phosphate kinase. (A) Typical examples of physical clustering between the AT1G78620 homologs in bacteria and Archaea (COG1836) and phytol kinase (PK) or other genes of polyprenyl metabolism. (B) Examples of domain fusions between the predicted phytyl-phosphate kinase and phytol kinase. Arrows of the same color represent homologous genes; genes not involved in conserved clustering are shown in gray. COG1836, predicted phytyl-phosphate kinase; DS, putative dehydrosqualene/phytoene desaturase; IPK, isopentenyl phosphate kinase; PPS, polyprenyl pyrophosphate synthetase, AT, Arabidopsis; C. tepid, Chlorobium tepidum TLS; E. lenta, Eggerthella lenta; Nostoc, Nostoc sp. 7120; P. arsen, Pyrobaculum arsenaticum.
Discussion
PlantSEED is a new resource to support functional annotation, comparative analysis, and modeling of plant genomes. It is based on a combination of genome annotation technologies, including a specially tailored set of plant-specific protein families, a subsystems approach to annotation, a rich biochemistry database, a model-driven annotation system, and a reference set of 10 plant genomes. The subsystems, genomes, metabolic models, and tools that comprise the PlantSEED resource are accessible through the PlantSEED gateway (http://plantseed.theseed.org). Information on navigating this gateway is given in The PlantSEED Gateway and Figs. S3 and S4.
The models that PlantSEED generates are functioning, predictive ones that include subcellular compartmentation and detailed qualitative biomass compositions. They compare favorably with published plant models and current metabolic databases. With essential gapfilling, the models correctly simulate heterotrophic plant growth. Our pathway gapfilling identified and resolved missing steps in metabolic pathways, proposed many candidate genes to be mapped to these missing steps, and provided predictions for subsequent experimental validation. Critically, the model-based gapfilling provided a means to correct omissions in annotations caused by our extremely conservative annotation propagation procedure. We propose this approach as a valuable breakthrough in eukaryotic genome annotation, because it provides a means of overcoming the major problem of overannotation without losing many correct annotations. These outcomes establish the efficacy of PlantSEED models in supporting model-driven annotation.
Comparative analysis of the PlantSEED reference genomes confirmed the high degree of conservation of primary metabolism and revealed annotation errors that then were corrected by gapfilling and curation. This work highlights the value of a comparative approach in rapidly improving annotations and demonstrates that consistent annotation is crucial to high throughput and accurate comparisons. Given the swelling flood of plant genomes and transcriptomes, this pipeline is likely to be of great value to the plant-research community.
Finally, we stress that PlantSEED is a work in progress, and various improvements are planned. These include (i) deeper curation of existing subsystems, protein families, biochemistry, and models; (ii) addition of reference genomes from algae and a wider range of land plants; (iii) development of subsystems that cover secondary metabolism and nonmetabolic systems; (iv) a development of an automated annotation pipeline that allows users to submit their own genomes and transcriptomes; and (iv) integration of tools that better exploit transcriptomics data to support modeling and model-driven annotation.
Methods
The results of curating associations between genes and reactions were captured in the subsystem spreadsheets. We organized subsystems around primary metabolic pathways found in AraCyc (PlantSEED Biochemistry and Pathways and Dataset S4). Each subsystem has its own web page on which any specific references for gene–reaction associations are listed. Subsystems are listed at http://plantseed.theseed.org (Fig. S3 and Dataset S4). The Ensembl Compara protein families were refined to remove weak orthologs (SI Text). In the interests of transparency, the PlantSEED website lists each gene and the corresponding gene trees that were used for every function in a subsystem.
Supplementary Material
Acknowledgments
We thank Kate Dreher for extensive discussions and support in the use of the AraCyc database and Joshua Stein for support in the use of the genomes and protein families. This work was supported by National Science Foundation Grant IOS-1025398, by an endowment from the C V Griffin Sr Foundation, and by the Office of Science, Office of Biological and Environmental Research, of the US Department of Energy (DOE) under Contract DE-ACO2-06CH11357, as part of the DOE Systems Biology Knowledgebase.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401329111/-/DCSupplemental.
