Significance
It has long been assumed that climate played a major role in the population history of the Central Andes. Although adaptations of the Andean populations to climatic changes such as the intensification of agriculture have been inferred from the archaeological record, evidence for demographic adaptations such as migration is missing so far. In this paper, ancient DNA data from populations that lived in southern Peru between 840 BC and 1450 AD provide evidence for two large-scale migrations in the Central Andes coincident with episodes of drought and increased climatic variability. These migrations led to a successive genetic homogenization of southern Peruvian populations generally attributed to intrusions by the late pre-Columbian highland empires such as the Wari, Tiwanaku, or Inca.
Keywords: ancient DNA, South America, population history
Abstract
Several archaeological studies in the Central Andes have pointed at the temporal coincidence of climatic fluctuations (both long- and short-term) and episodes of cultural transition and changes of socioeconomic structures throughout the pre-Columbian period. Although most scholars explain the connection between environmental and cultural changes by the impact of climatic alterations on the capacities of the ecosystems inhabited by pre-Columbian cultures, direct evidence for assumed demographic consequences is missing so far. In this study, we address directly the impact of climatic changes on the spatial population dynamics of the Central Andes. We use a large dataset of pre-Columbian mitochondrial DNA sequences from the northern Rio Grande de Nasca drainage (RGND) in southern Peru, dating from ∼840 BC to 1450 AD. Alternative demographic scenarios are tested using Bayesian serial coalescent simulations in an approximate Bayesian computational framework. Our results indicate migrations from the lower coastal valleys of southern Peru into the Andean highlands coincident with increasing climate variability at the end of the Nasca culture at ∼640 AD. We also find support for a back-migration from the highlands to the coast coincident with droughts in the southeastern Andean highlands and improvement of climatic conditions on the coast after the decline of the Wari and Tiwanaku empires (∼1200 AD), leading to a genetic homogenization in the RGND and probably southern Peru as a whole.
The role of transient or long-term climatic changes and resulting environmental impacts on human dispersals and demographic development is generally accepted (1, 2). Nevertheless, the exact mechanisms underlying such changes and the extent of their influence on local population histories of prehistoric humans remain poorly understood. Pre-Columbian Andean cultures can offer exceptional insights into such human–environment interactions. In fact, western South America underwent several climate changes following the Pleistocene–Holocene transition since the initial peopling ∼14,000 y ago (3, 4). The frequency of climatic changes seems to have been accelerated with the establishment of modern atmospheric conditions and increasing variability of the El Niño Southern Oscillation around the end of the Mid-Holocene (4, 5), resulting in multiple transient and protracted environmental impacts on the Central Andean region (6, 7). Several studies have pointed at the temporal coincidence of occasional environmental and catastrophic events (7, 8) and long-term climatic alterations (5, 9) with episodes of cultural transition and changes of socioeconomic structure throughout the pre-Columbian period, suggesting that climate has played a major role in the formation of the cultural and demographic complexity of the Central Andean area (5, 7, 8, 10). Adaptive responses of the Andean populations to environmental impacts (e.g., droughts) such as the intensification of irrigation and extensive terracing have been directly inferred from the archaeological record (7, 8, 10–12). However, demographic changes including migration in response to climatic alterations have so far only been inferred indirectly (7, 9, 12, 13).
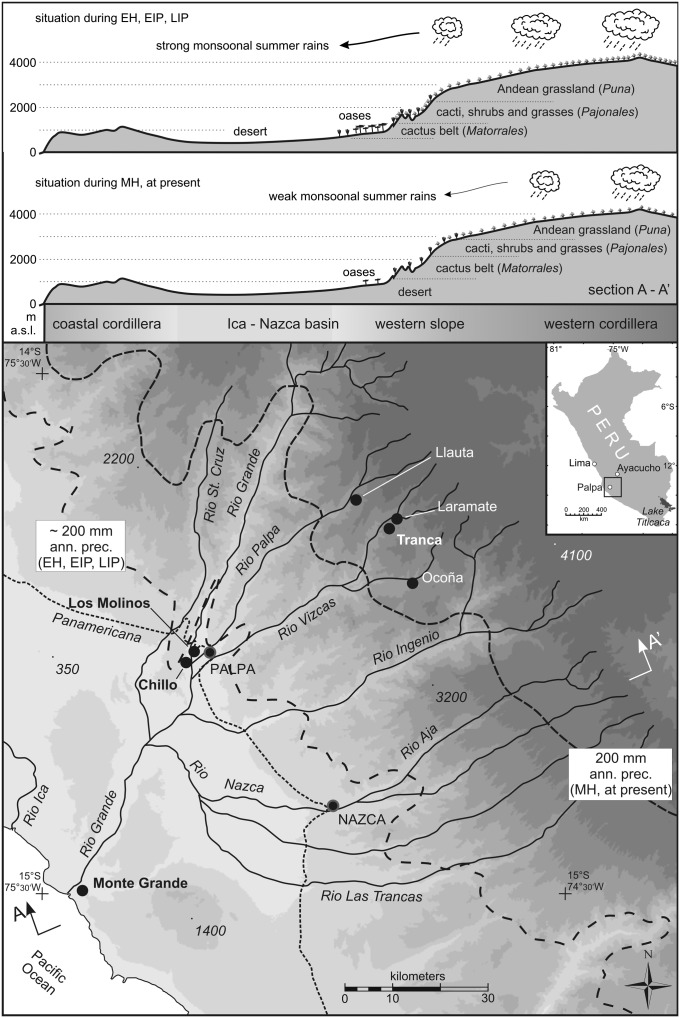
The heartland of most pre-Columbian coastal cultures, especially in southern Peru, was located in the river oases of the lower valleys of the western Andean foothills. These settlement areas can be described as low-resilience, fragile ecosystems (5). Situated on the boundary between the hyperarid coastal desert and the high-altitude Andes, slight hydrological oscillations can change the availability of water, thereby triggering the fertility of the landscape. Interestingly, these ecosystems were inhabited very early in Andean prehistory (14, 15). Today there is nearly no direct precipitation in the western Andean foothills of southern Peru. The main sources of water are the rivers fed by seasonal rainfall in the headwaters of the high-altitude Andes. Recent investigations in the area around the modern city of Palpa, situated in the northern Rio Grande de Nasca drainage (RGND) area in southern coastal Peru (Fig. 1), have shown that the climatic conditions in this settlement area oscillated between humid and dry conditions in the Late Holocene (5). The RGND region received a reliable level of