Significance
Retroviral infection of cells can be blocked by the action of the postentry restriction factors. The Trim5α and Fv1 factors do so by targeting the capsid that surrounds the viral core. The nature of the interaction of these factors with the viral assembly is unclear. We show that these factors form antiparallel dimers that display specificity domains spaced to target motifs on the capsid lattice surface. In doing so Fv1 and Trim5α take advantage of the regularly spaced array of binding sites on the capsid surface, generating avidity to aid recognition of retroviral pathogens.
Keywords: retrovirus, MLV, SAXS, X-ray crystallography
Abstract
Restriction factors (RFs) form important components of host defenses to retroviral infection. The Fv1, Trim5α, and TrimCyp RFs contain N-terminal dimerization and C-terminal specificity domains that target assembled retroviral capsid (CA) proteins enclosing the viral core. However, the molecular detail of the interaction between RFs and their CA targets is unknown. Therefore, we have determined the crystal structure of the B-box and coiled-coil (BCC) region from Trim5α and used small-angle X-ray scattering to examine the solution structure of Trim5α BCC, the dimerization domain of Fv1 (Fv1Ntd), and the hybrid restriction factor Fv1Cyp comprising Fv1NtD fused to the HIV-1 binding protein Cyclophilin A (CypA). These data reveal that coiled-coil regions of Fv1 and Trim5α form extended antiparallel dimers. In Fv1Cyp, two CypA moieties are located at opposing ends, creating a molecule with a dumbbell appearance. In Trim5α, the B-boxes are located at either end of the coiled-coil, held in place by interactions with a helical motif from the L2 region of the opposing monomer. A comparative analysis of Fv1Cyp and CypA binding to a preformed HIV-1 CA lattice reveals how RF dimerization enhances the affinity of interaction through avidity effects. We conclude that the antiparallel organization of the NtD regions of Fv1 and Trim5α dimers correctly positions C-terminal specificity and N-terminal effector domains and facilitates stable binding to adjacent CA hexamers in viral cores.
In the course of evolution, mammals have developed systems to prevent and contain infection by retroviral pathogens (1). These include the restriction factors (RFs) that target various stages of the retroviral lifecycle, including reverse transcription, integration, and viral egress (2). A particular subset of these factors includes the postentry RFs Fv1, Trim5α, and TrimCyp that act shortly after the retroviral core has entered the cytoplasm of the cell and prevent reverse transcription or integration of the viral genome (3).
The murine RF Fv1 blocks infection of murine leukemia virus (MLV) by a still poorly understood mechanism. Two major alleles of Fv1 have been identified, Fv1n, which can restrict infection by B-tropic MLV, and Fv1b, which restricts N-tropic MLV (4, 5). The Fv1 protein contains at least two functional regions, an N-terminal dimerization domain and a C-terminal domain that is required for capsid (CA) recognition (6). Previous biophysical studies have demonstrated that the N-terminal domain of Fv1 forms a tightly associated α-helical elongated dimer (7).
Whereas alleles of Fv1 are limited to different species of mice (8), Trim5α acts in a similar manner to restrict retroviral infection in primates and other mammalian species (9). The Trim5α protein contains an N-terminal tripartite motif consisting of a RING domain, a B-box2 domain, and a predicted coiled-coil region (RBCC) (10). The C terminus of the protein contains a B30.2 or PRY/SPRY domain connected to the RBCC motif through a linker region, L2 (11). Residues in the B30.2 domain are largely responsible for determining the range of retroviruses a particular Trim5α can restrict (12, 13). The rhesus macaque form of Trim5α inhibits HIV-1 replication, whereas the human homolog does not (14). However, a single amino acid change in the B30.2 domain of human Trim5α is sufficient to gain restriction of HIV-1 (15, 16). Retrotransposition events have on several occasions resulted in fusion of the cis-trans prolyl isomerase Cyclophilin A (CypA), a cellular factor capable of binding to HIV-1 CA (17), to the RBCC motif of Trim5α, resulting in TrimCyp RFs (18, 19). In a similar manner, fusion of CypA to the N-terminal dimerization domain of Fv1 yields an artificial RF, Fv1Cyp, that shares the characteristics of Fv1 restriction but is capable of blocking infection by HIV-1 (20).
Central to the mechanism of postentry restriction is the recognition of the intact retroviral capsid (21). Whereas structural and sequence comparisons demonstrate that the site of restriction, for both Fv1 and Trim5α, is located across the apical surface of the retroviral CA protein (22–24), abrogation experiments have clearly demonstrated that the intact retroviral capsid lattice is required for recognition, rather than individual capsid proteins (25, 26). Assembly of MLV CA on lipid nanotubes has demonstrated in vitro that a lattice is required for Fv1 binding and that perturbations in this lattice prevent recognition by Fv1 (27). Whereas the B30.2 domain determines the range of retroviruses a particular species’ Trim5α can restrict, an intact tripartite motif is required for efficient restriction, in part because individual interactions between B30.2 and CA are of low affinity (28). Avid binding of incoming virus therefore requires extensive Trim5α multimerization (29). Deletion of the coiled-coil region prevents stable binding of the retroviral CA protein (21), whereas the B-box promotes cooperative CA binding by mediating higher-order Trim5α self-association (30). Disruption or removal of the RING domain interferes with proteasome interactions, delaying the block in virus replication until after reverse transcription has occurred, but can also affect higher-order RF association (31).
