Significance
Mitochondria are organelles that perform many essential functions, including providing the energy to cells. Cells remove damaged mitochondria through a process called mitophagy. Mitophagy is considered a subset of a process called autophagy, by which damaged organelles are enwrapped and delivered to lysosomes for degradation. Implicit in the categorization of mitophagy as a subset of autophagy, which means “self-eating,” is the assumption that a cell degrades its own mitochondria. However, we show here that in a location called the optic nerve head, large numbers of mitochondria are shed from neurons to be degraded by the lysosomes of adjoining glial cells. This finding calls into question the assumption that a cell necessarily degrades its own organelles.
Keywords: mitophagy, phagocytosis
Abstract
It is generally accepted that healthy cells degrade their own mitochondria. Here, we report that retinal ganglion cell axons of WT mice shed mitochondria at the optic nerve head (ONH), and that these mitochondria are internalized and degraded by adjacent astrocytes. EM demonstrates that mitochondria are shed through formation of large protrusions that originate from otherwise healthy axons. A virally introduced tandem fluorophore protein reporter of acidified mitochondria reveals that acidified axonal mitochondria originating from the retinal ganglion cell are associated with lysosomes within columns of astrocytes in the ONH. According to this reporter, a greater proportion of retinal ganglion cell mitochondria are degraded at the ONH than in the ganglion cell soma. Consistently, analyses of degrading DNA reveal extensive mtDNA degradation within the optic nerve astrocytes, some of which comes from retinal ganglion cell axons. Together, these results demonstrate that surprisingly large proportions of retinal ganglion cell axonal mitochondria are normally degraded by the astrocytes of the ONH. This transcellular degradation of mitochondria, or transmitophagy, likely occurs elsewhere in the CNS, because structurally similar accumulations of degrading mitochondria are also found along neurites in superficial layers of the cerebral cortex. Thus, the general assumption that neurons or other cells necessarily degrade their own mitochondria should be reconsidered.
The number, half-life, and morphology of mitochondria vary widely across cell types and are regulated by both intrinsic and extrinsic mechanisms. Mitochondria number is controlled through regulated production (1) and degradation (2), as well as by the regulated fusion and fission of existing mitochondria (3). Damaged mitochondria are removed by mitophagy, a subtype of autophagy that involves the enwrapping of mitochondria in autophagosomes that subsequently fuse with lysosomes to become autophagolysosomes (2). Implicit in the categorization of mitophagy as a subtype of autophagy is the assumption that each cell degrades its own mitochondria.
Recently, we described a phenomenon at the optic nerve head (ONH) of WT mice, where evulsions originating from otherwise intact axons are engulfed and degraded by resident phagocytic astrocytes (4). Serial section-based 3D reconstructions obtained through serial block-face scanning electron microscopy (SBEM) revealed that the protrusions on axons and the evulsions near axons were common throughout the ONH in both the glial lamina, where retinal ganglion cell axons are unmyelinated, and in the adjacent myelination transition zone (MTZ). The axonal protrusions and evulsions were, on average, larger than the mean diameter of axons and contained membrane-bound organelles of unknown identity.
Results
Axonal Protrusions and Evulsions Within the ONH Contain Mitochondria.
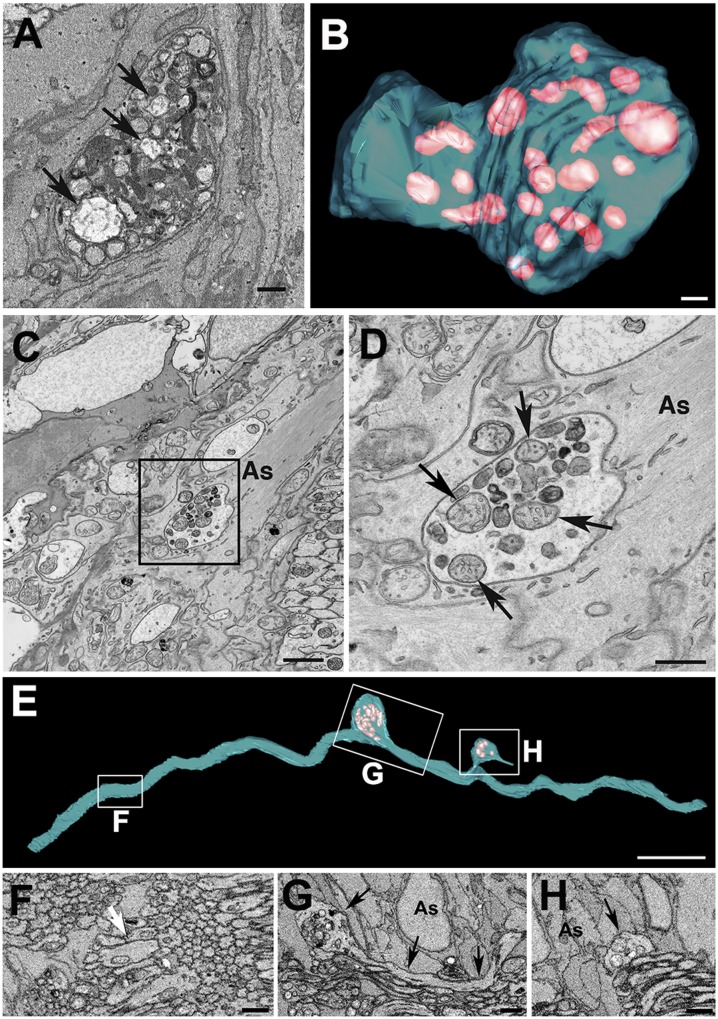
To determine the identity of the membranous material contained within the axonal evulsions at the ONH, a 3-mo-old WT C57BL/6J mouse was analyzed using 3D electron microscopic volumes produced by SBEM. Contents within the axonal evulsions that were fully surrounded by astrocyte processes (Movie S1) included organelles with the morphological characteristics of mitochondria, including the presence of cristae (Fig. 1 A and B and Movie S2). To determine whether the mitochondria within axonal evulsions were healthy or damaged and whether any of the other irregular membranous structures contained within axonal evulsions might be mitochondria remnants, the ONH of mice was examined at higher resolution by transmission EM (Fig. 1 C and D). Within the axonal evulsions, mitochondria with an intact cristae structure (arrows in Fig. 1D) were interspersed with irregular membranous bodies, some of which could be readily identified as fragments of mitochondria. The large mitochondrial clusters were found in 3- and 9-mo-old C57BL/6J mice at comparable densities: 1.9 and 4.8 per 10,000 μm3, respectively. At 9 mo of age, axonal protrusions averaged 11 ± 8 μm3 in volume and contained 29 ± 18 mitochondria (n = 12, ±SD). To exclude the possibility that axonal evulsions containing intact and degrading mitochondria were associated only with rare degenerating axons, fibers containing protrusions were reconstructed from SBEM data. Consistent with our previous study (4), protrusions and evulsions appeared to represent a continuum of morphologies. They all had in common the presence of subaxolemmal accumulations of mitochondria clustered within otherwise healthy axons (Fig. 1 E, G, and H and Movies S3 and S4. In all cases (n = 103), the axonal protrusions filled with mitochondria were found specifically at the sites of direct contact between the axons and astrocyte processes (arrows in Fig. 1 G and H). To determine what other axoplasmic components might be within the axonal evulsions, they were also examined by high-resolution EM tomography (Fig. S1). Microtubules were found in close proximity to the mitochondria, demonstrating that the evulsions shed from axons contain axoplasmic components other than just mitochondria.
