Significance
In eukaryotic cilia and flagella, outer arm dyneins (OADs) are bound to axonemal doublet microtubules every 24 nm; however, how this regular arrangement is produced remains unknown. To approach this problem, we studied the properties of the OAD-docking complex (ODA-DC), a three-subunit complex that functions as the OAD-docking site on the doublet. Using recombinant ODA-DC, we found that the ODA-DC has an ∼24-nm-long ellipsoidal shape and cooperatively binds to the axoneme in an end-to-end manner. These and other results indicate that cooperative association of the ODA-DC underlies the periodic OAD arrangement at specific positions on the doublets. These findings provide insight into how the regular axonemal repeat structure is produced.
Abstract
Outer arm dynein (OAD) in cilia and flagella is bound to the outer doublet microtubules every 24 nm. Periodic binding of OADs at specific sites is important for efficient cilia/flagella beating; however, the molecular mechanism that specifies OAD arrangement remains elusive. Studies using the green alga Chlamydomonas reinhardtii have shown that the OAD-docking complex (ODA-DC), a heterotrimeric complex present at the OAD base, functions as the OAD docking site on the doublet. We find that the ODA–DC has an ellipsoidal shape ∼24 nm in length. In mutant axonemes that lack OAD but retain the ODA-DC, ODA-DC molecules are aligned in an end-to-end manner along the outer doublets. When flagella of a mutant lacking ODA-DCs are supplied with ODA-DCs upon gamete fusion, ODA-DC molecules first bind to the mutant axonemes in the proximal region, and the occupied region gradually extends toward the tip, followed by binding of OADs. This and other results indicate that a cooperative association of the ODA-DC underlies its function as the OAD-docking site and is the determinant of the 24-nm periodicity.
Cilia and flagella of eukaryotic cells are organelles that generate fluid flow on the cell surface and/or sense chemical or mechanical stimuli from the external environment (1). Cilia/flagella beating is driven by outer arm dynein (OAD) and inner arm dyneins. The arrangement of dyneins on the axoneme has an overall periodicity of 96 nm, within which OAD binds every 24 nm; this 24-nm periodicity is completely conserved in essentially all eukaryotic organisms with “9 + 2” axonemes (2, 3) and even occurs in insect sperm flagella containing multiple rows of doublet microtubules arranged in a spiral configuration (4, 5). OAD is best characterized in Chlamydomonas. It is a very large protein complex of ∼2 MDa, comprising 3 heavy chains, 2 intermediate chains, and 11 distinct light chains. Most of the subunits are conserved from protists to mammals (6). OAD is the most abundant and most powerful axonemal dynein, generating about two-thirds of the total propulsive force of ciliary beating (7, 8). Human diseases due to ciliary motility defects [termed primary ciliary dyskinesia (PCD)] are caused most commonly by defects in OAD assembly (9–11). The assembly process and the in situ structure of the OAD complex in the axoneme have been well studied (3, 12, 13). However, the mechanism underlying the periodic binding of OAD to the doublet is poorly understood.
The outer dynein arm-docking complex (ODA-DC) has been identified as a complex that mediates OAD binding to the doublet (14, 15). In the flagella of Chlamydomonas mutants (e.g., outer-dynein-arm deficient oda6) retaining the ODA-DC but not OAD, the ODA-DC is observed by electron microscopy as a small projection linearly arrayed every 24 nm along the outer doublet (3, 14–17). It is composed of three subunits: DC1, ∼83 kDa, encoded by ODA3; DC2, ∼62 kDa, encoded by ODA1; and DC3, ∼21 kDa, encoded by ODA14 (18–20), which assemble in the cell cytoplasm and are transported into the flagella independently of OAD (16). DC1 and DC2 are coiled-coil proteins; they are essential for the OAD docking activity because mutants (oda1, oda3) lacking these proteins entirely lack OAD. In contrast, DC3 is a redox-sensitive Ca2+-binding protein and apparently is nonessential for OAD binding because a mutant (oda14) lacking it retains ∼40% of OADs (20, 21). Presumably, DC3 has an as yet unidentified regulatory function. DC2 is well conserved throughout evolution, and seems to have undergone duplication in humans (CCDC63 and CCDC114) (22, 23). Defects in CCDC114 are known to cause PCD and the patients lack outer dynein arms, suggesting that DC2 functions in OAD docking in humans also (22, 23). The ODA-DC binds OAD via at least three interactions: one between DC1 and the OAD intermediate chain IC1, one between DC1 and the other OAD intermediate chain IC2, and one between DC2 and the OAD light chain LC7b (24, 25). However, we still do not understand the architecture of the ODA-DC and the detailed manner of its binding to the doublet microtubules, which will be essential for understanding the molecular basis of OAD periodicity. In the present study, we analyzed the structure and microtubule-binding properties of the ODA-DC by using recombinant proteins as well as native proteins, and observed how it and OAD are incorporated in vivo into axonemes lacking one or both structures. We found that the ODA-DC itself is 24 nm in length and that it produces 24-nm periodicity by end-to-end association on the doublet microtubules. We propose that this periodicity determines the ordered arrangement of the OAD.
Results and Discussion
Properties of Recombinant ODA-DC.