To investigate the mechanism of restriction and the recognition of the retroviral capsid by the postentry RFs we have examined the core self-association module of Trim5α, the N-terminal dimerization domain of Fv1, and the hybrid RF Fv1Cyp by X-ray crystallography, small-angle X-ray scattering, electron microscopy, and surface plasmon resonance. These experiments reveal a strong structural similarity between all of the members of this class of RFs and show that they are molecules tailored to recognize the repeat spacing of CA in the retroviral capsid lattice. Moreover, we demonstrate that multivalency, a general property of these factors, generates avidity effects that contribute to specificity and potency of retroviral restriction.
Results
Fv1 and Trim5α RFs Form Tight Dimers.
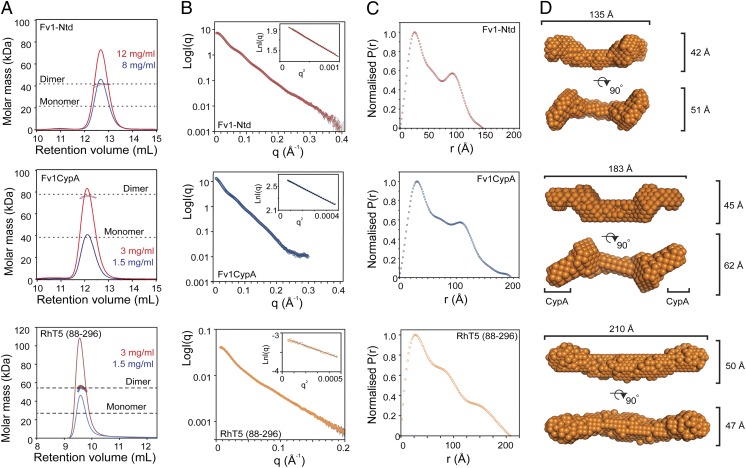
Size-exclusion chromatography coupled to multiangle laser light scattering (SEC-MALLS) was used to determine the oligomeric state of Fv1Ntd (Fv1 residues 20–200), Fv1Cyp (Fv1 residues 20–200 fused to CypA), and RhT5 (88–296) EK/RD, consisting of residues 88–296 of Rhesus macaque Trim5α (RhT5) encompassing the B-box, coiled-coil, and L2 core self-association region and containing the (E120K/R121D) aggregation suppression mutations (32). Fv1Ntd and Fv1Cyp elute from the SEC column as single symmetrical peaks. The MALLS analysis yields an invariant molecular weight of 42 kDa for Fv1NtD (concentration range of 1–12 mg/mL) and 80 kDa for Fv1Cyp (concentration range of 1.5–3 mg/mL), indicating both proteins form tight dimers in solution (Fig. 1A and Table S1). In a similar manner, SEC-MALLS analysis of this RhT5 (88–296) EK/RD construct (Fig. 1A and Table S1) gives a solution molecular mass of 54 kDa (concentration range of 0.5–5 mg/mL), revealing the core association domain of Trim5α is also a tight dimer.
Fig. 1.
SAXS analysis of Fv1Ntd, Fv1Cyp, and RhT5 (88–296) EK/RD. (A) SEC-MALLS analysis of Fv1-Ntd, Fv1-CypA, and RhT5 (88–296) EK/RD demonstrates that all proteins are dimers in solution. (B) Concentration-corrected merged SAXS curves. (Insets) Enlargement of the Guinier region and analysis. (C) Normalized pair-distribution functions. (D) Consensus ab initio models.
Small-Angle X-Ray Scattering.
To determine low-resolution structures for Fv1 and Trim5α RFs small-angle X-ray scattering (SAXS) experiments were performed. SAXS data were recorded from samples of Fv1Ntd and Fv1Cyp over a q range of 0.006–0.4 Å−1 and protein concentration of 1–4 mg/mL. Details of data collection parameters are presented in Table S1. Scattering curves for Fv1Ntd recorded between 1 and 3 mg/mL were coincident across the entire q range and show no sign of aggregation or any evidence of a structure factor in the very-low-q region. Therefore, these data were combined and the merged scattering curve was used for further analysis (Fig. 1B). Guinier analysis of the merged scattering curve (Fig. 1B, Inset) shows good linearity and gives an estimated radius of gyration (Rg) of 43.7 ± 0.6Å (1 SD) for the particle. Evaluation of the forward scatter compared with BSA gives a molecular mass of 46 kDa, consistent with the molecular mass of 43 kDa for the Fv1NtD dimer (Table S1). Scattering data collected for Fv1Cyp at >1 mg/mL showed a concentration-dependent signal in the low-q region and signs of aggregation. Therefore, further analysis was limited to data collected at 1 mg/mL. Guinier plots of these low-concentration data were linear (Fig. 1B, Inset) and gave an estimated Rg of 55.6 Å and molecular mass 88.3 kDa, consistent with an Fv1Cyp dimer. SAXS data were also recorded for RhT5 (88–296) EK/RD over a q range of 0–0.2 Å−1 at concentrations from 0.4 to 1.54 mg/mL. Guinier analysis gave an estimate of the Rg of 56.7 Å. The particle molar mass was derived from the scattering contrast after normalization to the scattering of water (33), giving a value of 55.4 kDa, also consistent with a dimer.
Fv1 and Trim5α RFs Are Elongated Molecules.