precipitation from the Early Horizon (EH; 840–260 BC) throughout the Early Intermediate Period (EIP; 260 BC–640 AD), culturally characterized by the successive thriving of Paracas and Nasca cultures (5, 15). According to local climate records, the variability of precipitation increased in the Late EIP (∼600 AD), resulting in frequent droughts and occasional flash floods (16, 17). During the Middle Horizon (MH; 640–1180 AD), climatic conditions continued to become more unfavorable, peaking in a hyperarid phase starting around 750 AD (5). These local observations match those of a high-resolution marine climate record from the Peruvian shelf west of Lima (18), which indicates episodes of extreme drought between 800 and 1250 AD, following a peak in El Niño activity with heavy rainfalls. The climatic changes are coincident with a much lower settlement density in the lower valleys of the RGND (12, 19–22). It has been hypothesized that the inhabitants of the lower RGND valley successively retreated to the upper valleys of the settlement area because of the increasing aridity (5, 12, 23, 24). However, recent research found that a first decrease of the settlement density in the northern RGND already started at the end of the Middle EIP (∼440 AD) (25). Interestingly, the marine climate record shows short periods of reduced El Niño activity and drought concurrent with this settlement decrease (18), although no alterations are noted in local records (17). The settlement density in the northern RGND recovered at the beginning of the Late Intermediate Period (LIP; 1180–1450 AD), reaching a maximum settlement density around 1250 AD (25), which roughly coincides with a period of high levels of precipitation starting around 1250 AD (5, 12, 15, 18). The increase of grass pollen from a peatland record 50 km east of Palpa shows that this humid period was preluded by a successive improvement of climatic conditions starting around 1150 AD in the northern RGND (17). The comparison of the Palpa climate record with ice-accumulation records from the Quelccaya ice cap in the southeastern Peruvian highlands (26, 27) and lake-level records of Lake Titicaca (10, 28) reveals that the oscillations between dry and humid conditions at the coast alternated with humid and dry spells in the southeastern Peruvian highlands (5). Contemporaneous with the coastal droughts during the Early MH, the area around Lake Titicaca saw the rise of the Tiwanaku culture and intensification of agriculture coinciding with more humid conditions (7, 10, 13). In the south-central highlands around Ayacucho, the rise and expansion of the Wari highland empire also began at the start of the MH (29–31). A period of severe aridity and cooler temperatures beginning in the early to mid-12th century (6, 32) has been identified as the proximate cause of the disintegration of the Tiwanaku state (33), contemporaneous with the period of increased humidity in the foothills of the RGND (5) and the demise of the Wari state (29).
Fig. 1.
Map of the Rio Grande de Nasca drainage area in southern Peru. The geographical profiles (Upper) show the maximum extend of monsoonal summer rains down the valleys into the desert and the resulting vegetation during the EH, EIP, and LIP (Upper profile) and MH (Lower profile). The map (Lower) shows the shift of the desert margin (broken line) between more humid (EH, EIP, LIP) and more arid (MH, at present) periods.
Here we address directly the impact of climatic changes on the spatial population dynamics of the Central Andes by reconciling changes in the paleogenetic record with marked climate fluctuations and cultural transitions in the past. We performed population genetic analysis on a large dataset of published (34, 35) and newly obtained pre-Columbian mitochondrial DNA sequences from sites in the northern RGND area, spanning ∼2,000 y. A series of alternative demographic scenarios was tested using serial coalescent analysis in an approximate Bayesian computational (ABC) framework.
Results and Discussion
Ancient DNA Analysis.
We obtained mitochondrial (mt) hypervariable region I (HVR I) sequences from 207 individuals of the northern RGND area deriving from coastal (coast and foothills) and highland (upper valleys) archaeological sites dating from the EH to the LIP. Of these, 157 HVR I sequences (mt haplotypes) had been published in previous studies (34, 35). We also reproducibly determined the mt haplotypes of 50 individuals from the coastal sites Monte Grande (EIP: n = 11; LIP: n = 11), Chillo (LIP: n = 11), and Los Molinos (MH: n = 5), and Tranca (MH: n = 12) in the highlands, to complement the geographical coverage in our research area (Fig. 1). The 207 sequences have been grouped into six populations depending on their chronological and ecogeographical origin (Table 1 and Dataset S1). The authenticity of the ancient DNA typing results has been ensured following the strict precautions common for ancient DNA analysis (Materials and Methods). Additionally, sequence data of selected individuals have been successfully reproduced in ancient DNA laboratories at the University of Adelaide and Yale University. All sequences have been deposited in the GenBank database under accession nos. KJ489100–KJ489306.