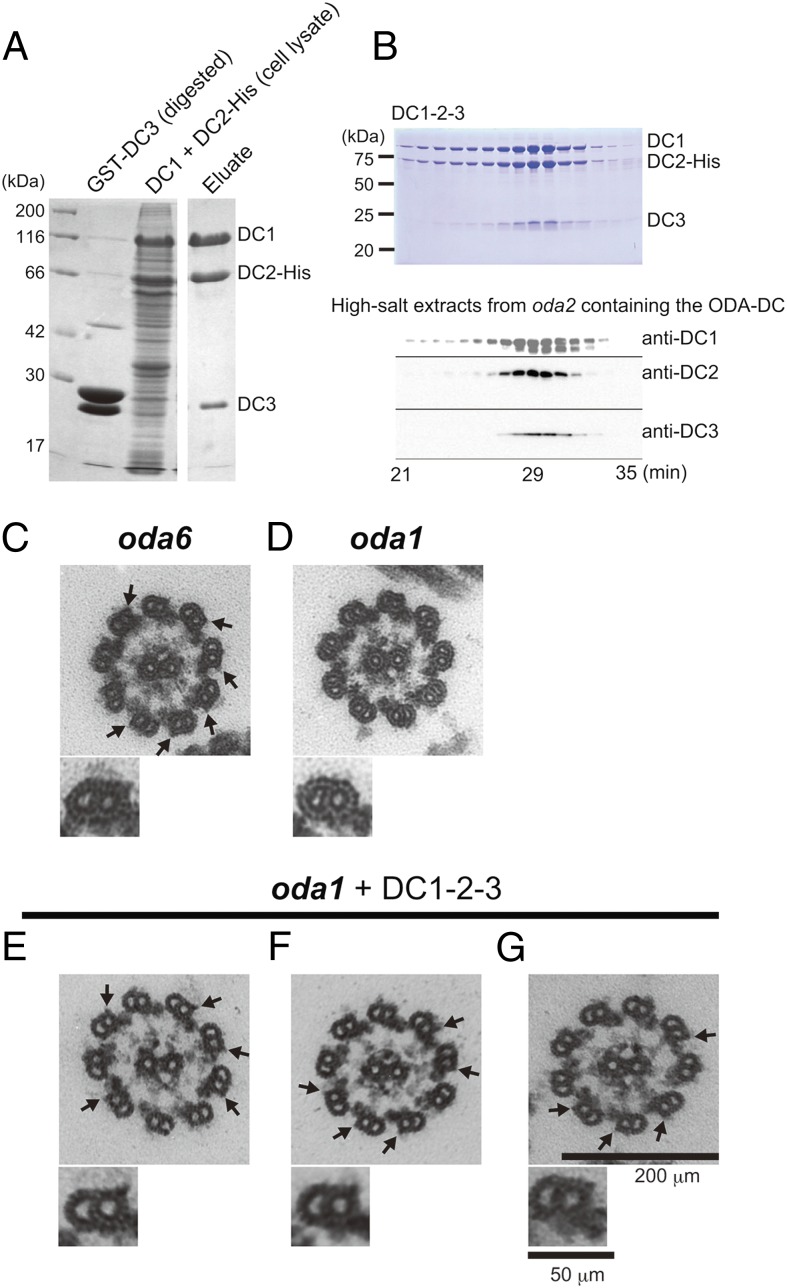
Recombinant ODA-DC (which is hereafter referred to as “DC1–2–3” to distinguish it from the native ODA-DC isolated from the axoneme) was prepared by combining bacterially expressed DC3 (20) with the DC1–DC2–6×His complex produced by coexpression of these proteins in cultured Sf21 insect cells (25), followed by purification on a Ni⋅NTA agarose column. The eluates contained DC1, DC2–6×His, and DC3, verifying that DC1–2–3 forms a single stable complex (Fig. 1A).
Fig. 1.
Expression of functional recombinant ODA-DC (DC1–2–3) in vitro. (A) SDS/PAGE of DC1–2–3 stained with Coomassie brilliant blue. DC1 and DC2–6×His coexpressed in Sf21 insect cells were mixed with bacterially expressed GST–DC3 after digestion with protease to cleave off GST, and then purified using a Ni⋅NTA column. The resultant DC1+DC2–6×His+DC3 complex (termed DC1–2–3) contains equimolar amounts of the three proteins (DC1:DC2:DC3 = 1.0:0.89:0.92). (B) Gel filtration column chromatography fractions of DC1–2–3 (Upper, Coomassie brilliant blue-stained gel) and the native ODA-DC extracted from oda2 axonemes (Lower, Western blot probed with antibodies for each subunit). For each sample, fractions from 21 to 35 min were subjected to SDS/PAGE or Western analysis. The elution patterns of DC1–2–3 and native ODA-DC were almost the same, indicating the structural similarity of the two complexes. (C–G) Thin-section electron microscopy images of axonemes: (C) oda6, lacking OAD but having the ODA-DC; (D) oda1, lacking both OAD and the ODA-DC; and (E–G) oda1 mixed with DC1–2–3. C–G, Lower show representative doublets. The projections (arrows) were observed at the same site on the doublets as the native ODA-DC on the oda6 axoneme (C, arrows).
By several criteria, the structure and functional properties of DC1–2–3 are identical to those of the native ODA-DC extracted from oda2 axonemes. First, the stoichiometry of the three subunits in DC1–2–3 estimated from the band intensities in the SDS/PAGE gel (assuming that all subunits have the same affinity for Coomassie brilliant blue) was 1:1:1, which is the same as that of the native ODA-DC (19). Second, the elution profile of DC1–2–3 from a gel filtration column is similar to that of the native ODA-DC, suggesting the structural similarity of the recombinant and native complexes (Fig. 1B). Third, DC1–2–3 rescued the phenotype of the oda3ida4 mutant (lacking the ODA-DC, OAD, and inner arm dyneins a, c, and d; nonmotile) when introduced by electroporation-mediated protein delivery: A few hours after introduction of proteins, ∼19% (n = 2,735) of cells became motile whereas all control cells (pulse applied without proteins) remained nonmotile (n = 862) (Movies S1 and S2). This motility recovery rate was much higher than when DC1–2 was used (<2%) (25), suggesting that DC3 may stabilize the ODA-DC. Finally, when DC1–2–3 was added to isolated axonemes of the oda1 mutant lacking the ODA-DC and OAD (Fig. 1D), it bound to the correct site on the doublet microtubules as judged by thin-section electron microscopy; the addition resulted in the appearance of projections (Fig. 1 E–G, arrows) identical to those observed in oda6 axonemes that lack ODA but retain the ODA-DC (Fig. 1C, arrows) (17).