MALLS and Guinier analysis of scattering profiles from Fv1Ntd, Fv1Cyp, and RhT5 (88–296) EK/RD reveals that these proteins are dimeric and have a large Rg compared with the molar mass, suggestive of an elongated nature. Therefore, to gain insight into the distribution of mass within these proteins the pair-distribution function [P(r)] was calculated from the scattering data measured for each particle (Fig. 1C). The P(r) function for Fv1Ntd is distinctly bimodal, containing a primary peak at 26 Å, a secondary peak at 95 Å, and a maximum dimension (Dmax) of 145 Å. The real-space Rg derived from the distribution is 46.7 Å, comparable to that from the Guinier analysis (Table S1). Similarly, the pair-distribution function for Fv1Cyp is also bimodal with a Dmax at 195 Å and containing a primary peak at 32 Å and a secondary peak at 105 Å. The strong secondary maxima in the pair-distribution functions are characteristic of a dumbbell-shaped molecule where the secondary maxima represent the center-to-center distance of the lobes (34). The pair-distribution function for RhT5 (88–296) EK/RD is also characteristic of an elongated molecule with Dmax at 208 Å. However, the distribution more closely resembles that from a cylindrical particle (34) with a single major peak at 27 Å and an asymmetrical fall-off to the maximum dimension of 208 Å that contains shoulders at 75 Å and 150 Å (Fig. 1C).
Ab initio modeling of the molecular envelopes of Fv1Ntd and Fv1Cyp was undertaken and produced molecules with extended shapes (Fig. S1). Moreover, application of P2 symmetry in the modeling procedure generated models that best fit the data and have a high degree of convergence (Table S1). Given the twofold symmetry, the consensus models for Fv1NtD and Fv1Cyp are distinctly antiparallel, comprising narrow central regions with globular lobes located at each end of the structure (Fig. 1D). The Fv1Ntd model is 135 Å in length and contains a 60-Å central spacer. The globular end lobes are oriented ∼115° to the central region with a spacing that accounts for the secondary maxima at 95 Å in the P(r) function. Fv1Cyp is substantially larger, encompassing a central region comparable to the Fv1Ntd envelope but with additional volume at each globular lobe that locates the CypA domains to each end of the molecule (Fig. S2) and accordingly shifts the secondary maximum in the P(r) function to 105 Å. These data reveal that Fv1NtD and Fv1Cyp are dumbbell-shaped molecules and that the secondary maxima of the P(r) function derive from the peripherally located globular lobes that contain CypA domains in Fv1Cyp.
A best-fit ab initio model for RhT5 (88–296) EK/RD was also generated with applied P2 symmetry (Table S1). The consensus model reveals an extended structure ∼210 Å in length comprising a 115-Å central spacer region and two small globular domains apparent at each end of the structure (Fig. 1D). Comparison of the globular domain at each end of the SAXS model with the B-box structures of Rhesus and human Trim5α (32) shows them to be of comparable molecular volume, suggesting that although not as pronounced as the dumbbell structure of the Fv1 factors the B-boxes of Trim5 are also arranged in an antiparallel fashion.
Crystal Structure of RhT5-T4 Lysozyme.
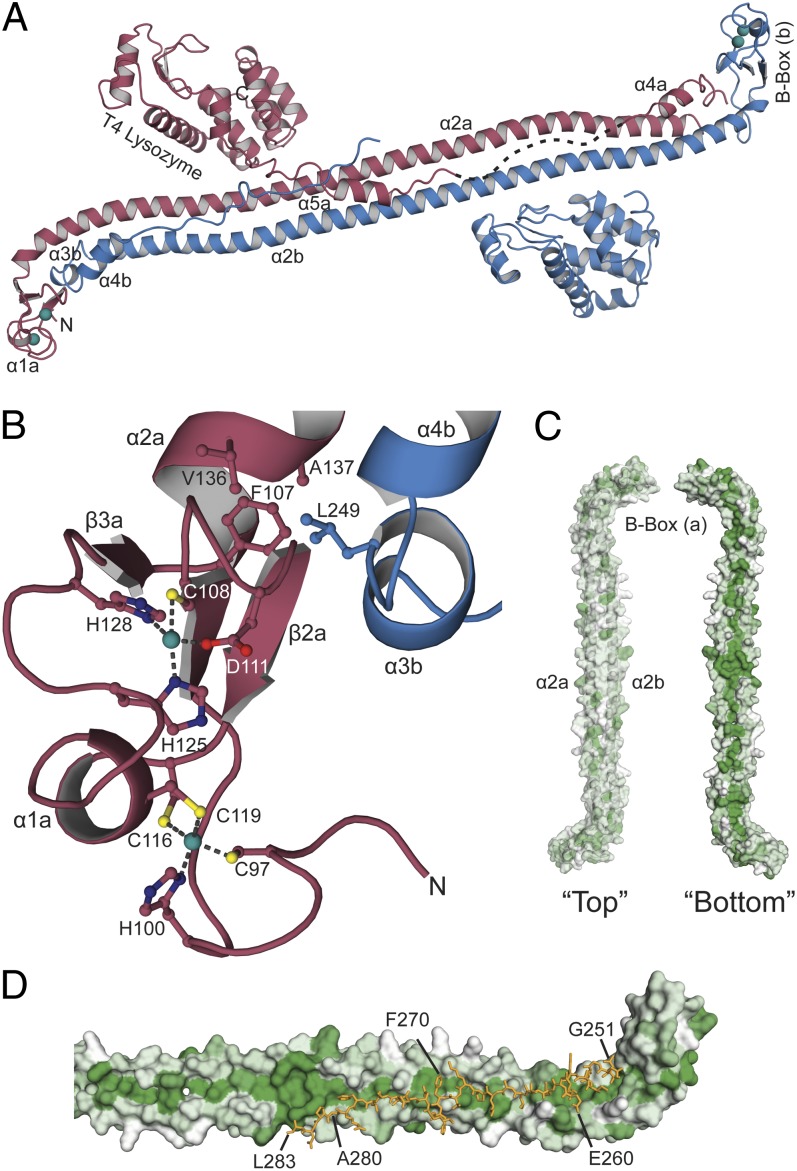
To aid crystallization and determine the structure of a Trim5α homodimer, the sequence for bacteriophage T4 lysozyme (T4L) was fused C-terminally to residue 291 in place of the CypA domains present RhT5Cyp but still incorporating the 12-aa upstream RhT5Cyp linker sequence (Fig. S3). Crystals of this chimera (RhT5-T4L) belong to the spacegroup C2 and contain one dimer in the asymmetric unit. The structure was determined using a combination of molecular replacement and single-wavelength anomalous dispersion and the model refined at a resolution of 3.2 Å (R/Rfree of 26%/32%, respectively). Details of data collection, phasing, and model refinement are presented in Table S2.