Table 1.
Summary genetic diversity statistics and mitochondrial haplogroup frequencies for the six analyzed ancient populations
| Summary statistics | Mt haplogroup frequencies | ||||||||
| Population* | n | H† | Hd | π | Td | A | B | C | D |
| C_EH | 31 | 17 | 0.9527 | 0.0088 | −1.208 | 0.06 | 0.00 | 0.20 | 0.74 |
| C_EIP | 66 | 42 | 0.9800 | 0.0117 | −1.196 | 0.03 | 0.14 | 0.33 | 0.50 |
| C_MH | 11 | 9 | 0.9455 | 0.0127 | −0.663 | 0.00 | 0.18 | 0.36 | 0.46 |
| C_LIP | 22 | 16 | 0.9654 | 0.0137 | −0.589 | 0.05 | 0.59 | 0.27 | 0.09 |
| H_MH | 39 | 23 | 0.9555 | 0.0142 | −0.805 | 0.05 | 0.56 | 0.28 | 0.11 |
| H_LIP | 38 | 25 | 0.9730 | 0.0160 | −0.578 | 0.05 | 0.53 | 0.32 | 0.10 |
Population: C, coast/foothills; H, highlands/upper valleys.
H, number of observed haplotypes; Hd, haplotype diversity; π, nucleotide diversity; Td, TajimaD.
All mt haplotypes could be assigned to one of the four Native American founding haplogroups A, B, C, or D and confirmed through the analysis of mt coding region polymorphisms. In total, 98 different mt haplotypes could be determined. Summary statistics are given in Table 1 and a detailed list of all haplotype mutation profiles is in Table S1. Haplotype comparison and temporal and spatial network analysis (Fig. S1) reveal that ∼61% of the observed mt haplotypes are unique to individuals, whereas 17% were shared among individuals within a population, 12% were shared between populations of the same ecogeographical area but different time periods, and the remaining 9% were matches between the highlands and coast (Table S1). Of the latter, most haplotypes were shared between the coastal LIP population and both MH and LIP highland populations (Table S1). These three populations also shared high frequencies of haplogroup B, as commonly observed in modern Central Andean populations (36), whereas the other coastal populations (C_EH, C_EIP, C_MH) exhibited low B frequencies and high haplogroup D frequencies (Table 1). Analysis of the genetic distances (FST) based on HVR I variation (Fig. 2) showed that temporally successive coastal populations from the EH to the MH were genetically close to each other (FST −0.025–0.024) as well as to successive MH and LIP highland populations (FST −0.009). When comparing coast to highland, EH to MH coastal populations were genetically distant from MH and LIP highland populations (FST 0.111–0.300). In contrast, the subsequent coastal LIP population is genetically close to both highland populations but shows a significant distance (P > 0.05, after Benjamini–Hochberg correction) from the preceding MH population and the other coastal populations (Fig. 2).
Fig. 2.
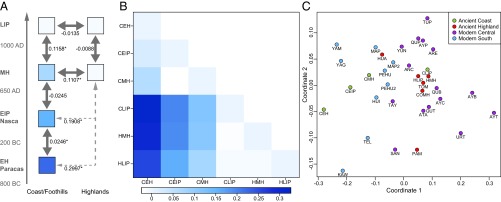
Genetic distances (pairwise FST) between the ancient populations from the RGND area and in comparison with other ancient and modern-day populations. (A and B) Genetic distances between the populations from the northern RGND. Significant FST values in A are marked with an asterisk, and increasing blue intensity on the heatmap indicates larger genetic distance (B). (C) The MDS plot is based on pairwise FST values and includes modern and ancient Andean populations (raw stress, 0.1088; names of populations are listed in Table S2).
Population Genetic Comparison with Other Andean Populations.
We further compared the analyzed ancient populations with several ancient and modern-day indigenous Andean populations (Table S2) to evaluate the population genetic relationships in the broader context of western South America. Genetic distances based on mitochondrial HVR I sequences were calculated and visualized using multidimensional scaling (MDS). The first coordinate of the MDS broadly distinguishes modern Central Andean from the modern Southern Andean and Fuegian populations. Both highland and coastal LIP populations from Palpa showed moderate to low genetic distances (FST 0.00–0.07) compared with most ancient and modern-day populations from the Central Andes and clustered broadly in the MDS (Fig. 2B). In contrast, the ancient coastal populations (EH and MH) exhibited significant, large genetic distances compared with most modern and ancient Central Andean populations, and are similar to indigenous populations from South Andean and southernmost South American populations (Fig. 2C).
Bayesian Serial SimCoal Analysis.