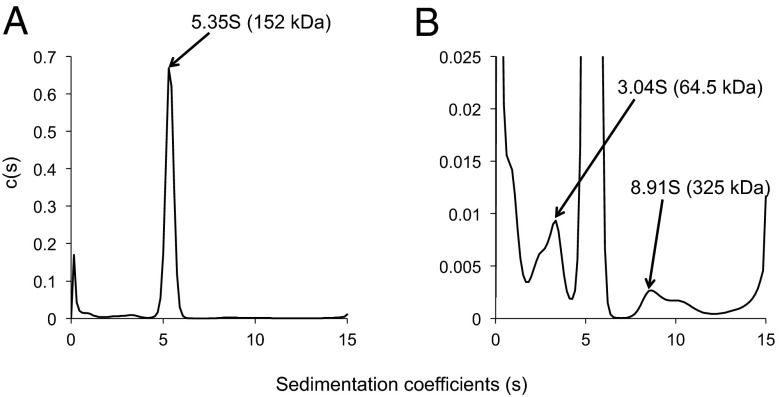
To obtain information about the shape of DC1–2–3, we carried out analytical ultracentrifugation and electron microscopy. Calculation of the molecular mass from the sedimentation velocity resulted in an estimate of ∼152 kDa (Fig. 2A and Fig. S1). This value is close to the sum of the predicted molecular masses of DC1, DC2, or DC3 (∼167 kDa), providing further evidence that DC1–2–3 is a heterotrimer. Low-angle rotary shadowing electron microscopy after fixation with 0.1% glutaraldehyde revealed that DC1–2–3 takes on an ellipsoidal shape (Fig. 3 A and B). The major axis length of these particles was normally distributed with a mean of ∼28 nm (Fig. 3C). Assuming that the thickness of the platinum coating was 2 nm on all sides, the actual mean length of DC1–2–3 is estimated to be ∼24 nm.
Fig. 2.
ODA-DC is a heterotrimer. (A) Sedimentation coefficient distribution function c(s) was obtained from sedimentation velocity data by SEDFIT (36). c(s) was converted to c(M) to estimate the molecular mass of DC1–2–3 to be ∼152 kDa. (B) There were two minor peaks in A; 2.74% of the solutes had an S value of 3.04S (64.5 kDa), which is close to the molecular mass of DC2 (62,204 Da). This peak most likely reflects DC2–6×His that was purified by Ni⋅NTA but did not form a complex with DC1 or DC3; 0.43% of the solutes had an S-value of 8.91S (325 kDa), which is close to twice the sum of the molecular masses of DC1, DC2, and DC3 (333,850 Da). This peak suggests that a small fraction of the DC1–2–3 complexes formed dimers.
Fig. 3.
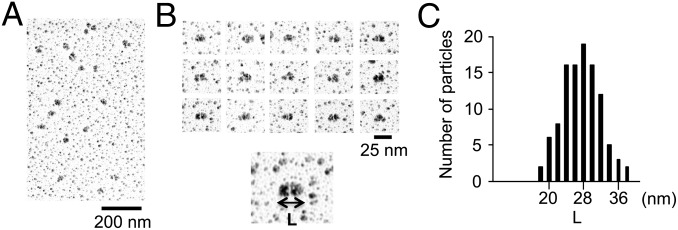
Structural analysis of DC1–2–3. (A) Low-magnification electron microscopy images of DC1–2–3 prepared by low-angle rotary shadowing. (B) Typical images of DC1–2–3. Each particle has an ellipsoidal shape. (C) Histogram of the major axis length of DC1–2–3 (L) (n = 108). Considering the thickness of platinum coating (2 nm on all sides), the peak length is ∼24 nm.
End-to-End Association of ODA-DC on Outer Doublets.
To examine subunit–subunit interactions either within the ODA-DC or between ODA-DCs, we used chemical cross-linking with 1,6-Bismaleimidohexane (BMH), a sulfhydryl–sulfhydryl cross-linker, under various conditions.
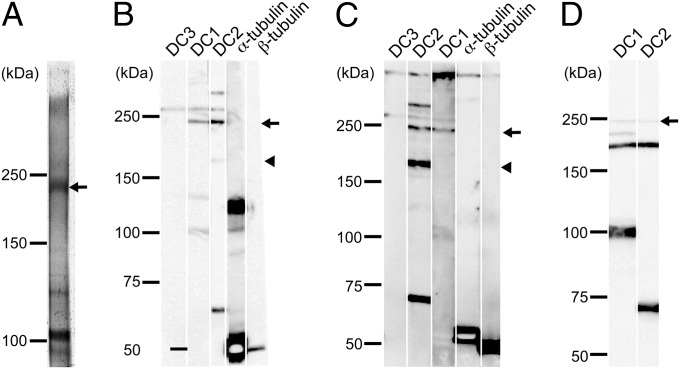
First, we treated isolated oda6 axonemes with BMH; the cross-linked products were then solubilized and immunoprecipitated with anti-DC1 antibody. The SDS/PAGE pattern of the immunoprecipitates showed three or four major bands above the band of DC1. Mass spectrometry revealed that the band at ∼230 kDa contained peptides from only DC1 and DC2 (Fig. 4A, arrow; and Fig. S2). Although the predicted molecular masses of DC1 and DC2 are 83 and 62 kDa, they migrate in SDS/PAGE with apparent masses of ∼105 and 70 kDa, respectively. Therefore, this product is most likely composed of one DC1 and two DC2 molecules (the combined molecular masses would be 207 kDa with an apparent mass in SDS/PAGE of ∼245 kDa). In support of this possibility, BMH reacts with Cys residues, and there are two Cys in DC1 and one Cys in DC2 (Fig. S2A); hence, DC1 can be cross-linked to two adjoining DC2s, and those DC2s then cannot be cross-linked to another protein. Western analysis of the high-salt extract from the cross-linked axonemes confirmed that the ∼230-kDa product contained both DC1 and DC2 but not α- or β-tubulin (Fig. 4B, arrow).
Fig. 4.
ODA-DCs associate with each other in situ. (A) Silver-stained gel of anti-DC1 immunoprecipitate of products from cross-linked oda6 axonemes. Isolated axonemes of oda6 were cross-linked with BMH, solubilized with SDS, and the solute subjected to immunoprecipitation using the anti-DC1 antibody. Because mass spectrometry revealed that the ∼230-kDa product contained only DC1 and DC2 peptides (Fig. S2), the product is likely to be composed of one DC1 and two DC2s. (B) oda6 axonemes or (C) cytoplasmic microtubules mixed with DC1–2–3 were chemically cross-linked with BMH and the products were analyzed by Western blotting using anti-DC1, -DC2, -DC3, –α-tubulin, and –β-tubulin antibodies. An ∼230-kDa product (arrow) was detected by anti-DC1 and -DC2 antibodies in both samples. Note that an ∼260-kDa product (presumably consisting of one DC1, two DC2s and one DC3) was also detected in both samples. (D) The DC1–2 complex in solution was chemically cross-linked with 1 μM BMH and the results analyzed in a Western blot probed with anti-DC1 and -DC2 antibodies. Two products (∼240 kDa and ∼190 kDa) were detected by both antibodies. Based on the apparent molecular weights of DC1 and DC2, it seems likely that the former is composed of one DC1 and two DC2s, and the latter is composed of one DC1 and one DC2.