Fig. 1.
Evulsions of retinal ganglion cell axons within the ONH contain mitochondria. An SBEM single section (A) and serial reconstruction (B) of axonal evulsion within the ONH of a 3-mo-old C57BL/6J mouse containing morphologically distinct mitochondria (arrows in A and pink volumes in B). Transmission EM (C) and enlarged view of the boxed area (D) showing mitochondria with normal morphology (arrows) and mitochondria remnants within the same evulsion. (E) SBEM-based reconstruction of a single axon displaying two protrusions (boxes G and H). (F–H) Sections through the areas boxed in E. The white arrow in F points to close apposition between axons without intervening glia, and the black arrows in G and H point to direct contacts between the axon and astrocyte processes (As). (Scale bars: A, B, and D, 0.5 μm; C, 1 μm; E, 5 μm; F–H, 2 μm.)
Retinal Ganglion Cell Mitochondria Are Degraded Within ONH Astrocytes.
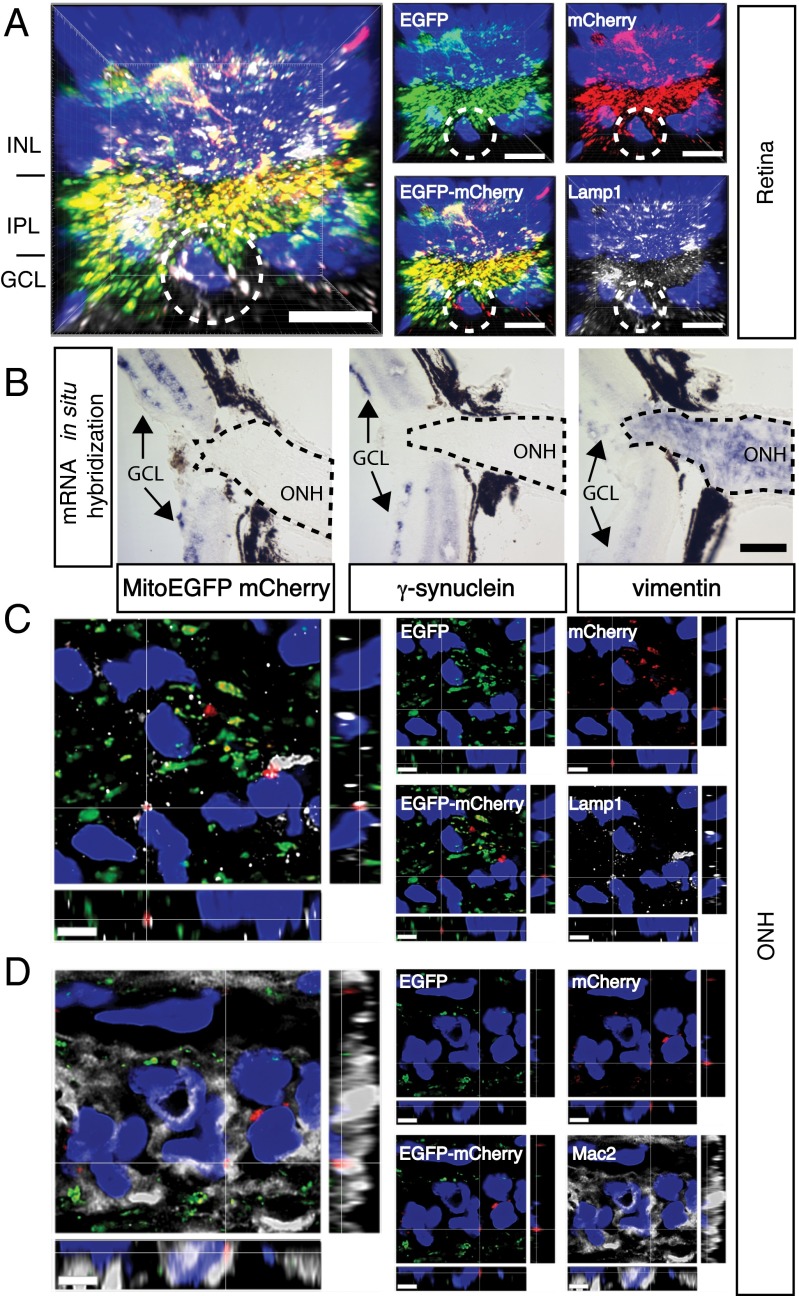
The presence of axonal protrusions containing mitochondria at sites of astrocyte contact and axonal evulsions also containing mitochondria fully surrounded by astrocytes suggested the possibility that some mitochondria from a neuron, the retinal ganglion cell, might be constitutively degraded outside that neuron. To test this possibility directly, a transgenic reporter was designed to track mitochondria degradation. We reasoned that because mitochondria are degraded within lysosomes, transgenes targeted to mitochondria expressing fluorophores that change spectral properties within the acidic environment of lysosomes might be used to monitor the sites of mitochondria degradation. Previously, a tandem fluorophore transgene containing the acid-sensitive fluorophore EGFP and the acid-resistant fluorophore mCherry linked to the autophagosomal protein LC3 has been used to distinguish autophagic vesicles before and after fusion with lysosomes (5). Here, a similar tandem fluorophore reporter of EGFP and mCherry was targeted to mitochondria by fusion to 29 amino acids of human cytochrome c oxidase subunit 8 (sequence provided in Fig. S2A) and was expressed in the retina by intravitreal injection of an adenoassociated virus 2 (AAV2) vector. AAV2::MitoEGFPmCherry infection of the mouse retina produced a single protein of the expected size, 57 kDa (Fig. S2B), as well as widespread expression in retinal ganglion cells and their axons (Fig. S2C). Most of the EGFP and mCherry fluorescence localized together in the retinal ganglion cell layer (GCL) and inner plexiform layer of the retina and colocalized with the mitochondria marker ATP synthase (Fig. S2D). A few discrete puncta with mCherry but not EGFP fluorescence were also found in the retinas of mice infected with AAV2::MitoEGFPmCherry. These mCherry-positive/EGFP-negative (mCherry-ONLY) puncta were found near the nuclei of cells in the retinal GCL and colocalized with lysosomal-associated membrane protein 1 (Lamp1) (Fig. 2A), confirming that the MitoEGFPmCherry transgene can be used to identify at least a subset of degrading