The structure (Fig. 2A) is dominated by two central 160-Å 30-turn antiparallel helices, α2A and α2B, that form a coiled-coil structure running the length of the protein. The N-terminal B-box domains are located at either end of the coil and pack against a pair of helices (α3–α4) from the L2 region of the opposing monomer. The remainder of the L2 region, residues 260–283, forms an extended chain (L2-E) that turns away from the B-box and runs down the coil toward the twofold axis at the center of the dimer. The structure is well-defined for both chains at residues comprising the B-boxes, coiled coil, α3–α4 regions of L2, and the T4Ls (Fig. S4) but with differing sections of L2-E visible in the A and B molecules. The B-box structure is essentially identical to that described for human Trim5α (32), comprising a three-stranded antiparallel β-sheet and short α-helix, α1 connected with a β1–β2–α1–β3 topology. Each B-box contains two zinc atoms coordinated in a tetrahedral fashion by residues H125, H128, C108, and D111 (Zn1) and C116, C119, C97, and H100 (Zn2) (Fig. 2B). The mutated residues, E120K/R121D, that aid solubility are located at the C terminus of α1 and have solvent-exposed side chains. At each end of the dimer, the antiparallel arrangement of the α2 helices places the B-box from one monomer in the proximity of the α3–α4 motifs from the opposing monomer. This arrangement facilitates the packing of α3–α4 against the opposing B-box and forms a hydrophobic core around the B-box β-sheet (Fig. 2B). The α2 helices also pack at only a shallow writhe angle. As a result the degree of twist along the entire coiled-coil is only small and gives rise to “top” and “bottom” surfaces. Analysis of the distribution of residue polarity along theses surfaces (Fig. 2C) reveals a top surface with a variegated residue distribution. However, the bottom surface contains a channel running the length of the structure lined with hydrophobic residues that interact with residues in the L2-E region that traverses toward the center of the molecule (Fig. 2D). At the center of the molecule the sequence that links to the T4L domains is largely disordered and only a single α5 helix from chain A of the structure is visible. This flexibility in the linker region is also apparent with respect to the T4L domains that do not obey the twofold symmetry of the coiled-coil, adopting different orientations with respect to the rest of the molecule (Fig. 2A).
Fig. 2.
Crystal structure of RhT5-T4L. (A) Cartoon representation of the RhT5-T4L structure with zinc ions shown as spheres. Residues in the L2-E region (between α4 and α5) are missing in chain A (red) and are represented as a dashed line but are present in the other monomer (chain B, blue). (B) Zn1 is coordinated by C108, D111, H125, and H128 and Zn2 is coordinated by C97, H100, C116, and C119. The B-box of one monomer packs with the α3–α4 region of the opposing monomer to form a hydrophobic core. (C) The “top” of the extended antiparallel helices has no distinct hydrophobic groove, whereas the “bottom” has a clear hydrophobic groove (green) that spans the length of the helices. (D) The L2-E region (orange sticks, labeled every 10th residue) closely follows the hydrophobic groove on the surface of the extended coil. Hydrophobicity of residues was calculated using AAindex (database code FASG890101) in PyMol where increasing hydrophobicity is proportional to the intensity of the green color.
Electron Microscopy Analysis of Fv1Cyp.