Second, a mixture of DC1–2–3 and porcine brain microtubules was cross-linked with BMH. In Western analysis of the high-salt extract of the specimen, a band pattern similar to that in Fig. 4B was observed (Fig. 4C, arrow). The resultant ∼240-kDa product reacted with anti-DC1 and -DC2 antibodies but not with anti-tubulin antibodies. Quantitative mass spectrometry of this product revealed that it was mostly composed of DC1 and DC2 and that the ratio of DC1 to DC2 was 0.6. Small amounts of DC3 (∼4%) and β-tubulin (<1%) were also present (Table S1). From these results, it seems most likely that this ∼240-kDa product contains one DC1 and two DC2s.
A band at ∼180-kDa was detected only by anti-DC2 antibody (Fig. 4C, arrowhead). Mass spectrometry showed that the product was composed mostly of DC2 (with small amounts of DC1 (∼1%), DC3 (<1%), and β-tubulin (<0.1%) (Table S2). These results suggest that the ∼180-kDa band contains at least two DC2s. A similar ∼180-kDa band was also detected by the anti-DC2 antibody in cross-linked axonemes (Fig. 4B, arrowhead), although its amount relative to other products was less than that in Fig. 4C. The difference may be due to the fact that binding of the ODA-DC is constrained to a specific site on the doublet microtubules, whereas it can bind to multiple sites around cytoplasmic microtubules.
Intriguingly, upon cross-linking the DC1–2 complex with BMH in the absence of microtubules, a product containing one DC1 and two DC2s was also detected (Fig. 4D, arrow). This result indicates that a small fraction of the DC1–2 complex dimerizes even in solution. [The slight differences in apparent masses of the 1:2 complex of DC1–2 in Fig. 4 A–D might be caused by different gel sizes (SI Materials and Methods)]. Consistent with this idea, in the sedimentation distribution of DC1–2–3, there was a minor peak of ∼325 kDa, suggesting that a small fraction of DC1–2–3 forms dimers in solution (Fig. 2B).
Because the sedimentation analysis (Fig. 2) showed that the ODA-DC is a heterotrimer (i.e., containing only one molecule each of DC1, DC2, and DC3), the appearance of cross-linked products containing two DC2s indicates that ODA-DCs associate with each other on the doublet. Furthermore, these results also suggest that self-association of the ODA-DCs does not require the presence of axonemal components other than microtubules.
Quantitative Binding Assay.
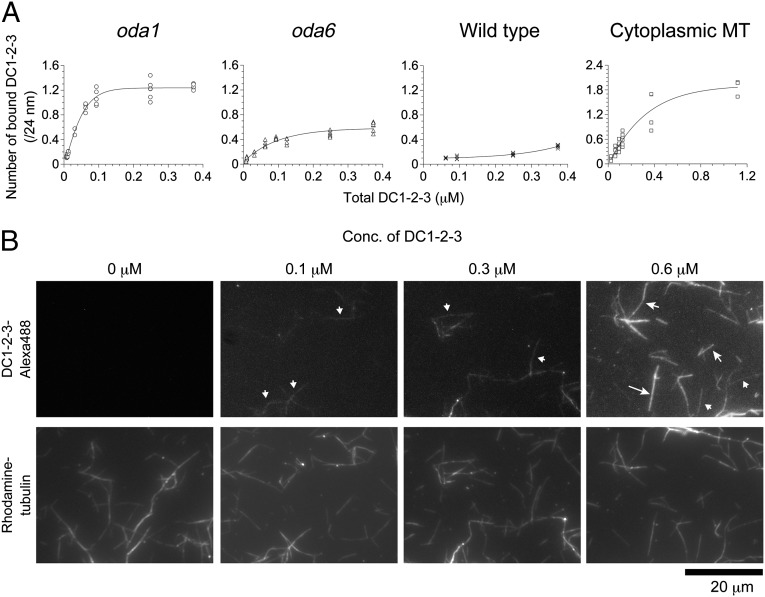
The apparent self-association of the ODA-DCs suggests potential cooperative binding to microtubules. To explore this possibility, we quantified ODA-DC–microtubule binding by assaying cosedimentation of DC1–2–3 with either porcine brain cytoplasmic microtubules or three kinds of Chlamydomonas axonemes: axonemes from wild type (in which the binding sites for the ODA-DC are fully occupied), from oda6 (∼70% occupied; Fig. S3), and from oda1 (all sites empty). The amount (B) of DC1–2–3 bound to microtubules and axonemes shows a dependence on the DC1–2–3 concentration that can be approximated by the Hill equation B = Bmax × Fn/(Kd + Fn), where F is the concentration of free DC1–2–3, Bmax is the saturated amount of bound DC1–2–3, n is the Hill coefficient (a measure of cooperativity), and Kd is the apparent dissociation constant (Fig. 5A). The saturated amounts of bound DC1–2–3 were in the order of cytoplasmic microtubules > oda1 axonemes > oda6 axonemes > wild-type axonemes, reflecting the availability of “empty” sites for the ODA-DC. From Scatchard plots of the data, the number of ODA-DCs that bind to a 24-nm segment of microtubule were estimated to be ∼0.46 for wild type, ∼0.71 for oda6, ∼1.3 for oda1, and ∼2.0 for cytoplasmic microtubules (Table 1 and Fig. S4 A–D). We may consider the binding to wild-type axonemes as nonspecific and subtract this value from all data. With this correction the amount of ODA-DC bound to the oda1 axoneme would be close to 0.9, therefore ODA-DC binding to oda1 axonemes saturates at the wild-type level (in Chlamydomonas, one of the nine doublets has no ODA-DC and OAD, hence the number of native ODA-DC molecules on wild-type axonemes is ∼0.9 per 24 nm) (26). The results are consistent with the electron microscopy observations that the ODA-DC preferentially binds to a specific site on the doublet microtubules. This site presumably is specified by some additional structure(s) on the microtubules.