mitochondria. Based on the mRNA distribution of the MitoEGFPmCherry transgene relative to the retinal ganglion cell-enriched gene γ-synuclein and the optic nerve astrocyte-expressed gene vimentin, intravitreal AAV2 injection produced widespread MitoEGFPmCherry mRNA expression only within the retina and no expression within the ONH (Fig. 2B). Therefore, AAV2-mediated delivery of the MitoEGFPmCherry transgene into the retina was deemed a suitable method to determine whether some retinal ganglion cell mitochondria were being degraded within the ONH outside of retinal ganglion cells. To examine this possibility, longitudinal sections of the ONHs from mice that received AAV2::MitoEGFPmCherry intravitreal injections were examined together with markers for lysosomes and for astrocytes by confocal microscopy, optical deconvolution, and 3D reconstruction. In all 29 naive WT mice examined by AAV2:MitoEGFPmCherry intravitreal injection, mCherry-ONLY puncta reporting acidified mitochondria were common within the ONH (Fig. 2 C and D). These mCherry-ONLY puncta were associated with and often fully surrounded by vesicles containing Lamp1 (Fig. 2C and Movie S5). To determine whether these degrading mitochondria were within axons or astrocytes, their position was assessed relative to expression of Mac2, a marker of phagocytic activity (6) abundant in the cytoplasm of MTZ astrocytes (4) (Fig. S3). The majority of the mCherry-ONLY puncta at the MTZ were found not on the axon bundles but within the intervening glial columns, and they colocalized with Mac2 (Fig. 2D and Movie S6). Therefore, expression of the MitoEGFPmCherry reporter within the retina established that some retinal ganglion cell mitochondria are degraded within astrocytes in the ONH, specifically those astrocytes that express high levels of Mac2 (Mac2High).
Fig. 2.
AAV2::MitoEGFPmCherry infection of the retina demonstrates that some retinal ganglion cell mitochondria are degraded within ONH astrocytes. (A) mCherry-ONLY puncta are within the retinal GCL of AAV2:MitoEGFPmCherry-infected retinas and colocalize with the lysosomal marker Lamp1. Circles surround a GCL cell with prominent mCherry-ONLY puncta. INL, inner nuclear layer; IPL, inner plexiform layer. (B) AAV::MitoEGFPmCherry mRNA is found only in the retina, including the GCL, where ganglion cells are labeled by γ-synuclein mRNA, but not in the ONH, where astrocytes are labeled by vimentin mRNA. Dotted traces demarcate the ONH. (C) Within the ONH, common mCherry-ONLY puncta overlap with or are surrounded by the lysosomal marker Lamp1. A single mCherry-ONLY punctum surrounded by multiple vesicles containing Lamp1 is shown in the xyz projection. (D) Within the ONH MTZ, the mCherry-ONLY puncta localize largely separate from axon bundles and within the glial columns, and colocalize with the MTZ astrocyte cytoplasmic marker Mac2. Single mCherry-ONLY punctum surrounded by Mac2-labeled cytoplasm is shown in the xyz projection. All images show nuclei labeled by DAPI as a reference. (Scale bars: A, C, and D, 5 μm; B, 100 μm.)