The observed genetic distance patterns (Fig. 2), haplotype sharing (Fig. S1 and Table S1), and changes in haplogroup composition suggest a genetic discontinuity between the MH and LIP periods on the coast and an apparent affinity of the coastal LIP population to both highland populations. We used Bayesian serial SimCoal (BayeSSC) analysis (37) to test whether the observed population genetic summary statistics (FST, haplotype diversity, and TajimaD) could be explained by a demographic scenario involving large-scale migrations between the ecogeographical areas in the settlement chamber as hypothesized by archaeological research. We tested several demographic models based on the observed genetic, archaeological, and paleoclimate data, and used BayeSSC to compute a wide range of values for a number of priors in the parameter space (Fig. S2 and Dataset S2). The ABC framework allowed us to explore priors for deme size in the past and dependent growth rate as well the timing of the migration and divergence events to maximize the credibility of the final results. We subsequently compared simulated values with the observed summary statistics to assess which model was the most likely, given the data. The Akaike information criterion (AIC) was used to evaluate a goodness-of-fit value of the range of models in light of the observed summary statistics. In addition, a relative likelihood estimate for each of the 10 basic models and their variations (SI Materials and Methods and Fig. S2) given the data was calculated via Akaike weights (ω). High AIC values, and therefore a poor fit, were obtained for the null model (H0), which represented the coastal and highland populations as one large single population and assumed continuity at the coast from the EH to the LIP. The best goodness-of-fit value and highest relative likelihood value (68%) were retrieved for model H5a (Table 2 and Dataset S2). This model allowed for a split of the coastal deme and the highland demes before the EH and two main unidirectional gene-flow events: a moderate-to-high emigration (10%) from the coast to the highlands, followed by a massive reimmigration (25%) of people from the highlands to the coast in the Early-to-Middle LIP. Models H6–H9, which explored different proportions of migrants and alternatively in reverse directions, returned low relative likelihood values. This means that the percentage of migrants per deme and the directionality are driving the models rather than a particular ratio, which confirms the usefulness of exploring the parameter space (here priors for deme sizes and their dependent growth rates). For models involving directed migration from one deme to another, we explored the timing of the migration events by using uniform priors spanning the time between the six ancient population pools for which a gene flow between coast and highlands has been assumed (Fig. S2). The event time posteriors for model H5a with the best support were 29.3 generations before 1400 AD (∼668 AD) for the 10% migration event from the coast to the highlands, and 9.9 generations (∼1153 AD) for the 25% back-migration to the coast. The next highest relative likelihood (20%) was retrieved for H1 assuming no migration between the two demes since the population split, whereas all other alternative models again returned negligible likelihood values (Table 2). Interestingly, the two models returned a posterior value of 103 (∼1175 BC) and 102 generations, respectively, for the divergence time of the coastal and highland populations.
Table 2.
AIC goodness-of-fit estimates, resulting model probabilities via Akaike weights, and estimated migration event times of the demographic models analyzed with BayeSSC
| Model | AIC | Akaike weight, ω | Event time 1 (EIP) | Event time 2 (LIP) |
| H5a | 81.46 | 0.678197 | 29.3 | 9.93 |
| H1 | 83.87 | 0.203759 | n/a | n/a |
| H9b | 85.81 | 0.077213 | 26 | 6 |
| H5c | 87.92 | 0.026822 | 32 | 10 |
| H6a | 89.82 | 0.010399 | 30 | 12 |
| H7a | 91.95 | 0.003575 | 47 | 6 |
| H9a | 102.07 | 2.276E-05 | 49 | 5 |
| H0 | 103.31 | 1.219E-05 | n/a | n/a |
| H5b | 108.21 | 1.052E-06 | 37 | 6 |
| H6b | 111.44 | 2.100E-07 | 31 | 7 |
| H8b | 111.57 | 1.961E-07 | 21 | 9 |
| H7b | 118.50 | 6.160E-09 | 50 | 3 |
| H4 | 125.69 | 1.686E-10 | n/a | 12 |
| H8a | 136.32 | 8.311E-13 | 36 | 4 |
| H7c | 141.63 | 5.824E-14 | 21 | 7 |
| H6c | 151.42 | 4.368E-16 | 30 | 4 |
| H3 | 166.94 | 1.863E-19 | n/a | n/a |
| H2 | 179.14 | 4.177E-22 | n/a | n/a |
Further result details can be found in Dataset S2. n/a, not applicable.
The overall low support for all tested scenarios assuming other migration proportions and directions confirms the directionality and timing of migration processes hypothesized in H5 in agreement with the archaeological record, and provides important estimates regarding the demographic dimension of these population dynamic processes. However, we are aware of the fact that our models represent oversimplifications and cannot consider all possible demographic scenarios. We also caution that our simulations are limited to the maternal demographic history. Archaeological and ethnohistorical evidence suggests that maternal and paternal demographic histories might have differed substantially (25). In addition, our simulations are also limited by a restricted genetic diversity due to the bottleneck during the initial peopling of the Americas. A refinement of the nuances of past demographic scenarios will be possible once nuclear genome data from past populations (especially before the MH) become available.
Synthesis of Paleogenetic, Archaeological, and Ecological Data.
The Nasca developed a complex social and demographic structure in the RGND (20, 38). The increase in settlement density and the advent of a hierarchic settlement structure observed in the EIP suggest that large populations accumulated in the lower valleys in the Nasca period (12, 39), highly dependent on the availability of water and complex irrigation systems (5, 12). The observed increasing climate variability at the end of the EIP would have decreased the predictability of agricultural productivity, and subsequent extremely dry conditions in the MH lowered the hydrological discharge from the highlands, inflicting a critical tail-off in yield. The settlement density in the RGND was already low when the climate instabilities started to increase (25), resulting in abandonment of the northern RGND during the hyperarid phase starting around 750 AD (12). Model H5a suggests that around 10% of the lower-valley effective population moved into the high-altitude upper valleys and highland regions and admixed with the local populations at ∼665 AD, also marking the end of the EIP and of the Nasca culture in the northern RGND. The 10% emigration to the highlands alone cannot explain the abandonment of the research area observed in the MH, but is a positive indicator that migrations out of the northern RGND coincided with the climatic alterations. It also suggests that people might have migrated to other destinations, such as southern extensions of the Nasca region, where an increasing settlement density was reported during the EIP–MH transition (19, 39). However, based on genetic data alone, we caution that alternative demographic factors such as mass mortality cannot be excluded at this stage.