Fig. 5.
Cooperative binding of DC1–2–3 to microtubules. (A) Binding between DC1–2–3 and axonemes or cytoplasmic microtubules was analyzed by cosedimentation assay. Purified DC1–2–3 was mixed with axonemes of oda1, oda6, and wild-type cells, and with porcine brain cytoplasmic microtubules. The amount of bound DC1–2–3 was calculated as the number of molecules bound to microtubules per 24 nm (y axis), and plotted against total DC1–2–3 (x axis). The saturated amount of bound DC1–2–3 was in the order of cytoplasmic microtubules > oda1 > oda6 > wild type, reflecting the available sites for ODA-DC binding. (B) Different concentrations of Alexa-488–labeled DC1–2–3 were mixed with rhodamine-labeled microtubules and observed by fluorescence microscopy. At 0.1 µM DC1–2–3, some microtubules were labeled whereas others were not at all. At 0.3 µM, more microtubules were labeled but some remained unlabeled. At 0.6 µM, almost all microtubules were labeled. (B, Upper) The lengths of the arrows represent the fluorescence intensity, and microtubules marked with the same size of arrows have similar intensities. Shortest arrows are ∼30 a.u., mid-size arrows ∼60 to ∼70 a.u., and the longest arrow ∼87 a.u.
Table 1.
Parameters of DC1–2–3 binding to microtubules
| Microtubules used for cosedimentation | Kd, M | Bmax, μM (no. of molecules per 24 nm) |
| oda1 axonemes | 7.8 × 10−9 | 0.069 (1.3) |
| oda6 axonemes | 6.1 × 10−8 | 0.037 (0.71) |
| Wild-type axonemes | 2.5 × 10−7 | 0.024 (0.46) |
| Cytoplasmic microtubules | 1.6 × 10−7 | 0.26 (2.0) |
A previous study showed by electron microscopy of microtubule cross-sections that up to about four rows of dynein arms become attached to a cytoplasmic microtubule upon addition of high-salt extracts from axonemes (containing OADs and ODA-DCs) (27). In contrast, in our experiments, the saturating amount of DC1–2–3 bound to microtubules was about two per 24 nm. Although the reason for this apparent discrepancy is not understood, it may be that the saturating level of DC1–2–3 was underestimated in our experiments because it was technically difficult to measure microtubule binding with high concentrations of DC1–2–3. Moreover, a mixture of OADs and ODA-DCs is likely to have a much higher microtubule-binding affinity than ODA-DCs alone, because the OAD intermediate chain IC1 binds directly to tubulin (28–30); in such a mixture microtubule binding would saturate at lower concentrations.
The dissociation constant between DC1–2–3 and oda1 axonemes was calculated to be 7.8 × 10−9, which is nearly 20 times lower than that between DC1–2–3 and cytoplasmic microtubules, 1.6 × 10−7 (Table 1 and Fig. S4 A–D). Therefore, the microtubule–ODA-DC affinity must be strengthened by yet unidentified factors on the doublet. The Hill coefficient of the binding curve was 2.8 for the oda1 axonemes and 1.4 for cytoplasmic microtubules, indicating that the binding is positively cooperative (Fig. S4 E and F).
In Vitro Cooperative Binding to Microtubules Observed by Microscopy.
Cooperative binding between DC1–2–3 and microtubules could be directly observed by light microscopy. In the experiment shown in Fig. 5B, rhodamine-labeled cytoplasmic microtubules were mixed with different concentrations of Alexa-488–labeled DC1–2–3 and observed by fluorescence microscopy. At each concentration of labeled DC1–2–3, two classes of microtubules appeared: Microtubules in one class were fully decorated with DC1–2–3 along their lengths and those in the other class were totally devoid of decoration. Thus, the binding occurred in an all-or-none manner. The number of decorated microtubules varied with the DC1–2–3 concentration; ∼34% microtubules were decorated at 0.1 μM DC1–2–3, whereas ∼82% at 0.6 μM DC1–2–3. This concentration dependence is consistent with the binding curve (Fig. 5A) and confirms that binding occurred with high cooperativity.
ODA-DC Binding to Axonemes in Vivo.
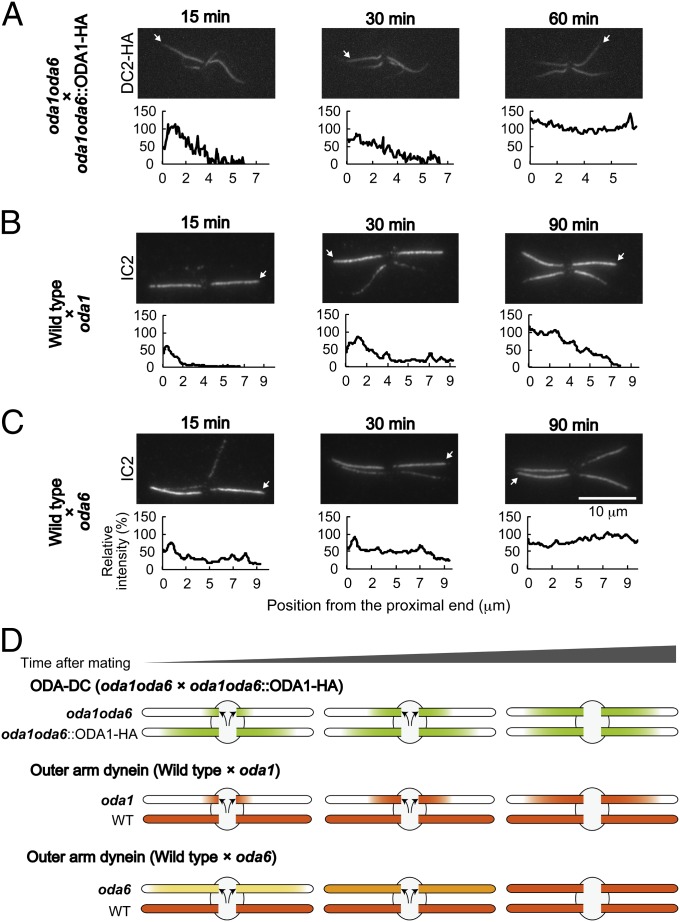
The process by which the native ODA-DC binds in vivo was observed by the immunofluorescence microscopy of Chlamydomonas zygotes. We first mated oda1oda6 with a transformant of oda1oda6 expressing HA-tagged DC2 to examine the manner of binding of DC2-HA to the oda1oda6 axonemes that initially lacked the ODA-DC. We found that the signal of DC2-HA first appeared in the proximal part and then proceeded to the distal part of the oda1oda6 flagella (Fig. 6A).