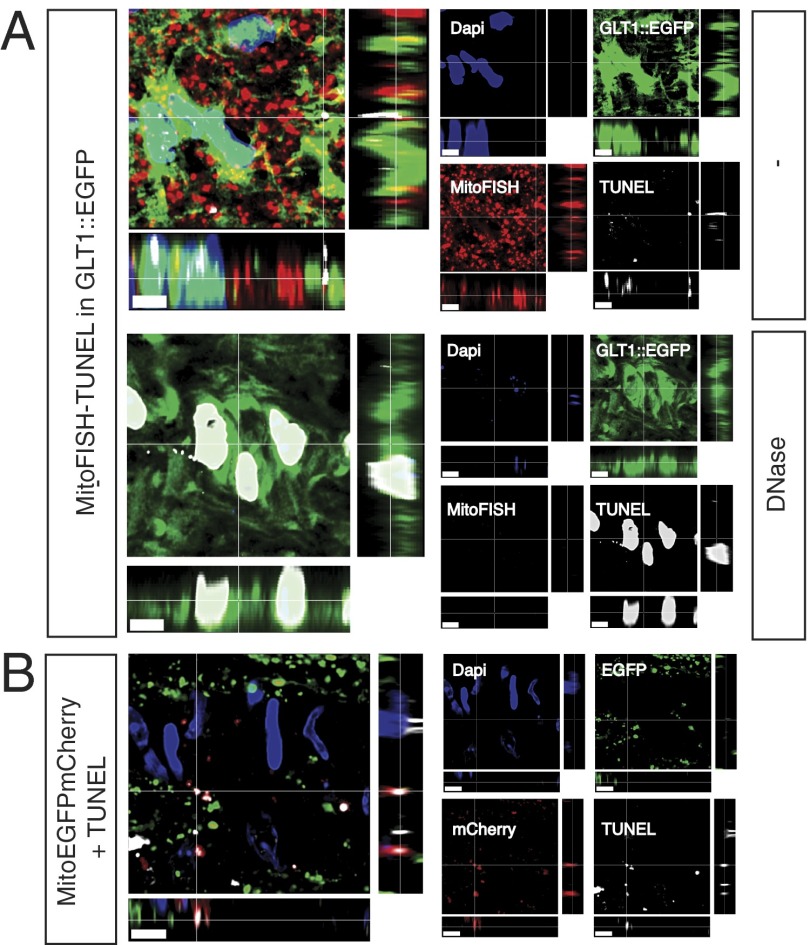
Given the large amount of mCherry-ONLY signal within the ONH, we reasoned that this extent of mitochondrial degradation might also be detectable by a sensitive TUNEL assay used in concert with FISH of mtDNA (MitoFISH). As expected, MitoFISH by itself detected discrete puncta within the elongated or spherical mitochondria detected by either the MitoEGFPmCherry transgene or ATP synthase immunoreactivity (Fig. S3 A and B). The combined MitoFISH-TUNEL protocol revealed the presence of clusters of TUNEL-positive mitochondria within the ONH (Fig. 3). The TUNEL-positive, MitoFISH-positive signal was within the soma and cell processes of ONH astrocytes, as identified by the glutamate transporter 1 (GLT1)::EGFP transgene (Fig. 3A and Movie S7). In GLT1::EGFP BAC-transgenic mice (7), EGFP is expressed in the cytoplasm of all GFAP-expressing cells within the ONH, both the Mac2Low astrocytes of the glial lamina and the Mac2High astrocytes of the MTZ (Fig. S3). Much of the TUNEL signal overlapped with the MitoFISH signal, presumably marking mitochondria at early stages of degradation. The remainder of the TUNEL signal, particularly that near regions of MitoFISH-TUNEL overlap, most likely represents mtDNA fragments too small to hybridize to the MitoFISH probes but long enough to seed nucleotide incorporation by terminal transferase (8). In a control experiment designed to confirm that the MitoFISH-TUNEL signal was due to mtDNA, some sections were treated with DNaseI. Nonnuclear TUNEL signal and the MitoFISH signal were eliminated by pretreatment of the samples with DNaseI, and nuclei became highly labeled by TUNEL even in regions that were too degraded to be detectable by the intercalating dye DAPI (Fig. 3A). Finally, to determine whether the ONH TUNEL signal was due to the degradation of internalized axonal mitochondria rather than the astrocytes’ own mitochondria, TUNEL labeling was performed in the optic nerves of C57BL/6J mice whose retinal ganglion cell mitochondria were previously labeled by AAV2::MitoEGFPmCherry intravitreal injection (Fig. 3B and Movie S8). The ONH TUNEL signal largely colocalized with the mCherry-ONLY puncta, confirming that AAV2::MitoEGFPmCherry reports degrading mitochondria and that surprisingly large amounts of retinal ganglion cell mitochondria are degraded by astrocytes within the ONH.
Fig. 3.
Combination of TUNEL and MitoFISH identifies mitochondria degraded by astrocytes within the ONH and confirms that some of these degrading mitochondria are derived from axons. (A) In sections of ONH where astrocytes are labeled by a GLT1::EGFP BAC transgene and mtDNA is detected by MitoFISH, TUNEL labels degrading mtDNA within astrocyte soma and processes; after DNase treatment, TUNEL only labels nuclei remnants. (B) In the ONH of C57BL/6J mice whose retinal ganglion mitochondria are labeled by the AAV2::MitoEGFPmCherry transgene, the TUNEL signal colocalizes with the mCherry-ONLY puncta. (Scale bars: 5 μm.)
Large Fraction of Retinal Ganglion Cell Mitochondria Are Degraded Within the ONH.
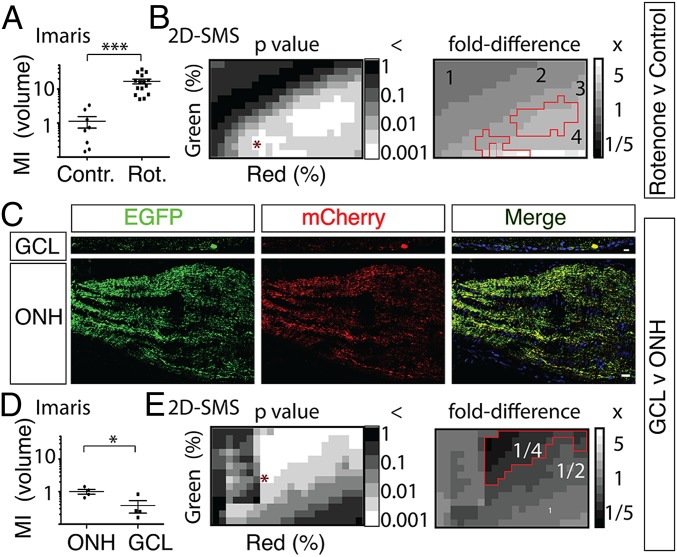
To estimate the contribution of this pathway to the overall degradation of retinal ganglion cell mitochondria, the proportion of mCherry-ONLY signal to total mCherry signal [here called the mitophagy index (MI)] was quantified in confocal 3D image stacks and single 2D optical sections of the retinal GCL and ONH (Fig. 4 and Fig. S4). First, to assess whether the MI could report differences in mitochondria degradation, the mitochondrial complex I disruptor rotenone was injected intravitreally at doses one order of magnitude lower than those previously used to kill retinal ganglion cells (9). Using subjective thresholding in Imaris (Bitplane, Inc.) 3D volume-based image analysis software, the MI by volume (MIvol) was increased 16-fold (P < 0.001, Student t test) by rotenone (Fig. 4A and Fig. S4A). Using similar subjective thresholding in Imaris to compare the amount of mCherry-ONLY signal in multiple (n = 4) z-stacks of the GCL and ONH of one eye revealed a MIvol in the ONH ∼2.7-fold higher (P < 0.05, Student t test) than in the GCL (Fig. 4D and Fig. S4B). To confirm this highly unexpected finding and to assess the dependence of this result on the thresholding parameters used, a quantification and analysis protocol termed 2D segmentation matrix statistics (2D-SMS; Materials and Methods) was also used to measure mCherry-ONLY signal (Fig. 4 B and E and Fig. S4 B–D). When 2D-SMS was applied to single confocal sections extracted from the middle of the z-stacks from the same datasets as above to calculate the MI by area (MIarea), rotenone was shown to produce a mean MIarea fold difference of 3.4 ± 0.9 (Fig. 4B and Fig. S4C; P < 0.001, Student t test), compared with DMSO vehicle control, whereas the GCL was shown to produce a mean MIarea fold difference of 0.12 ± 0.06 relative to the ONH, although this difference failed to reach statistical significance (0.1 < P < 0.05, Student t test). Thus, although generating a more conservative statistical comparison, 2D-SMS MIarea measurements generally confirmed the Imaris MIvol measurements by also showing that rotenone increases mitochondria degradation and supporting that there might be more degradation of retinal ganglion cell mitochondria in the ONH than in the retinal ganglion cell soma.