The lack of significant signs of Wari presence in most of the lower valleys in the MH (12, 38) and the substantial genetic distance between the coastal and highland MH populations (FST 0.1107; Fig. 2) suggest that the decline of the Nasca might not have resulted from invasion and replacement by the Wari, as had been suggested before (40). We propose instead that the increased fragmentation and alteration of the spatial settlement pattern of the Nasca led to a reorganization of the population to smaller organizational units and eventually to the decline of the culture. The loss of a centralized management for storage and distribution of staple goods would have increased the vulnerability of the society to crop failures. This suggests that the emigration of the remaining populations of the northern RGND at the EIP–MH transition might have been triggered by the increasing climate/precipitation variability.
Our results cannot explain the first decrease of settlement density in the northern RGND beginning at ∼440 AD. The archaeological record shows slightly increased settlement activities in the northern extension of the Nasca region around the Ica Valley (25), but there is no direct evidence for larger migrations from the RGND to the north. Given that we explored the parameter space covering the proposed 440 AD event, we assume that our data will only support the strongest signal, which appears to be the emigration reaching 10% after 665 AD. In addition, previous studies showed that the coastal populations of the Nasca culture region from the Paracas Peninsula to Nasca are very homogeneous (34), which indicates that migrations from the RGND to the northern or southern extensions would be difficult to detect with HVR I data alone.
During the LIP, the observed low genetic distances (Fig. 2), shared haplotypes (Table 2), and identical haplogroup frequencies of populations from both ecogeographical regions (Table 1) point to a process of genetic homogenization between highland and coastal populations during this period. The best-supported model, H5a, indeed indicates an immigration of ∼25% highland people to the coast around 1150 AD (Table 2). This is in line with the archaeological record showing a rapidly increasing settlement density in the lower valleys in the mid-12th century coinciding with a transient phase of increased humidity in this region and protracted droughts in the southern Peruvian highlands (5, 7, 12).
It is plausible that the enhanced hydrological conditions of the lower valleys in the LIP may have boosted the carrying capacity of the agronomy acting as a primary pull factor for this migration (5), with the increasing social and demographic complexity as a secondary pull factor (34, 41). In turn, aridity and droughts in the southern highlands in the mid-12th century and the decline of the highland empires are possible push factors. The large population size in the highlands remaining after the decline of the empires and the increasing demand of space to compensate the declining agricultural productivity would have resulted in demographic pressures. These would have forced formerly urbanized populations in the highlands to become more mobile and decrease population densities (7). Parts of the populations from the southern Peruvian highlands probably migrated to more favorable environments, namely the lower valleys of the western Andean slopes. This scenario is supported by the observed high genetic affinity of the ancient highland and LIP coastal populations from the RGND to other pre-Columbian highland and modern indigenous southern Central Andean populations (Fig. 2). However, we caution that the sample size of the ancient populations remains small and the geographical distribution is limited in the case of the coastal populations.
Depending on the type of migration, mobility can be sex-specific (41). Directed colonization and elite dominance, which presumably were used by the Wari and Tiwanaku in the MH to increase their sphere of political influence (42), mostly involve limited numbers of dominating males (41). Mass movements or folk migration are alternative forms of migration involving equal proportions of males and females (41). The fact that we find support for migration from mtDNA data, describing the female population history, therefore further corroborates our interpretation of the past population dynamic processes: Demographic pressure at the end of the EIP and in the LIP forced large numbers of people regardless of their sex to migrate.
Interestingly, ethnohistoric sources and the archeological record point to a cultural distinction of foothill and upper-valley populations in the LIP and Late Horizon in contrast to the observed genetic homogenization of both ecogeographical areas (25, 43, 44). Cultural traditions are distributed and inherited in a far more complex way than genes, and cultural borders do not reflect biological borders. Persisting strong cultural traditions in the remaining coastal populations but also the necessary economic adaptation of immigrating highland people to the different ecological conditions are only two possible explanations for the observed cultural differences but mitochondrial/matrilineal genetic identity.
Conclusion
Our integration of heterochronous population genetic data into the framework of an interdisciplinary dataset endorses prehistoric migration processes on the western slopes of the South Peruvian Andes. The supported migration events coincide with observed climatic alterations, which must have led to a genetic homogenization in the RGND over time. Most studies analyzing the impact of climate on the mobility and demography of ancient populations have concentrated on foraging societies. Here we reveal climatic impacts on socially complex, agricultural societies. Despite a range of anticipatory agricultural adaptations observed in the archaeological record, the instability of the coastal and highland ecosystems in southern Peru exceeded critical points in the past in which migration became inevitable to sustain the population.
Materials and Methods
Samples and Ancient DNA Analysis.