Fig. 6.
The ordered binding of the OAD to the axoneme is dependent on the ODA-DC. (A–C, Upper) Immunofluorescence staining of temporary dikaryons: (A) oda1oda6×oda1oda6::ODA1–HA expressing DC2–3×HA, (B) wild type×oda1, and (C) wild type×oda6. The time after mating is noted above each panel. Dikaryons were demembranated and labeled with (A) anti-HA or (B and C) anti-IC2 antibody. (A–C, Lower) The fluorescence intensity of DC2-HA or IC2 in axonemes originally lacking those proteins was normalized respectively with that of oda1oda6::ODA1–HA (A, arrows) or wild-type (B and C, arrows) axonemes in the same dikaryon. (A) Binding of the HA-tagged ODA-DC proceeded from the proximal part of the oda1oda6 axoneme to the distal part. (B) Similarly, binding of OAD proceeded from proximal to distal in the oda1 axoneme. (C) In contrast, binding of OAD gradually increased over the whole length of the oda6 axoneme. The difference is that the ODA-DC was already present on the doublets of the oda6 axonemes but not on the doublets of the oda1 axonemes; hence, ODA assembly was constrained to follow the pattern of ODA-DC assembly in the oda1 axonemes. (D) Schematic diagrams illustrating patterns of labeling for the ODA-DC (green) and the OAD (shades of orange) in the experiments of A–C.
The unidirectional extension of the bound portion indicates that newly transported ODA-DCs add next to preexisting ODA-DCs on the axoneme, supporting the hypothesis that binding of the ODA-DC to the axoneme is cooperative. In wild-type × oda1 zygotes, the progression of OAD binding to the axoneme, as monitored by the signal of the OAD intermediate chain IC2, was similar to that of DC2–HA (Fig. 6B). Because the ODA-DC and OAD are transported separately within the flagellum (16), these results suggest that OAD can attach to the axoneme only where the ODA-DC is already bound. In contrast, in zygotes of wild-type × oda6, the IC2 signal rapidly reached the flagellar tip and, with time, its intensity increased evenly along the length of the flagellum (Fig. 6C). A similar pattern of OAD recovery has previously been observed in wild-type × oda2 (31). These data suggest that when OAD is transported into flagella that already have ODA-DC bound to the axoneme, OAD binds to the axoneme without any preferred position along the length. In other words, OAD binding on axonemes containing the ODA-DC does not proceed in a single direction; this is in strong contrast to the binding of OAD on mutant axonemes lacking the ODA-DC, which always proceeds from base to tip.
A Model for Construction of OAD Periodicity.
Haimo et al. demonstrated that when a high-salt extract from Chlamydomonas wild-type axonemes is mixed with cytoplasmic microtubules, OAD orderly attaches to the microtubules with a periodicity of 24 nm (28). The 24-nm periodicity and the ordered arrangement were thought to be produced by interaction of adjacent OADs, which are ∼24 nm long. However, it must be noted that a high-salt extract from the wild-type axonemes contains both OAD and ODA-DCs. Our present results suggest that the ODA-DC provides the basis for cooperative binding on the outer doublet. It is likely that the ODA-DC also was responsible for the ordered arrangement of OAD on the cytoplasmic microtubules.
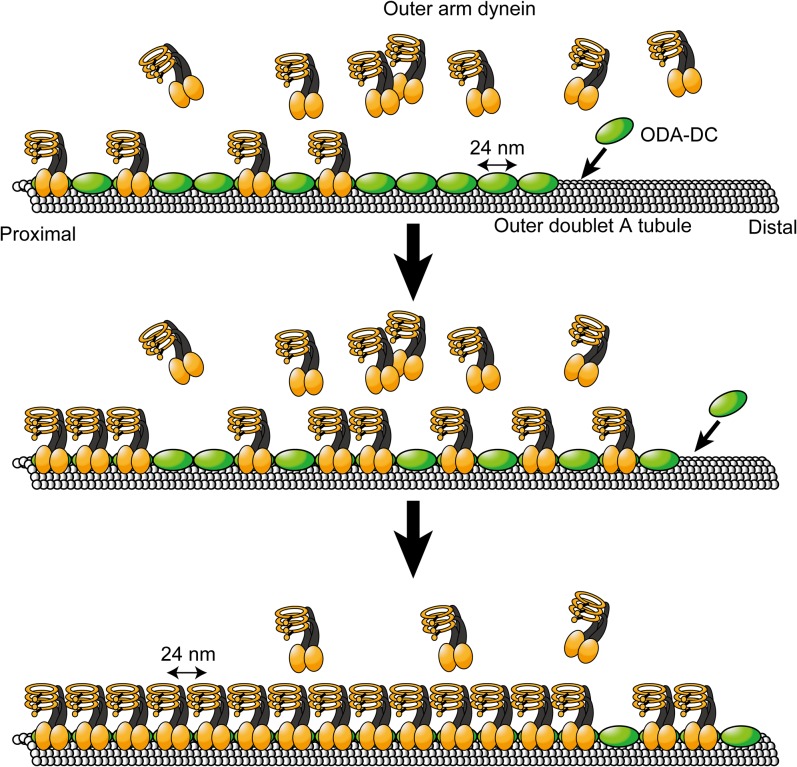
The data obtained in our present and previous studies lead to a model that explains the periodic arrangement of OAD on the outer doublet (Fig. 7). First, OAD and the ODA-DC are separately assembled in the cytoplasm and separately transported to the flagella (12, 16). After entry into the flagella, the ODA-DC, 24 nm in length, starts binding to the appropriate site at the proximal end of the doublet microtubule. The ODA-DC molecules cooperatively bind to the doublet in an end-to-end manner and establish a 24-nm periodicity. Subsequently, OAD binds to the outer doublet through its association with the ODA-DCs on the doublet, and this interaction is stabilized through the direct interaction of IC1 with the microtubule, thereby establishing the 24-nm periodicity in the OAD arrangement (24, 25, 29, 30).