Fig. 4.
Three-dimensional and 2D quantitative assays based on the Cherry-ONLY signal after AAV2::MitoEGFPmCherry intraocular injection demonstrate that a higher proportion of retinal ganglion cell mitochondria are degraded in the ONH relative to the GCL. (A) Intravitreal injection of 0.5 μL of 3.1 mM rotenone (Rot.) produces a 16-fold increase (***P < 0.001, Student t test) in the MI by volume [MI (volume)] within the ONH, as measured by subjectively set volume thresholding in Imaris software. Representative images are shown in Fig. S4A. Contr., control. (B) Two-dimensional SMS, which analyses 2D images at multiple segmentation values across two color channels, also shows that rotenone produces significant increases in MIarea, which range from 2.3- to 6.4-fold increases depending on the segmentation used but average a 3.4 ± 0.9-fold increase at the P < 0.001 significance level (Student t test). Representative images binarized at the segmentation pair denoted by an asterisk in P value distribution are shown in Fig. S4C. MIarea P values and fold differences for a scrambled control are shown in Fig. S4E. (C) Representative images of MitoEGFPmCherry labeling in the GCL and ONH from the same eye. (Scale bar: 20 μm.) (D) MI (volume) is lower (*P < 0.05, Student t test) in the GCL relative to the ONH, as determined by comparing ONH sections (n = 4) with retina flat-mount images (n = 4) of one mouse using subjective thresholding in Imaris software. (E) Comparing sections of the GCL and ONH from multiple animals (n = 10) using 2D-SMS also shows significantly lower MIarea in the GCL relative to the ONH, with MI fold differences ranging from 0.03 to 0.30 depending on the segmentation used and averaging a 0.22 ± 0.16-fold difference at the P < 0.001 level (paired Student t test). Representative images of GCL and ONH binarized at the segmentation pair denoted by an asterisk in P value distribution are shown in Fig. S4D. MIarea P values and fold differences for a scrambled control are shown in Fig. S4F.
To test rigorously whether the ONH really has a greater proportion of acidified mitochondria than the GCL, 2D-SMS was then used to compare these two regions in a larger set of animals (n = 10). Compared with the ONH, the GCL was shown to produce a mean MIarea fold difference of 0.22 ± 0.03 (Fig. 4E and Fig. S4D; P < 0.001, paired Student t test), which represents a 4.6-fold greater amount of mCherry-ONLY area in the ONH relative to the GCL. To confirm that 2D-SMS was not just finding statistical significance by virtue of so many comparisons, two control protocols were then implemented. First, the most statistically significant segmentation ranges identified by 2D-SMS in the rotenone and ONH vs. GCL datasets were applied to their respective datasets, and representative images were binarized according to the segmentation coordinates that represent the minimum Pythagorean distance from the origin of segmentation (Fig. 4 B and E and Fig. S4 C and D). In both datasets, these combinations of EGFP and mCherry segmentation thresholds identified by 2D-SMS selectively masked the mCherry pixels that colocalized with EGFP and detected only the mCherry-ONLY signals that report degrading mitochondria, thus confirming that the significance-driven segmentation carried out by 2D-SMS identifies the biological process being measured, namely, degrading mitochondria. In the second control protocol, both datasets were scrambled, and the MIarea P values and fold differences were analyzed. As expected, both statistical significance and fold differences were ablated (Fig. S4 E and F). Thus, analyses based on 3D datasets by Imaris and on 2D datasets by 2D-SMS agreed in showing that, at least as reported by the AAV2::MitoEGFPmCherry transgene, degradation of retinal ganglion cell mitochondria at the ONH constitutes a major pathway for the degradation of retinal ganglion cell mitochondria.
Similar Degradation of Mitochondria Likely Occurs in Superficial Layers of Cerebral Cortex.
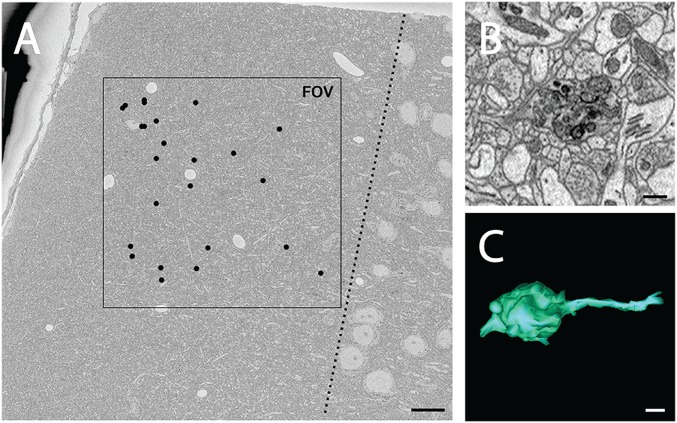
To determine whether axonal mitochondria might be degraded by a similar mechanism elsewhere in the CNS, SBEM was used to analyze locations where there were spherical bodies on and near axons labeled by an antibody that recognizes protease-resistant γ-synuclein, because this labeling was the first evidence that there was astrocyte phagocytosis of axonal elements within the ONH (4). Spherical bodies labeled by γ-synuclein were found in superficial layers of cerebral cortex on or near serotonergic axons, as identified in a Slc64a BAC transgenic mouse model where these axons are labeled by EGFP (Fig. S5). At this location in a naive 3-mo-old C57BL/6J mouse, numerous protrusions were found (Fig. 5A) that highly resemble those seen in the ONH by SBEM (Fig. 5 B and C and Movie S9), although whether they were present on these serotonergic axons or different neurites was not determined.