References
- 1.Edwards D, Batley J, Snowdon RJ. Accessing complex crop genomes with next-generation sequencing. Theor Appl Genet. 2013;126(1):1–11. doi: 10.1007/s00122-012-1964-x. [DOI] [PubMed] [Google Scholar]
- 2.McCormack JE, Hird SM, Zellmer AJ, Carstens BC, Brumfield RT. Applications of next-generation sequencing to phylogeography and phylogenetics. Mol Phylogenet Evol. 2013;66(2):526–538. doi: 10.1016/j.ympev.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Mutz KO, Heilkenbrinker A, Lönne M, Walter JG, Stahl F. Transcriptome analysis using next-generation sequencing. Curr Opin Biotechnol. 2013;24(1):22–30. doi: 10.1016/j.copbio.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Pagani I, et al. The Genomes OnLine Database (GOLD) v.4: Status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2012;40(Database issue):D571–D579. doi: 10.1093/nar/gkr1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang P, et al. Creation of a genome-wide metabolic pathway database for Populus trichocarpa using a new approach for reconstruction and curation of metabolic pathways for plants. Plant Physiol. 2010;153(4):1479–1491. doi: 10.1104/pp.110.157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mueller LA, Zhang P, Rhee SY. AraCyc: A biochemical pathway database for Arabidopsis. Plant Physiol. 2003;132(2):453–460. doi: 10.1104/pp.102.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tatusova T, Smith-White B, Ostell J. A collection of plant-specific genomic data and resources at NCBI. Methods Mol Biol. 2007;406:61–87. doi: 10.1007/978-1-59745-535-0_3. [DOI] [PubMed] [Google Scholar]
- 8.Van Bel M, et al. Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 2012;158(2):590–600. doi: 10.1104/pp.111.189514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sucaet Y, Wang Y, Li J, Wurtele ES. MetNet Online: A novel integrated resource for plant systems biology. BMC Bioinformatics. 2012;13:267. doi: 10.1186/1471-2105-13-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schnable JC, Springer NM, Freeling M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci USA. 2011;108(10):4069–4074. doi: 10.1073/pnas.1101368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seaver SM, Henry CS, Hanson AD. Frontiers in metabolic reconstruction and modeling of plant genomes. J Exp Bot. 2012;63(6):2247–2258. doi: 10.1093/jxb/err371. [DOI] [PubMed] [Google Scholar]
- 12.Henry CS, et al. Connecting genotype to phenotype in the era of high-throughput sequencing. Biochim Biophys Acta. 2011;1810(10):967–977. doi: 10.1016/j.bbagen.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Feist AM, Palsson BO. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat Biotechnol. 2008;26(6):659–667. doi: 10.1038/nbt1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douglas AE. Nutritional interactions in insect-microbial symbioses: Aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol. 1998;43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 15.Joyce AR, et al. Experimental and computational assessment of conditionally essential genes in Escherichia coli. J Bacteriol. 2006;188(23):8259–8271. doi: 10.1128/JB.00740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett CL, Herring CD, Reed JL, Palsson BO. The global transcriptional regulatory network for metabolism in Escherichia coli exhibits few dominant functional states. Proc Natl Acad Sci USA. 2005;102(52):19103–19108. doi: 10.1073/pnas.0505231102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satish Kumar V, Dasika MS, Maranas CD. Optimization based automated curation of metabolic reconstructions. BMC Bioinformatics. 2007;8:212. doi: 10.1186/1471-2105-8-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reed JL, et al. Systems approach to refining genome annotation. Proc Natl Acad Sci USA. 2006;103(46):17480–17484. doi: 10.1073/pnas.0603364103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry CS, Zinner JF, Cohoon MP, Stevens RL. iBsu1103: A new genome-scale metabolic model of Bacillus subtilis based on SEED annotations. Genome Biol. 2009;10(6):R69. doi: 10.1186/gb-2009-10-6-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar VS, Maranas CD. GrowMatch: An automated method for reconciling in silico/in vivo growth predictions. PLOS Comput Biol. 2009;5(3):e1000308. doi: 10.1371/journal.pcbi.1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka K, et al. Building the repertoire of dispensable chromosome regions in Bacillus subtilis entails major refinement of cognate large-scale metabolic model. Nucleic Acids Res. 2013;41(1):687–699. doi: 10.1093/nar/gks963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poolman MG, Miguet L, Sweetlove LJ, Fell DA. A genome-scale metabolic model of Arabidopsis and some of its properties. Plant Physiol. 2009;151(3):1570–1581. doi: 10.1104/pp.109.141267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Oliveira Dal’Molin CG, Quek LE, Palfreyman RW, Brumbley SM, Nielsen LK. AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol. 2010;152(2):579–589. doi: 10.1104/pp.109.148817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha R, Suthers PF, Maranas CD. Zea mays iRS1563: A comprehensive genome-scale metabolic reconstruction of maize metabolism. PLoS ONE. 2011;6(7):e21784. doi: 10.1371/journal.pone.0021784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mintz-Oron S, et al. Reconstruction of Arabidopsis metabolic network models accounting for subcellular compartmentalization and tissue-specificity. Proc Natl Acad Sci USA. 2012;109(1):339–344. doi: 10.1073/pnas.1100358109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Oliveira Dal’Molin CG, Quek LE, Palfreyman RW, Brumbley SM, Nielsen LK. C4GEM, a genome-scale metabolic model to study C4 plant metabolism. Plant Physiol. 