Fig. 7.
A model for the ordered assembly of OADs based on the cooperative binding of the ODA-DC. The ODA-DC cooperatively binds to the outer doublet from the proximal part of the axoneme to the distal part. Subsequently OAD is arranged on the “rail” of the ODA-DC. The 24-nm periodicity of OAD is based on the size of the ODA-DC.
Our study suggests that the ODA-DC functions not only as the docking site for OAD, but that its self-association during its assembly onto the axoneme forms a scaffold of 24-nm periodicity that ensures the ordered assembly of the OAD along the doublet microtubule. If the ODA-DC bound to doublet microtubules independently of other bound ODA-DCs, then the 24-nm ODA-DC would be able to bind to any three longitudinally aligned tubulin dimers, which together span 8 × 3 nm. In such a case, there would be a high probability of producing a gap of one or two tubulin dimers between adjacent ODA-DCs and hence between adjacent OADs. Ultimately, such gaps would perturb the regular arrangement of OAD in the 96-nm axonemal repeat and disrupt interactions between OAD and inner arm dyneins that are necessary for the coordination of force generation by the different dyneins (2, 32). The cooperativity in the ODA-DC binding would prevent such gap formation.
This study has yielded insight into the molecular mechanism that establishes the long-range periodicity of outer dynein arms along the doublet microtubules of the flagellar axoneme. A similar mechanism may underlie the periodic binding of other axonemal structures, such as the radial spokes and the projections of the central pair of microtubules.
Materials and Methods
Strains and Culture of Chlamydomonas Cells.
A C. reinhardtii wild-type strain (CC124) and the following mutants lacking outer dynein arms (17) were used: oda1, with a mutation in the structural gene of DC2, lacking the ODA-DC (19); oda2, with a mutation in the structural gene of the OAD γ-HC, lacking OAD; oda6, with a mutation in the structural gene of the ODA intermediate chain IC2, lacking OAD (33). Double mutants were produced by standard procedures (34). All cells were grown in Tris⋅acetate⋅phosphate medium (35) with aeration at 25 °C, on a 12:12 h light:dark cycle.
Generation and Purification of Recombinant DC1–2–3 and DC1–2.
Bacterially expressed GST–DC3 (20) was purified with glutathione Sepharose 4B (17-0756-01; GE Healthcare,) and treated with PreScission Protease (27-0843-01; GE Healthcare) to cleave off GST. A mixture of GST–DC3 and protease was mixed with insect culture cell lysate containing DC1 and DC2–6×His produced by the baculovirus system (25), followed by purification on a Ni-NTA agarose column. (For DC1–2 purification, GST–DC3 was not added.)
See SI Materials and Methods for further information.
Supplementary Material
Acknowledgments
We thank Dr. Masafumi Hirono (The University of Tokyo) for helping to prepare the dikaryons, Dr. Haru-aki Yanagisawa (The University of Tokyo) for assistance in generating the transformant-expressing HA-tagged DC2, Drs. Masahito Hayashi (Nagoya University) and Takahiro Ide (Tokyo Institute of Technology) for assistance in the protein introduction by electroporation, Dr. John Leszyk [University of Massachusetts Medical School (UMMS)] for mass spectrometry analysis, and Dr. Yuqing Hou (UMMS) for helpful discussion. M.O. is a recipient of a fellowship from the Japan Society for Promotion of Sciences for Young Scientists. S.M.K. is supported by National Institutes of Health (NIH) Grant GM051293. G.B.W. is supported by NIH Grant GM030626 and by the Robert W. Booth Endowment at UMMS. K.W. is supported by Japan Society for Promotion of Sciences Grants 25113507, 25117506, 25291058, and 26650093; and a grant from the Yamada Science Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.L.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403101111/-/DCSupplemental.
References
- 1.Bloodgood RA. Sensory reception is an attribute of both primary cilia and motile cilia. J Cell Sci. 2010;123(Pt 4):505–509. doi: 10.1242/jcs.066308. [DOI] [PubMed] [Google Scholar]
- 2.Nicastro D, et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313(5789):944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 3.Ishikawa T, Sakakibara H, Oiwa K. The architecture of outer dynein arms in situ. J Mol Biol. 2007;368(5):1249–1258. doi: 10.1016/j.jmb.2007.02.072. [DOI] [PubMed] [Google Scholar]
- 4.Lupetti P, Mencarelli C, Rosetto M, Heuser JE, Dallai R. Structural and molecular characterization of dynein in a gall-midge insect having motile sperm with only the outer arm. Cell Motil Cytoskeleton. 1998;39(4):303–317. doi: 10.1002/(SICI)1097-0169(1998)39:4<303::AID-CM5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 5.Mencarelli C, et al. Molecular structure of dynein and motility of a giant sperm axoneme provided with only the outer dynein arm. Cell Motil Cytoskeleton. 2001;50(3):129–146. doi: 10.1002/cm.10004. [DOI] [PubMed] [Google Scholar]
- 6.King SM. Composition and assembly of axonemal dyneins. In: King SM, editor. Dyneins: Structure, Biology and Disease. London: Elsevier; 2011. pp. 209–243. [Google Scholar]
- 7.Minoura I, Kamiya R. Strikingly different propulsive forces generated by different dynein-deficient mutants in viscous media. Cell Motil Cytoskeleton. 1995;31(2):130–139. doi: 10.1002/cm.970310205. [DOI] [PubMed] [Google Scholar]
- 8.Brokaw CJ. Control of flagellar bending: A new agenda based on dynein diversity. Cell Motil Cytoskeleton. 1994;28(3):199–204. doi: 10.1002/cm.970280303. [DOI] [PubMed] [Google Scholar]
- 9.Afzelius BA. Cilia-related diseases. J Pathol. 2004;204(4):470–477. doi: 10.1002/path.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zariwala MA, Knowles MR, Omran H. Genetic defects in ciliary structure and function. Annu Rev Physiol. 2007;69:423–450. doi: 10.1146/annurev.physiol.69.040705.141301. [DOI] [PubMed] [Google Scholar]