Fig. 5.
Protrusions containing degrading mitochondria are found in superficial layers of cerebral cortex of a young WT mouse. (A) Low-magnification view of a region of cortical layer 1 of a 3-mo-old C57BL/6J mouse imaged with SBEM, where black dots indicate the locations of protrusions found within volume. Total volume dimensions were 200 μm × 200 μm and 32.8 μm in depth. The dotted line indicates the boundary between layers 1 and 2. (B) Higher magnification views of a protrusion reveal morphology similar to that seen in the ONH, consisting of large, round, membrane-bound bodies filled with debris particles, including mitochondrial fragments. (C) Three-dimensional reconstruction of the same protrusion. (Scale bars: A, 15 μm; B and C, 0.5 μm.)
Discussion
According to the MitoEGFPmCherry mitochondria degradation reporter, a significantly greater proportion of retinal ganglion cell mitochondria are degraded in the ONH than at the cell soma, which is the site where most mitochondrial degradation is presumed to occur. Extrapolations based on the data presented in this study lead us to estimate that there is approximately a 10-fold greater amount of retinal ganglion cell mitochondria in the retinal GCL than in the ONH but that the total amounts of retinal ganglion cell mitochondria being degraded at both locations are comparable. It is, of course, possible that the MitoEGFPmCherry transgene provides a skewed report of mitochondria degradation, for example, by just reporting on large clusters of degrading mitochondria. Regardless, the MitoEGFPmCherry transgene demonstrates that the ONH is a major site of retinal ganglion cell degradation. This conclusion based on the MitoEGFPmCherry reporter is supported by EM ultrastructural investigations of the ONH. Based on the frequency of axonal protrusions and evulsions, we estimate that within the ONH of adult WT mice, most axons contain at least one such structure filled with mitochondria at various stages of degeneration. Further, taking into account the number of mitochondria per protrusion/evulsion, also measured by SBEM, we then estimate that approximately 1 million retinal ganglion cell mitochondria are being degraded at the ONH of mice at any one time.
Here, we present three independent corroborating lines of evidence that at this second major site of retinal ganglion cell degradation, at least some of the axonal mitochondria are not degraded cell-autonomously within retinal ganglion cell axons by traditional mitophagy but rather by the resident astrocytes through a process we term transcellular degradation of mitochondria, or transmitophagy. First, SBEM shows structures that represent a continuum: mitochondria clustered within axons at sites contacted by astrocyte processes, axonal protrusions filled with mitochondria being pinched off from axons, and membrane-enclosed evulsions filled with mitochondria fully surrounded by astrocyte cytoplasm. Second, acidified retinal ganglion cell mitochondria, as detected by ONH mCherry-ONLY signal after MitoEGFPmCherry transgene delivery to the retina, are found at the ONH specifically in columns of astrocytes within the astrocyte cytoplasm and surrounded by lysosomes. Third, large clusters of degrading mtDNA, as detected by a combination of TUNEL and MitoFISH, are also found within astrocytes specifically at this location, and at least some of these mitochondria are of axonal origin as demonstrated by colocalization with the retina-delivered MitoEGFPmCherry transgene.
Many questions remain. It is unknown whether the axonal mitochondria removed at the ONH are only mitochondria resident within axons at this location or whether other axonal mitochondria fated to undergo degradation are actively transported to the ONH for disposal. Why axonal mitochondria are degraded selectively at the ONH is also not known. One possibility is that this pathway is activated by focal axonal damage that is insufficient to cause axonal loss. Another possibility is that it is too dangerous or energetically demanding to transport axonal mitochondria through the narrow lamina region back to the soma for degradation. However, a third possibility is that the ONH contains a subset of astrocytes with extreme phagocytic activity, as might be suggested by their high expression of molecular markers such as Mac2. A previous study (10) described prominent focal swellings on axons and dystrophic neurites filled with mitochondria within the ONH glial lamina as the earliest sign of glaucomatous pathology in DBA/2J mice, an age-dependent glaucoma mouse model. Thus, a high priority will be to examine whether normal shedding of mitochondria from axons at the ONH is affected in glaucoma before the onset of axon loss. Because morphologically similar structures filled with degrading mitochondria are also found in the cortex at a 10-fold lower density, it is possible that transmitophagy of axonal mitochondria by astrocytes is a widespread phenomenon in the CNS. Recently, astrocytes have been shown to phagocytose whole synapses elsewhere in the visual pathway (11). Because synapses usually contain mitochondria, it is quite possible that the molecular machinery used by astrocytes in the phagocytosis of synapses may also be used in the phagocytosis of axonal mitochondrial evulsions in the ONH and elsewhere.
Although this description of a transmitophagy process is new for the nervous system, transcellular degradation of mitochondria is also seen upon fertilization, because sperm mitochondria are actively degraded by oocytes, through molecular machinery that is also used in autophagy (12). Similar autophagy-assisted phagocytosis is also involved in the turnover of photoreceptor outer segments by retinal pigment epithelial cells (13), and thus may be a common mechanism used in the nervous system. It is noteworthy that mtDNA is highly immunogenic (14); for example, mtDNA that is not properly degraded by autophagy can be released and cause cardiomyopathy (15). Because of the prominent roles that mitochondria play in axonal health (16) and in varied neurodegenerative disorders (17), a priority should be to determine whether specific defects in transmitophagy contribute to neuronal dysfunction or disease.
Materials and Methods
Mice.