2010;154(4):1871–1885. doi: 10.1104/pp.110.166488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poolman MG, Kundu S, Shaw R, Fell DA. Responses to light intensity in a genome-scale model of rice metabolism. Plant Physiol. 2013;162(2):1060–1072. doi: 10.1104/pp.113.216762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henry CS, et al. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol. 2010;28(9):977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karp PD, Riley M, Paley SM, Pellegrini-Toole A. The MetaCyc Database. Nucleic Acids Res. 2002;30(1):59–61. doi: 10.1093/nar/30.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33(17):5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vilella AJ, et al. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19(2):327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aziz RK, et al. SEED servers: High-performance access to the SEED genomes, annotations, and metabolic models. PLoS ONE. 2012;7(10):e48053. doi: 10.1371/journal.pone.0048053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aziz RK, et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Devoid S, et al. Automated genome annotation and metabolic model reconstruction in the SEED and Model SEED. Methods Mol Biol. 2013;985:17–45. doi: 10.1007/978-1-62703-299-5_2. [DOI] [PubMed] [Google Scholar]
- 36.Jankowski MD, Henry CS, Broadbelt LJ, Hatzimanikatis V. Group contribution method for thermodynamic analysis of complex metabolic networks. Biophys J. 2008;95(3):1487–1499. doi: 10.1529/biophysj.107.124784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerdes S, et al. Plant B vitamin pathways and their compartmentation: A guide for the perplexed. J Exp Bot. 2012;63(15):5379–5395. doi: 10.1093/jxb/ers208. [DOI] [PubMed] [Google Scholar]
- 38.Sonnhammer EL, Koonin EV. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18(12):619–620. doi: 10.1016/s0168-9525(02)02793-2. [DOI] [PubMed] [Google Scholar]
- 39.Jensen RA, Gu W. Evolutionary recruitment of biochemically specialized subdivisions of Family I within the protein superfamily of aminotransferases. J Bacteriol. 1996;178(8):2161–2171. doi: 10.1128/jb.178.8.2161-2171.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang WK, Liu YL, Xia EH, Gao LZ. Prevalent role of gene features in determining evolutionary fates of whole-genome duplication duplicated genes in flowering plants. Plant Physiol. 2013;161(4):1844–1861. doi: 10.1104/pp.112.200147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang H, Bowers JE, Wang X, Paterson AH. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc Natl Acad Sci USA. 2010;107(1):472–477. doi: 10.1073/pnas.0908007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Youens-Clark K, et al. Gramene database in 2010: Updates and extensions. Nucleic Acids Res. 2011;39(Database issue):D1085–D1094. doi: 10.1093/nar/gkq1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Q, et al. PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res. 2009;37(Database issue):D969–D974. doi: 10.1093/nar/gkn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caspi R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40(Database issue):D742–D753. doi: 10.1093/nar/gkr1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becker SA, et al. Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox. Nat Protoc. 2007;2(3):727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- 46.Latendresse M, Krummenacker M, Trupp M, Karp PD. Construction and completion of flux balance models from pathway databases. Bioinformatics. 2012;28(3):388–396. doi: 10.1093/bioinformatics/btr681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leustek T, et al. Siroheme biosynthesis in higher plants. Analysis of an S-adenosyl-L-methionine-dependent uroporphyrinogen III methyltransferase from Arabidopsis thaliana. J Biol Chem. 1997;272(5):2744–2752. doi: 10.1074/jbc.272.5.2744. [DOI] [PubMed] [Google Scholar]
- 48.Sønderby IE, Geu-Flores F, Halkier BA. Biosynthesis of glucosinolates—gene discovery and beyond. Trends Plant Sci. 2010;15(5):283–290. doi: 10.1016/j.tplants.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 49.Yizhak K, Benyamini T, Liebermeister W, Ruppin E, Shlomi T. Integrating quantitative proteomics and metabolomics with a genome-scale metabolic network model. Bioinformatics. 2010;26(12):i255–i260. doi: 10.1093/bioinformatics/btq183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.May P, et al. Metabolomics- and proteomics-assisted genome annotation and analysis of the draft metabolic network of Chlamydomonas reinhardtii. Genetics. 2008;179(1):157–166. doi: 10.1534/genetics.108.088336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ischebeck T, Zbierzak AM, Kanwischer M, Dörmann P. A salvage pathway for phytol metabolism in Arabidopsis. J Biol Chem. 2006;281(5):2470–2477. doi: 10.1074/jbc.M509222200. [DOI] [PubMed] [Google Scholar]
- 52.Valentin HE, et al. The Arabidopsis vitamin E pathway gene5-1 mutant reveals a critical role for phytol kinase in seed tocopherol biosynthesis. Plant Cell. 2006;18(1):212–224. doi: 10.1105/tpc.105.037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valentin HE, Qi Q. Biotechnological production and application of vitamin E: Current state and prospects. Appl Microbiol Biotechnol. 2005;68(4):436–444. doi: 10.1007/s00253-005-0017-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.