Samples from new archaeological sites in the area have been obtained for this study in addition to previously analyzed samples (18, 19). The new sites included two sites from the lower valleys and the coast, Chillo (n = 25) and Monte Grande (n = 20), dating to the LIP and not represented in previous studies. Other samples from coastal/foothill sites derived from the EIP phase of Monte Grande (n = 20) and the MH phase of Los Molinos (n = 12). Additionally, 18 samples came from the MH archaeological site Tranca, located in the upper valleys (Fig. 1). Sample preparation and DNA extractions were performed using methods similar to those in ref. 45. For each individual, we generated DNA extracts from at least two independent samples.
Mitochondrial haplogroups for the samples were determined by using a multiplex single-base extension assay described in ref. 19. To determine the mitochondrial haplotypes of the individuals, we analyzed a 388-bp fragment of the mitochondrial hypervariable region I [nucleotide position (np)16,021–np16,408 (rCRS)] using four overlapping primer pairs as described in ref. 18. Protocols and primer sequences used for the PCRs, DNA quantification by quantitative PCR, and other downstream applications are described elsewhere (34, 35, 46). Amplicons were analyzed by direct sequencing of the heavy and light strands. All four overlapping fragments were amplified at least twice from all extracts to define consensus sequences. The sequence data were manually verified with SeqMan software (Lasergene 4.05; DNAStar) to control for analysis and reading errors (45).
All pre-PCR analyses were carried out in a laboratory entirely dedicated to ancient DNA analysis. All analyses were carried out according to the precautions and contamination prevention strategies described in refs. 34, 35, and 46. Additionally, some of the mitochondrial HVR I sequences were independently replicated in the Ancient DNA Laboratory at the Yale Institute of Biospheric Studies and the Australian Centre for Ancient DNA at the University of Adelaide using methods similar to those in ref. 47.
Bayesian Coalescent Simulations.
We used the software program BayeSSC to determine the demographic scenario that would most likely support the observed population genetic diversity in ancient South American populations. Our goal was to test whether the observed summary statistics could be explained by genetic drift (null hypothesis) or by population dynamic events between structured subpopulations (alternative scenarios). The 10 main demographic models (H0–H9), plus several variants of specific models (a, b, c), were tested following various hypotheses put forward by archaeologists (5, 12). BayeSSC analysis was performed in accordance with previous publications (47–49) with the following parameters of sequence evolution for all simulations: a generation time of 25 y, a fixed mutation rate of 7.5 × 10−6 substitutions per site per generation (50), a transition:transversion ratio of 0.9841 (51), and a gamma distribution of rates with shape parameters of 0.205 (theta) and 10 (kappa) (50). We applied a uniform distribution to estimate the effective population size (Ne) for a Holocene source population (5–1,000 females) in our region before the LIP [448 generations (H0) or 81–500 generations (H1–H9)] exploring the time since the peopling of South America (3), and one (panmixia model H0) or two (alternative coast vs. highland models H1–H9) South American demes (1,000–1,000,000) at time 0, which is set here at approximately the late LIP (1400 AD) based on the youngest ages of our LIP samples. Migration events were set to explore the number of generations between each of our ancient population pools. For further information on the analyses and models, refer to SI Materials and Methods and Fig. S2.
Population Genetic Analysis.
We calculated population-specific pairwise genetic distance, haplotype diversity, and Tajima’s D in Arlequin version 3.5 (52) using 207 388-bp HVR I sequences (np16,021–np16,408) assigned to one of six populations (Table 1). FST values were estimated using the Kimura two-parameter model (51) using a gamma distribution with shape parameter of 0.205 (50). We used Arlequin version 3.5 to identify shared haplotypes between the analyzed populations. To visualize the patterns of haplotype sharing, we calculated temporal networks based on the sequence data using TempNet (53) and median-joining networks for the spatial comparison of synchronous populations using Network 4.612 (54) (Fig. S1).
For the population genetic comparison, mitochondrial HVR I sequence data from a number of modern and ancient populations from the Central Andes and Southern Andes were collected from the literature (Table S2) to evaluate the population genetic relationships of the pre-Columbian Palpa populations in the broader context of western South America. Population-specific pairwise genetic distances were calculated as described above. To visualize genetic relationships, we performed multidimensional scaling based on the distance matrices using Statistica 10 (StatSoft).
Supplementary Material
Acknowledgments
We thank the German Research Foundation for funding this research (Grant FE 1161/1-1) and the Australian Research Council for funding granted to W.H. and B.L. (DP1095782 and DP130102158). We also thank the Instituto Nacional de Cultura del Perú for granting us the exportation permission for the sample material (Resolucion Directoral Nacional 1346).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KJ489100–KJ489306).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403466111/-/DCSupplemental.