Experiments were carried out in accordance with animal protocols approved by Institutional Animal Care and Use Committees at The Johns Hopkins University School of Medicine and University of California, San Diego. GLT1::EGFP mice (7) and gene expression nervous system atlas (18) Slc6a4::EGFP mice were obtained from Jeff Rothstein and David Linden, respectively (both from The Johns Hopkins University School of Medicine).
SBEM and Electron Tomography.
For SBEM, the ONHs of two male C57BL/6J mice (aged 3 mo and 9 mo) were processed with procedures modified from Wilke et al. (19), essentially as previously described by Nguyen et al. (4). Volumes were processed and analyzed using IMOD software (http://bio3d.colorado.edu/imod) (20). Electron tomography specimens from the ONH of a 9-mo-old C57BL/6J mouse processed for SBEM staining as described above were sectioned with a diamond knife at a thickness of 300 nm. Sections were coated on both sides with carbon. Following glow discharge, 5- and 10-nm diameter colloidal gold particles were deposited on each side to serve as fiducial markers. Data were generated with an FEI Titan microscope operating at 300 kV; the micrographs were produced using a 4k × 4k Gatan CCD camera. For reconstruction, single tilt series of images were recorded between −60° and +60° at regular increments of 0.5°. Fine alignment of projections and 3D reconstruction were performed using the transform-based tracking, bundle adjustment, and reconstruction package (21) in conjunction with IMOD. Examples presented in Fig. 1 and quantitative data described in the main text are derived from two mice; similar structures have been observed by SBEM in four additional 3-mo-old C57BL/6J mice, as well as in three mice of the DBA/2J background. The electron tomography data are derived from an additional 9-mo-old C57BL/6J mouse.
MitoEGFPmCherry Detection of Degrading Mitochondria.
The MitoEGFPmCherry construct was first made in a pCS2+ vector by in-frame fusion of amino acids 1–29 of human (and other primate) cytochrome c subunit 8 to EGFP, followed by a flexible glycine linker, followed by mCherry (sequence provided in Fig. S2). The resulting cDNA was subcloned into MluI and KpnI sites of pENN.AAV.CB7.CI to produce AAV2::MitoEGFPmCherry. Virus production and purification were carried out at the University of Pennsylvania Vector Core. For ocular delivery, 2 μL of AAV2 (2.36 × 1012 genome copies per milliliter) were delivered over the course of 1 min into the vitreous space using a micromanipulator-guided 10-μL Nanofil syringe loaded onto a microinjector (World Precision Instruments).
Intravitreal Injection of Rotenone.
At least 1 mo after the AAV2::MitoEGFPmCherry intravitreal injection, 0.5 μL of 3.1 mM rotenone (Sigma–Aldrich) in DMSO (Sigma–Aldrich) or the DMSO vehicle alone was injected into the vitreous space over the course of 1 min in a similar fashion as the AAV injections. Mice were killed for analyses 24 h after the ocular injections of rotenone or vehicle.
MitoFISH-TUNEL Detection of Degrading Mitochondria in Cryosections.
To produce mouse mtDNA probes, C57BL/6J mouse liver mitochondria were purified by differential centrifugation (22). Over 12 kb of the mitochondria genome was amplified with Expand Long Polymerase (Roche) by three primer pairs each amplifying ∼4 kb: AATTCCATAATTTACACCAACTTCCTA and CTCGTGTGTCTACATCTAATCCTACT, ATTACTATACTACTAACAGACCGCAA and AACATGGGCTTTTGGTAGTCATAGGT, and TTATCATTCACAACACACACCTTAGAC and CTTTGGGTGCTGGTGGTGG. Gel-purified PCR bands were labeled with digoxigenin-11–dUTP by nick translation (Roche) and purified by MicroSpin G-50 columns (Amersham Pharmacia Biotech, Inc.). TUNEL detection by BrdUTP incorporation was performed by modified procedures based on the method of Darzynkiewicz et al. (23). After TUNEL and immunohistochemistry, MitoFISH was detected by modified procedures based on the methods of van de Corput et al. (24) and Janes et al. (25). A detailed protocol is provided in SI Materials and Methods.
Immunohistochemistry, mRNA in Situ Hybridization, and Image Acquisition.
Antibodies used were a rat anti-mouse Mac2 (American Type Culture Collection), rabbit anti-human Lamp1 (Abcam), mouse anti-rat ATP synthase (Millipore), chicken anti-GFP (Aves Labs), and rat anti-BrdU (Abcam). Goat secondary antibodies used were anti-rabbit IgG Cy5, anti-mouse IgG Cy5, and anti-chicken IgY Cy2 (all from Jackson Immunoresearch), as well as anti-rat IgG Cy5 (Invitrogen). The slides were stained with DAPI (Invitrogen) to visualize nuclei. Riboprobes for γ-synuclein and vimentin and procedures for mRNA in situ hybridization have been described previously (26). The probe for MitoEGFPmCherry was made by first cloning the cDNA into pBluescript. Fluorescence image stacks of retinas and optic nerves were collected with a Fluoview 500 confocal laser-scanning microscope (Olympus) using a 1.42-N.A. PlanApoN 60× oil immersion objective (Olympus) and acquired at a 0.4-μm z-interval for all image stacks other than those presented in Fig. S4B and used in the quantification shown in Fig. 4D. The latter images were acquired using a Leica SPE II inverted confocal microscope (Leica Microsystems GmbH) with a 1.3.-N.A. ACS Apo 63× oil immersion objective and collected at a 0.25-μm z-interval. For 2D-SMS comparisons of the ONH and retina in larger sets of animals, single confocal images were acquired with the Fluoview 500 microscope using a 1.0-N.A. UPlanApo 40× oil immersion objective (Olympus).
Three-Dimensional and 2D Image Quantification.