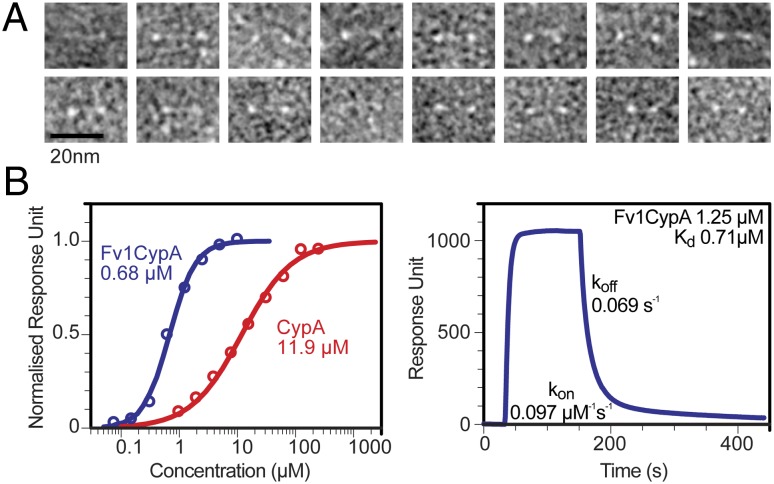
To obtain an independent measurement of the shape and dimensions, negatively stained Fv1Cyp particles were analyzed by transmission electron microscopy. In agreement with the SAXS analysis, the images (Fig. 3A and Fig. S5A) showed elongated particles with globular domains separated by greater than 100 Å (Fig. S5B). Thin connections of density between the globular domains were also observed, consistent with a narrow flexible region between the C-terminal globular features that include the CypA domains.
Fig. 3.
EM and Biacore analysis of Fv1Cyp. (A) Gallery of negatively stained EM images of Fv1Cyp. Images are the sums of three different images of the same particle recorded in 0.5-μm steps (Fig. S5A), revealing thin connections between the globular domains. (B) The interaction of Fv1Cyp and monomeric CypA with a HIV-1 CA-p2 lattice immobilized on a Ni-NTA tagged lipid monolayer analyzed by Biacore. Equilibrium affinity measurements derived from sensorgram plateau values (Left) show Fv1Cyp exhibits an ∼18-fold higher affinity. Analysis of the kinetics for individual sensorgram binding curves gives comparable values for equilibrium dissociation constants (Right).
Surface Plasmon Resonance Analysis of Fv1Cyp–CA Interactions.
Given the arrangement of the CypA recognition domains within Fv1Cyp the effect of dimerization on recognition and affinity for the capsid lattice was investigated using Biacore surface plasmon resonance (SPR). To do this a lattice of HIV-1 capsid was first constructed on the surface of hydrophobic Biacore chip. We have previously shown that the capsid protein from MLV forms a capsid-like hexagonal array when immobilized on lipid nanotubes by a C-terminal his-tag (27). Repeating these experiments with the CA of HIV-1 demonstrated that a construct containing CA followed by the P2 spacer peptide (CA-P2) also coated lipid nanotubes and produced regular arrays (Fig. S6). To test binding of Fv1Cyp to an intact capsid lattice we reproduced this lattice by doping 1,2-dioleoyl-sn-glycero-3-phosphocholine liposomes with 30% 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (DGS-NTA) to create a Ni-NTA containing lipid layer on a hydrophobic Biacore SPR HPA chip. HIV-1 CA-P2 was then loaded onto the surface saturating the Ni2+ sites on the lipid monolayer, producing a stable baseline. To evaluate our approach we first tested the binding of free monomeric CypA. To account for potential nonspecific interaction with the lipid layer and SPR chip a second channel containing B-tropic MLV was included as a non-CypA binding control. Analysis of equilibrium binding curves from CypA results in a dissociation constant of 11.9 µM (Fig. 3B), in good agreement with the reported affinity in solution from previous experiments (35, 36). The experiment was then repeated with Fv1Cyp over a concentration range of 75 nM–10 µM. Analysis of these equilibrium binding curves gave an equilibrium dissociation constant of 0.68 µM. As expected for a weak interaction, the CypA-binding kinetics were rapid and could not be analyzed reliably. However, kinetic analysis of the Fv1-Cyp binding curves yielded an on rate of 0.097 µM−1⋅s−1 and off rate of 0.069 s−1 for the 1.25 µM injection of Fv1Cyp, resulting in a Kd of 0.71 µM (Fig. 3B). These data reveal a dimerization-conferred avidity where dimerization and spacing of CypA monomers by the Fv1NtD results in a 17.5-fold increase in apparent affinity of CypA for an HIV-1 CA array.
Discussion
Recognition of the retroviral capsid protein in the context of the intact capsid lattice is required for restriction by the postentry RFs Fv1 and Trim5α. Although not related by sequence or homology both proteins share significant similarities in their mode of action and recognition of the retroviral CA. Key features of Fv1 and Trim5α are the formation of dimers through N-terminal coiled-coil regions and C-terminal specificity domains that determine the repertoire of viruses that can be restricted, B30.2 in Trim5α (12, 13) and the Fv1CtD of Fv1n and Fv1b (6). In addition, the Trim5α N-terminal region also contains RING and B-box domains that promote higher-order associations required for recognition and stable binding of the retroviral capsid (37). It is likely that Fv1 also contains one or more association-promoting regions but located in the CtD (7). To understand how the N-terminal regions of Fv1 and Trim5α contribute to recognition of the retroviral capsid we examined the size and shape of the self-association domains by SAXS and determined the structure of the N-terminal B-box and coil region from Trim5α. These data clearly show that the N-terminal domains of Fv1 and Trim5α comprise antiparallel coiled-coil proteins that form elongated dimers with dimensions sufficient to span the interhexamer distances on the outer surface of the retroviral capsid.