References
- 1.Eriksson A, et al. Late Pleistocene climate change and the global expansion of anatomically modern humans. Proc Natl Acad Sci USA. 2012;109(40):16089–16094. doi: 10.1073/pnas.1209494109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stephenson J, Newman K, Mayhew S. Population dynamics and climate change: What are the links? J Public Health (Oxf) 2010;32(2):150–156. doi: 10.1093/pubmed/fdq038. [DOI] [PubMed] [Google Scholar]
- 3.Dillehay TD. Monte Verde: A Late Pleistocene Settlement in Chile. Vol 1. Washington, DC: Smithsonian Inst Press; 1989. [Google Scholar]
- 4.Sandweiss DH, et al. Variation in Holocene El Niño frequencies: Climate records and cultural consequences in ancient Peru. Geology. 2001;29(7):603–606. [Google Scholar]
- 5.Mächtle B, Eitel B. Fragile landscapes, fragile civilizations—How climate determined societies in the pre-Columbian south Peruvian Andes. Catena. 2012;103:62–73. [Google Scholar]
- 6.Thompson LG. In: in Abrupt Climate Change: Mechanisms, Patterns, and Impacts. Rashid H, Polyak L, Mosley-Thompson E, editors. Washington, DC: Am Geophys Union; 2011. pp. 215–233. [Google Scholar]
- 7.Dillehay TD, Kolata AL. Long-term human response to uncertain environmental conditions in the Andes. Proc Natl Acad Sci USA. 2004;101(12):4325–4330. doi: 10.1073/pnas.0400538101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandweiss DH, Solís RS, Moseley ME, Keefer DK, Ortloff CR. Environmental change and economic development in coastal Peru between 5,800 and 3,600 years ago. Proc Natl Acad Sci USA. 2009;106(5):1359–1363. doi: 10.1073/pnas.0812645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marquet PA, et al. Emergence of social complexity among coastal hunter-gatherers in the Atacama Desert of northern Chile. Proc Natl Acad Sci USA. 2012;109(37):14754–14760. doi: 10.1073/pnas.1116724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binford MW, et al. Climate variation and the rise and fall of an Andean civilization. Quat Res. 1997;47:235–248. [Google Scholar]
- 11.Williams PR. Rethinking disaster-induced collapse in the demise of the Andean highland states: Wari and Tiwanaku. World Archaeol. 2002;33(2):361–374. [Google Scholar]
- 12.Reindel M. In: in New Technologies for Archaeology: Multidisciplinary Investigations in Palpa and Nasca, Peru. Reindel M, Wagner G, editors. Berlin: Springer; 2009. pp. 439–462. [Google Scholar]
- 13.Paulsen AC. Environment and empire: Climatic factors in prehistoric Andean culture change. World Archaeol. 1976;8(2):121–132. [Google Scholar]
- 14.Gorbahn H. The Middle Archaic site of Pernil Alto, southern Peru: The beginnings of horticulture and sedentariness in Mid-Holocene conditions. Diálogo Andin. 2013;41:61–82. [Google Scholar]
- 15.Unkel I, et al. A comprehensive numerical chronology for the pre-Columbian cultures of the Palpa valleys, south coast of Peru. J Archaeol Sci. 2012;39(7):2294–2303. [Google Scholar]
- 16.Hesse R, Baade J. Irrigation agriculture and the sedimentary record in the Palpa Valley, southern Peru. Catena. 2009;77:119–129. [Google Scholar]
- 17.Schittek K, et al. Holocene environmental changes in the highlands of the southern Peruvian Andes (14° S) and their impact on pre-Columbian cultures. Clim Past Discuss. 2014;10:1707–1746. [Google Scholar]
- 18.Rein B, et al. A major Holocene ENSO anomaly during the Medieval period. Geophys Res Lett. 2004;31:L17211. [Google Scholar]
- 19.Conlee CA, Schreiber KJ. 2006. in Intermediate Elites in Pre-Columbian States and Empires, eds Elson CM, Covey RA (Univ of Arizona Press, Tucson, AZ), pp 94–111.
- 20.Silverman H, Proulx DA. The Nasca. Oxford: Blackwell; 2002. [Google Scholar]
- 21.Unkel I, Kromer B, Reindel M, Wacker L, Wagner G. A chronology of the pre-Columbian Paracas and Nasca cultures in south Peru based on AMS C-14 dating. Radiocarbon. 2007;49(2):551–564. [Google Scholar]
- 22.Maechtle B. Geomorphologische-bodenkundliche Untersuchungen zur Rekonstruktion der holozaenen Umweltgeschichte in der noerdlichen Atacam im Raum Palpa. Heidelberg: Univ Heidelberg; 2007. [Google Scholar]
- 23.Eitel B, Maechtle B. In: in New Technologies for Archaeology: Multidisciplinary Investigations in Palpa and Nasca, Peru. Reindel M, Wagner G, editors. Berlin: Springer; 2009. pp. 17–37. [Google Scholar]
- 24.Reindel M, Isla-Cuadrado J. Cambio climático y patrones de asentamiento en la vertiente occidental de los Andes del sur del Perú. Diálogo Andin. 2013;41:83–99. [Google Scholar]
- 25.Sossna V. Los patrones de asentamiento del Periodo Intermedio Temprano en Palpa, costa sur del Perú. Zeitschrift Archaeologie Aussereuropaeischer Kult. 2012;4:207–280. [Google Scholar]
- 26.Thompson LG, Mosley-Thompson E, Bolzan JF, Koci BR. A 1500-year record of tropical precipitation in ice cores from the Quelccaya ice cap, Peru. Science. 1985;229(4717):971–973. doi: 10.1126/science.229.4717.971. [DOI] [PubMed] [Google Scholar]