For mCherry-ONLY volume comparisons, stacks of confocal images were deconvoluted with Autoquant X3 software (Media Cybernetics, Inc.) and rendered as 3D reconstructions in Imaris analysis software (Bitplane, Inc.). Imaris was used to identify mCherry-ONLY and MitoFISH-positive, TUNEL-positive signals. To compare mitochondria degradation in the ONH and GCL, and for the rotenone-positive control, volumes of mCherry-ONLY were measured using Imaris, and the results were statistically analyzed and graphed using GraphPad Prism. For mCherry-ONLY measurements that did not depend on subjective thresholding values, raw or deconvoluted individual confocal images were analyzed using a custom IPlab (Becton Dickinson) script called 2D-SMS. The mCherry-ONLY signal was quantified by zeroing pixels in EGFP segments and counting the remaining mCherry segments. Multiple ascending segmentation thresholds in both color channels were used to quantify single confocal optical sections, generating matrices of values where the coordinates of individual values reflected the segmentation thresholds of the two color channels. Performing statistical analysis on values in congruent segmentation coordinates generated a 2D distribution of P values. Statistics were then computed and graphed using Excel software (Microsoft), and final graphs were produced using IPlab. The MI is defined as an area ratio in the 2D-SMS analyses and a volume ratio in the Imaris analyses, and it is defined as mCherry-ONLY signal/total mCherry signal. MI fold differences were calculated by MIRotenone/MIVehicle in the rotenone dataset and by MIGCL/MIONH in the GCL vs. ONH dataset.
Supplementary Material
Acknowledgments
This work was supported by Grant R01 EY022680 (to N.M.-A. and M.H.E.); Grant R01 EY019960, and a “Catalyst for a Cure” grant from the Glaucoma Research Foundation and the Melza M. and Frank Theodore Barr Foundation (to N.M.-A.). This work was also supported by National Center for Research Resources Grant 5P41RR004050, National Institute on Drug Abuse Human Brain Project Grant DA016602, National Institute of General Medical Sciences (NIGMS) Grants 5R01GM82949 and 5P41GM103412-25 (to M.H.E.), NIGMS Training Grant 5T32GM07814, and National Science Foundation Grant DGE-1232825 (to N.A.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404651111/-/DCSupplemental.
References
- 1.Scarpulla RC, Vega RB, Kelly DP. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol Metab. 2012;23(9):459–466. doi: 10.1016/j.tem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20(1):31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337(6098):1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen JV, et al. Myelination transition zone astrocytes are constitutively phagocytic and have synuclein dependent reactivity in glaucoma. Proc Natl Acad Sci USA. 2011;108(3):1176–1181. doi: 10.1073/pnas.1013965108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 6.Rotshenker S. The role of Galectin-3/MAC-2 in the activation of the innate-immune function of phagocytosis in microglia in injury and disease. J Mol Neurosci. 2009;39(1-2):99–103. doi: 10.1007/s12031-009-9186-7. [DOI] [PubMed] [Google Scholar]
- 7.Regan MR, et al. Variations in promoter activity reveal a differential expression and physiology of glutamate transporters by glia in the developing and mature CNS. J Neurosci. 2007;27(25):6607–6619. doi: 10.1523/JNEUROSCI.0790-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato KI, Gonçalves JM, Houts GE, Bollum FJ. Deoxynucleotide-polymerizing enzymes of calf thymus gland. II. Properties of the terminal deoxynucleotidyltransferase. J Biol Chem. 1967;242(11):2780–2789. [PubMed] [Google Scholar]
- 9.Zhang X, Jones D, Gonzalez-Lima F. Mouse model of optic neuropathy caused by mitochondrial complex I dysfunction. Neurosci Lett. 2002;326(2):97–100. doi: 10.1016/s0304-3940(02)00327-0. [DOI] [PubMed] [Google Scholar]
- 10.Howell GR, et al. Axons of retinal ganglion cells are insulted in the optic nerve early in DBA/2J glaucoma. J Cell Biol. 2007;179(7):1523–1537. doi: 10.1083/jcb.200706181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung WS, et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 2013;504(7480):394–400. doi: 10.1038/nature12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334(6059):1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- 13.Kim JY, et al. Noncanonical autophagy promotes the visual cycle. Cell. 2013;154(2):365–376. doi: 10.1016/j.cell.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464(7285):104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oka T, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485(7397):251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Court FA, Coleman MP. Mitochondria as a central sensor for axonal degenerative stimuli. Trends Neurosci. 2012;35(6):364–372. doi: 10.1016/j.tins.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Schon EA, Przedborski S. Mitochondria: The next (neurode)generation. Neuron. 2011;70(6):1033–1053. doi: 10.1016/j.neuron.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heintz N. Gene expression nervous system atlas (GENSAT) Nat Neurosci. 2004;7(5):483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- 19.Wilke SA, et al. Deconstructing complexity: Serial block-face electron microscopic analysis of the hippocampal mossy fiber synapse. J Neurosci. 2013;33(2):507–522. doi: 10.1523/JNEUROSCI.1600-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence A, Bouwer JC, Perkins G, Ellisman MH. Transform-based backprojection for volume reconstruction of large format electron microscope tilt series. J Struct Biol. 2006;154(2):144–167. doi: 10.1016/j.jsb.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 22.Vermulst M, Bielas JH, Loeb LA. Quantification of random mutations in the mitochondrial genome. Methods. 2008;46(4):263–268. doi: 10.1016/j.ymeth.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darzynkiewicz Z, Galkowski D, Zhao H. Analysis of apoptosis by cytometry using TUNEL assay. Methods. 2008;44(3):250–254. doi: 10.1016/j.ymeth.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van de Corput MP, et al. Detection of mitochondrial DNA deletions in human skin fibroblasts of patients with Pearson’s syndrome by two-color fluorescence in situ hybridization. J Histochem Cytochem. 1997;45(1):55–61. doi: 10.1177/002215549704500108. [DOI] [PubMed] [Google Scholar]
- 25.Janes MS, et al. Rapid analysis of mitochondrial DNA depletion by fluorescence in situ hybridization and immunocytochemistry: Potential strategies for HIV therapeutic monitoring. J Histochem Cytochem. 2004;52(8):1011–1018. doi: 10.1369/jhc.3A6209.2004. [DOI] [PubMed] [Google Scholar]
- 26.Soto I, et al. Retinal ganglion cells downregulate gene expression and lose their axons within the optic nerve head in a mouse glaucoma model. J Neurosci. 2008;28(2):548–561. doi: 10.1523/JNEUROSCI.3714-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]