- 11.Fliegauf M, Benzing T, Omran H. When cilia go bad: Cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8(11):880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 12.Fowkes ME, Mitchell DR. The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol Biol Cell. 1998;9(9):2337–2347. doi: 10.1091/mbc.9.9.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Omran H, et al. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature. 2008;456(7222):611–616. doi: 10.1038/nature07471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takada S, Kamiya R. Functional reconstitution of Chlamydomonas outer dynein arms from alpha-beta and gamma subunits: Requirement of a third factor. J Cell Biol. 1994;126(3):737–745. doi: 10.1083/jcb.126.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takada S, Kamiya R. Beat frequency difference between the two flagella of Chlamydomonas depends on the attachment site of outer dynein arms on the outer-doublet microtubules. Cell Motil Cytoskeleton. 1997;36(1):68–75. doi: 10.1002/(SICI)1097-0169(1997)36:1<68::AID-CM6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 16.Wakabayashi K, Takada S, Witman GB, Kamiya R. Transport and arrangement of the outer-dynein-arm docking complex in the flagella of Chlamydomonas mutants that lack outer dynein arms. Cell Motil Cytoskeleton. 2001;48(4):277–286. doi: 10.1002/cm.1015. [DOI] [PubMed] [Google Scholar]
- 17.Kamiya R. Mutations at twelve independent loci result in absence of outer dynein arms in Chylamydomonas reinhardtii. J Cell Biol. 1988;107(6 Pt 1):2253–2258. doi: 10.1083/jcb.107.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koutoulis A, et al. The Chlamydomonas reinhardtii ODA3 gene encodes a protein of the outer dynein arm docking complex. J Cell Biol. 1997;137(5):1069–1080. doi: 10.1083/jcb.137.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takada S, Wilkerson CG, Wakabayashi K, Kamiya R, Witman GB. The outer dynein arm-docking complex: Composition and characterization of a subunit (oda1) necessary for outer arm assembly. Mol Biol Cell. 2002;13(3):1015–1029. doi: 10.1091/mbc.01-04-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casey DM, et al. DC3, the 21-kDa subunit of the outer dynein arm-docking complex (ODA-DC), is a novel EF-hand protein important for assembly of both the outer arm and the ODA-DC. Mol Biol Cell. 2003;14(9):3650–3663. doi: 10.1091/mbc.E03-01-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casey DM, Yagi T, Kamiya R, Witman GB. DC3, the smallest subunit of the Chlamydomonas flagellar outer dynein arm-docking complex, is a redox-sensitive calcium-binding protein. J Biol Chem. 2003;278(43):42652–42659. doi: 10.1074/jbc.M303064200. [DOI] [PubMed] [Google Scholar]
- 22.Knowles MR, Daniels LA, Davis SD, Zariwala MA, Leigh MW. Primary ciliary dyskinesia. Recent advances in diagnostics, genetics, and characterization of clinical disease. Am J Respir Crit Care Med. 2013;188(8):913–922. doi: 10.1164/rccm.201301-0059CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onoufriadis A, et al. UK10K Splice-site mutations in the axonemal outer dynein arm docking complex gene CCDC114 cause primary ciliary dyskinesia. Am J Hum Genet. 2013;92(1):88–98. doi: 10.1016/j.ajhg.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DiBella LM, Sakato M, Patel-King RS, Pazour GJ, King SM. The LC7 light chains of Chlamydomonas flagellar dyneins interact with components required for both motor assembly and regulation. Mol Biol Cell. 2004;15(10):4633–4646. doi: 10.1091/mbc.E04-06-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ide T, Owa M, King SM, Kamiya R, Wakabayashi K. Protein-protein interactions between intermediate chains and the docking complex of Chlamydomonas flagellar outer arm dynein. FEBS Lett. 2013;587(14):2143–2149. doi: 10.1016/j.febslet.2013.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoops HJ, Witman GB. Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J Cell Biol. 1983;97(3):902–908. doi: 10.1083/jcb.97.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haimo LT, Fenton RD. Microtubule crossbridging by chlamydomonas dynein. Cell Motil. 1984;4(5):371–385. doi: 10.1002/cm.970040506. [DOI] [PubMed] [Google Scholar]
- 28.Haimo LT, Telzer BR, Rosenbaum JL. Dynein binds to and crossbridges cytoplasmic microtubules. Proc Natl Acad Sci USA. 1979;76(11):5759–5763. doi: 10.1073/pnas.76.11.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King SM, Patel-King RS, Wilkerson CG, Witman GB. The 78,000-M(r) intermediate chain of Chlamydomonas outer arm dynein is a microtubule-binding protein. J Cell Biol. 1995;131(2):399–409. doi: 10.1083/jcb.131.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King SM, Wilkerson CG, Witman GB. The Mr 78,000 intermediate chain of Chlamydomonas outer arm dynein interacts with alpha-tubulin in situ. J Biol Chem. 1991;266(13):8401–8407. [PubMed] [Google Scholar]
- 31.Piperno G, Mead K, Henderson S. Inner dynein arms but not outer dynein arms require the activity of kinesin homologue protein KHP1(FLA10) to reach the distal part of flagella in Chlamydomonas. J Cell Biol. 1996;133(2):371–379. doi: 10.1083/jcb.133.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oda T, Yagi T, Yanagisawa H, Kikkawa M. Identification of the outer-inner dynein linker as a hub controller for axonemal dynein activities. Curr Biol. 2013;23(8):656–664. doi: 10.1016/j.cub.2013.03.028. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell DR, Kang Y. Identification of oda6 as a Chlamydomonas dynein mutant by rescue with the wild-type gene. J Cell Biol. 1991;113(4):835–842. doi: 10.1083/jcb.113.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris EH. The Chlamydomonas Sourcebook. San Diego: Academic; 1989. [Google Scholar]
- 35.Gorman DS, Levine RP. Cytochrome f and plastocyanin: Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci USA. 1965;54(6):1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78(3):1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]