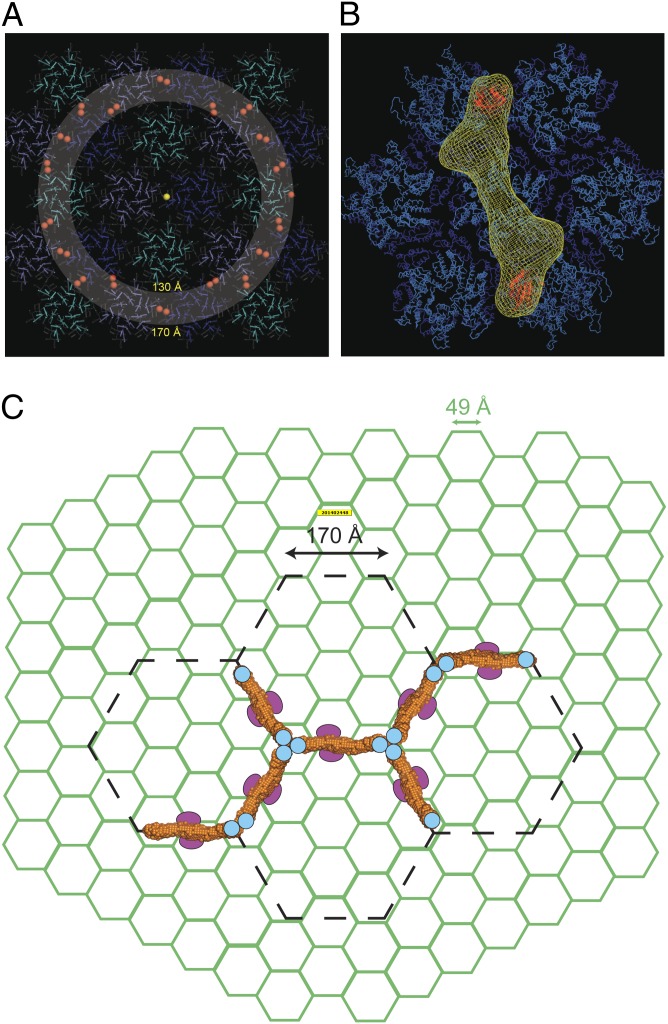
In the CypA-containing factors, Fv1Cyp and TrimCyp, the interaction with the HIV-1 capsid is mediated although binding of CypA domains to residues in the CA-Cyp loops displayed on the outer surface of the capsid (17, 38). Moreover, mutations in the HIV-Cyp loop that diminish or abolish CypA binding restore cell permissiveness by making HIV-1 resistant to restriction by the CypA-containing factors (39). Our data reveal the Fv1Cyp molecule contains two globular CypA-containing domains around 150 Å apart separated by a narrow spacer observed both directly in negative-stain EM experiments (Fig. 3A) and as the 2° maxima in the distance distribution function derived from SAXS data (Fig. 1C). Calculation of the distribution of interatomic vectors between pairs of residues residing in HIV-1 Cyp loops across the surface of the reconstructed HIV CA shell (38) reveals that pair distances of 130–170 Å are highly represented (Fig. 4A). This striking correlation between the lobe spacing in Fv1Cyp and the spacing of Cyp loops in the HIV-1 capsid suggests a model where the extended Fv1 and Trim5α dimers have evolved to facilitate the spacing of specificity domains to optimize their interaction with many distal binding sites on the retroviral CA. In this model, an Fv1Cyp dimer with a CypA domain located on one CA is able to sample greater than 30 other Cyp loops located in adjacent or more distal CA hexamers at ∼150 Å spacing to make a productive interaction (Fig. 4B). This notion is further supported by our Biacore analysis of the Fv1Cyp–CA lattice interaction that reveals strong avidity effects likely resulting from the many ways Fv1Cyp can simultaneously associate with pairs of Cyp-loop binding sites on the CA lattice. Moreover, it has been demonstrated that TrimCyp efficiently inhibits HIV-1 even when only 25% of the CA in the capsid is competent to bind to CypA (40), suggesting that only one to two Cyp-loop binding sites per hexamer are actually required for a productive RF engagement.
Fig. 4.
RF CA–lattice interactions. (A) A layer of CA lattice taken from 3J34. Red spheres mark the Cyp loops and the gray region highlights those that are spaced at 130–170 Å from the central Cyp loop (yellow) and accessible to bind the second CypA of Fv1Cyp. (B) The SAXS envelope of Fv1Cyp dimer is positioned with one CypA domain located on the Cyp loop of a central hexamer and the second spaced between 140–150 Å located on the Cyp loop of an adjacent CA hexamer. (C) A schematic representation of the HIV-CA lattice (green) with the Trim5-21R lattice, from ref. 37, is overlaid as dashed lines (black). The SAXS model for the RhT5 (88–296) EK/RD dimer shown in orange surface representation has dimensions equating to the length of a single edge of the hexamer in the Trim5-21R lattice and sufficient to span adjacent hexamers in the CA lattice. The B-boxes are shown as cyan circles at the interface between Trim5α dimers and the B30.2 domains are represented by the ovals toward the Trim5-21R twofold axis.