- 27.Shimada I, Schaaf CB, Thompson LG, Mosley-Thompson E. Cultural impacts of severe droughts in the prehistoric Andes: Application of a 1,500-year ice core precipitation record. World Archaeol. 1991;22(3):247–270. [Google Scholar]
- 28.Rowe HD, Dunbar RB. Hydrologic-energy balance constraints on the Holocene lake-level history of Lake Titicaca, South America. Clim Dyn. 2004;23(3-4):439–454. [Google Scholar]
- 29.Isbell WH. In: in Handbook of South American Archaeology. Silverman H, Isbell WH, editors. New York: Springer; 2008. pp. 731–760. [Google Scholar]
- 30.Ploudre AM, Stanish C. In: in Andean Archaeology III. North and South. Isbell WH, Silverman H, editors. Springer, New York: 2008. pp. 237–257. [Google Scholar]
- 31.Kolata AL. In: in Environmental Disaster in the Archaeology of Human Response. Bawden G, Reycraft RM, editors. Albuquerque: Univ of Mexico Press; 2000. pp. 163–178. [Google Scholar]
- 32.Abbott MB, Anderson L. In: in Encyclopedia of Paleoclimatology and Ancient Environments. Gornitz V, editor. Heidelberg: Springer; 2009. pp. 489–492. [Google Scholar]
- 33.Seltzer GO, Hastorf CA. Climatic change and its effect on prehispanic agriculture in the Central Peruvian Andes. J Field Archaeol. 1990;17:397–414. [Google Scholar]
- 34.Fehren-Schmitz L, Reindel M, Cagigao ET, Hummel S, Herrmann B. Pre-Columbian population dynamics in coastal southern Peru: A diachronic investigation of mtDNA patterns in the Palpa region by ancient DNA analysis. Am J Phys Anthropol. 2010;141(2):208–221. doi: 10.1002/ajpa.21135. [DOI] [PubMed] [Google Scholar]
- 35.Fehren-Schmitz L, et al. Diachronic investigations of mitochondrial and Y-chromosomal genetic markers in pre-Columbian Andean highlanders from south Peru. Ann Hum Genet. 2011;75(2):266–283. doi: 10.1111/j.1469-1809.2010.00620.x. [DOI] [PubMed] [Google Scholar]
- 36.Bisso-Machado R, Bortolini MC, Salzano FM. Uniparental genetic markers in South Amerindians. Genet Mol Biol. 2012;35(2):365–387. doi: 10.1590/S1415-47572012005000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson CN, Ramakrishnan U, Chan YL, Hadly EA. Serial SimCoal: A population genetics model for data from multiple populations and points in time. Bioinformatics. 2005;21(8):1733–1734. doi: 10.1093/bioinformatics/bti154. [DOI] [PubMed] [Google Scholar]
- 38.Isla-Cuadrado J, Reindel M. 2006. in Andean Archaeology III. North and South, eds Isbell WH, Silverman H (Springer, New York), pp 374–400. [Google Scholar]
- 39.Kellner CM, Schoeninger MJ. Wari’s imperial influence on local Nasca diet: The stable isotope evidence. J Anthropol Archaeol. 2008;27:226–243. [Google Scholar]
- 40.Allison MJ. Paleopathology in Peru. Nat Hist. 1979;88(2):74–82. [PubMed] [Google Scholar]
- 41.Sanjek R. Rethinking migration, ancient to future. Glob Netw. 2003;3(3):315–336. [Google Scholar]
- 42.Schreiber KJ. 1992. Wari Imperialism in Middle Horizon Peru (Mus Anthropol, Univ of Michigan, Ann Arbor)
- 43.Abraham S. 2010. Provincial life in the Inca empire: Continuity and change at Pulapuco, Peru. PhD dissertation (Univ of California, Santa Barbara)
- 44.Schreiber K. In: in Provincial Inca: Archaeological Identification of the Impact of the Inca State. Malpass M, editor. Iowa City, IA: Univ of Iowa Press; 1993. pp. 77–116. [Google Scholar]
- 45.Salas A, Carracedo A, Macaulay V, Richards M, Bandelt HJ. A practical guide to mitochondrial DNA error prevention in clinical, forensic, and population genetics. Biochem Biophys Res Commun. 2005;335(3):891–899. doi: 10.1016/j.bbrc.2005.07.161. [DOI] [PubMed] [Google Scholar]
- 46.Seidenberg V, et al. A new miniSTR heptaplex system for genetic fingerprinting of ancient DNA from archaeological human bone. J Archaeol Sci. 2012;39(10):3224–3229. [Google Scholar]
- 47.Haak W, et al. Members of the Genographic Consortium Ancient DNA from European early neolithic farmers reveals their Near Eastern affinities. PLoS Biol. 2010;8(11):e1000536. doi: 10.1371/journal.pbio.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bramanti B, et al. Genetic discontinuity between local hunter-gatherers and central Europe’s first farmers. Science. 2009;326(5949):137–140. doi: 10.1126/science.1176869. [DOI] [PubMed] [Google Scholar]
- 49.Ghirotto S, et al. Inferring genealogical processes from patterns of Bronze-Age and modern DNA variation in Sardinia. Mol Biol Evol. 2010;27(4):875–886. doi: 10.1093/molbev/msp292. [DOI] [PubMed] [Google Scholar]
- 50.Ho SY, Endicott P. The crucial role of calibration in molecular date estimates for the peopling of the Americas. Am J Hum Genet. 2008;83(1):142–146. doi: 10.1016/j.ajhg.2008.06.014. author reply 146–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 52.Excoffier L, Lischer HE. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10(3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 53.Prost S, Anderson CNK. TempNet: A method to display statistical parsimony networks for heterochronous DNA sequence data. Methods Ecol Evol. 2011;2(6):663–667. [Google Scholar]
- 54.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16(1):37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.