The SAXS analysis of RhT5 (88–296) EK/RD and crystal structure of the RhT5-T4L demonstrates that, like Fv1, Trim5α also comprises an antiparallel extended dimer but with the B-box effector domains displayed at either end. Examination of the B-box spacing gives a distance of 172 Å (distance between the equivalent Zn atoms). Notably, this is equivalent to one edge of the Trim5α hexagonal lattice observed in previous EM studies and proposed to contain four Trim5α molecules arranged as two pairs of parallel dimers (37). Our data now suggest an alternative model (Fig. 4C) that places a single antiparallel Trim5α dimer on each hexagon edge with B-box–(α3–α4) and RING domains located at each threefold vertex and B30.2 domains connected by L2-E regions located toward the dimer twofold axis. Given that the 170-Å length constraint is already accommodated by the B-box and coil regions, the model predicts the B30.2 and RING domains would locate outside the plane of the assembly, plausibly with the B30.2 domain below facing toward the capsid lattice and RING domain above to be accessible to components of the ubiquitination machinery. This model is supported by the recently published structure of the coiled-coil domain from the related protein Trim25 (41). This structure shows the same elongated antiparallel assembly of two central helices with low writhe, and the similarity with the Trim5α structure now suggests antiparallel flattened coil structures are likely a general feature of the whole Trim family. However, in Trim25 structure the B-boxes are absent, so it is unclear whether the packing we observe between B-box and opposing (α3–α4) regions is also found in other Trims or is specific to Trim5α to support a B-box conformation required for Trim5α higher-order assembly and capsid recognition.
At the present time we can only speculate about the positioning of the CA binding domains of Trim5α and Fv1. However, it seems reasonable to hypothesize that the Fv1 C-terminal domains would locate similarly to the CypAs in Fv1Cyp. In this case as well as recognition of the MLV capsid further Fv1-CtD self-association could mediate higher-order assembly similar to the B-boxes in Trim5α in Fig. 4C. In contrast, given the location of the Trim5α L2-E C terminus in the crystal structure and the electron density observed at the center of the coil in the Trim5α hexagonal assembly (37) it is likely the B30.2 domains locate close to the twofold axis of the coiled-coil. In support of this notion, alignment of the Trim5α B30.2 structure (PDB ID code 43BN) with equivalent residues in the short helix α5 of Trim5α BCC places the SPRY domain at the twofold axis of the coiled-coil and orients the variable capsid recognition loops in an appropriate position to recognize the retroviral capsid (Fig. S7).
In TrimCyp factors, there are additional residues resulting from CypA fusion into helix α5 (18). These form an unstructured linker between α5 and T4L in the Rh5-T4L structure. Modeling of TrimCyp by replacement of the T4L with CypA (Fig. S7) then positions the CypA domains around 120 Å apart and given the local disorder around α5 affords them a large degree of conformational flexibility. In this way, TrimCyp would have the capacity to sample multiple hexamers, as seen in Fv1Cyp, for a productive interaction with the HIV-1 capsid lattice.
In summary, recognition of the retroviral capsid by Trim5α and Fv1 factors is mediated by a combination of avidity generated by primary dimerization/higher-order association and the weak intrinsic CA-binding site affinity of individual B30.2 or CypA domains. The elongated antiparallel structure of these factors plays a key role in providing the optimal spacing and orientation for specificity domains to sample and promote a productive interaction with the CA lattice and for RING and B-box effector domains to recruit the components of ubiquitination and immune innate signaling pathways. Notably, comparison of the structure, evolution, and mode of action of Trim5α and the antiviral proteins MxA/MxB (42) also reveals a number of common design features. These include (i) a virus interaction domain showing signatures of positive selection, (ii) one or more multimerization domains, and (iii) an effector domain. Together they allow recognition and restriction of multiple agents with repeated structures characteristic of viruses and may describe the properties of multiple IFN-stimulated genes.
Materials and Methods
Protein Expression and Purification.
Fv1Ntd (residues 20–200 of Fv1) and Fv1Cyp consisting of Fv1-Ntd fused to CypA were expressed with N-terminal His-tags in Escherichia coli BL21(DE3). HIV CA-P2 and MLV CA were expressed with C-terminal His-tags. Proteins were purified by immobilized nickel affinity and SEC. CypA was expressed as a GST fusion in E. coli BL21(DE3) and purified by glutathione affinity and SEC. RhT5 (88–296) EK/RD and Rh5-T4L were expressed from pET47b with an N-terminal His-tags and purified by immobilized nickel affinity chromatography and SEC. For structure determination, selenium was incorporated into Rh5-T4L by supplementing culture media with seleno-methionine combined with inhibition of methionine biosynthesis. Details of protein expression constructs and protein production procedures are provided in SI Materials and Methods.
SEC-MALLS.
SEC-MALLS was used to determine the solution molecular weight of Fv1NtD, Fv1Cyp, and RhT5 (88–296) EK/RD and to assess protein heterogeneity. Data for Fv1NtD and Fv1Cyp were recorded using a Superdex 200 10/300 GL column mounted on a Jasco HPLC. Scattered light intensities and protein concentration data were measured using a DAWN HELEOS laser photometer and an OPTILAB-rEX differential refractometer (Wyatt Instruments). SEC-MALLS analysis of RhT5 (88–296) EK/RD was carried out using Superdex 200 10/300 GL column, on a Dionex HPLC with a PSS SLD7000 7 angle MALLS detector and Shodex RI-101 differential refractive index detector. Details of experimental procedures and data analysis are provided in SI Materials and Methods.
SAXS.
SAXS data for Fv1NtD and Fv1Cyp were recorded on European Synchrotron Radiation Facility beamline 14-3 on a PILATUS 1M detector at a wavelength of 0.931Å and camera length of 2.43 m covering a momentum transfer of 0.006 < q < 0.6Å−1 [q = 4πsin(θ)/λ]. Data for RhT5 (88-296) EK/RD were collected at the Australian Synchrotron SAXS/WAXS beamline at a wavelength of 1.13 Å with a camera length of 3 m covering a momentum transfer range of 0.0 < q < 0.3Å−1 [q = 4πsin(θ)/λ]. Details of sample preparation, experimental procedures, data processing, and analysis are provided in SI Materials and Methods.
Crystallization and Structure Determination.
Crystals of Rh5-T4L were grown by vapor diffusion at 18 °C in drops consisting of 1 µL of Rh5-T4L at a concentration of 3 mg/mL and 0.5 µL of reservoir solution (10% PEG8000, 20% ethylene glycol, and 0.1 M bicine/Tris⋅HCl, pH 8.5). X-ray diffraction data were collected on Australian Synchrotron beamline MX2 at a wavelength of 0.9793 Å. Details of data processing, structure determination, and refinement are provided SI Materials and Methods.
Negative-Stain Electron Microscopy.
Samples were absorbed to carbon-coated grids and negatively stained with 1% sodium silicotungstate, pH 7.0. The grids were viewed with an FEI Spirit TWIN microscope operated at 120 kV with a tungsten filament source. Images were recorded on an Eagle 2K camera (FEI) at a magnification of 52K (4.3Å per pixel) using a range of defocuses.
SPR.
Binding of Fv1Cyp and CypA to a preformed HIV-1 CA lattice was performed using SPR. Data were recorded on a Biacore 2000 instrument using the hydrophobic HPA biosensor (GE Healthcare). Details of sample and chip preparation, experimental procedure, and data analysis are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We gratefully acknowledge the European Synchrotron Radiation Facility (Grant MX1153) and the New Zealand synchrotron group for access to small-angle X-ray scattering beamlines. This work was supported by UK Medical Research Council Grants U117565647 (to I.A.T.), U117581334 (to P.B.R.), and U117512710 (to J.P.S.) and by a Rutherford Discovery Fellowship from the New Zealand government administered by the Royal Society of New Zealand (to D.C.G.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4TN3).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402448111/-/DCSupplemental.
References
- 1.Stoye JP. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol. 2012;10(6):395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- 2.Zheng YH, Jeang KT, Tokunaga K. Host restriction factors in retroviral infection: Promises in virus-host interaction. Retrovirology. 2012;9:112. doi: 10.1186/1742-4690-9-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanz-Ramos M, Stoye JP. Capsid-binding retrovirus restriction factors: Discovery, restriction specificity and implications for the development of novel therapeutics. J Gen Virol. 2013;94(Pt 12):2587–98. doi: 10.1099/vir.0.058180-0. [DOI] [PubMed] [Google Scholar]
- 4.Lilly F. Fv-2: Identification and location of a second gene governing the spleen focus response to Friend leukemia virus in mice. J Natl Cancer Inst. 1970;45(1):163–169. [PubMed] [Google Scholar]
- 5.Hartley JW, Rowe WP, Huebner RJ. Host-range restrictions of murine leukemia viruses in mouse embryo cell cultures. J Virol. 1970;5(2):221–225. doi: 10.1128/jvi.5.2.221-225.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop KN, Bock M, Towers G, Stoye JP. Identification of the regions of Fv1 necessary for murine leukemia virus restriction. J Virol. 2001;75(11):5182–5188. doi: 10.1128/JVI.75.11.5182-5188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop KN, et al. Characterization of an amino-terminal dimerization domain from retroviral restriction factor Fv1. J Virol. 2006;80(16):8225–8235. doi: 10.1128/JVI.00395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan Y, Buckler-White A, Wollenberg K, Kozak CA. Origin, antiviral function and evidence for positive selection of the gammaretrovirus restriction gene Fv1 in the genus Mus. Proc Natl Acad Sci USA. 2009;106(9):3259–3263. doi: 10.1073/pnas.0900181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson WE, Sawyer SL. Molecular evolution of the antiretroviral TRIM5 gene. Immunogenetics. 2009;61(3):163–176. doi: 10.1007/s00251-009-0358-y. [DOI] [PubMed] [Google Scholar]
- 10.Reymond A, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20(9):2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Javanbakht H, et al. Characterization of TRIM5alpha trimerization and its contribution to human immunodeficiency virus capsid binding. Virology. 2006;353(1):234–246. doi: 10.1016/j.virol.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 12.Ohkura S, Yap MW, Sheldon T, Stoye JP. All three variable regions of the TRIM5alpha B30.2 domain can contribute to the specificity of retrovirus restriction. J Virol. 2006;80(17):8554–8565. doi: 10.1128/JVI.00688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song B, et al. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits lineage-specific length and sequence variation in primates. J Virol. 2005;79(10):6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 15.Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;15(1):73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 16.Song H, et al. A single amino acid of the human immunodeficiency virus type 2 capsid affects its replication in the presence of cynomolgus monkey and human TRIM5alphas. J Virol. 2007;81(13):7280–7285. doi: 10.1128/JVI.00406-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamble TR, et al. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87(7):1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 18.Stoye JP, Yap MW. Chance favors a prepared genome. Proc Natl Acad Sci USA. 2008;105(9):3177–3178. doi: 10.1073/pnas.0800667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malfavon-Borja R, Wu LI, Emerman M, Malik HS. Birth, decay, and reconstruction of an ancient TRIMCyp gene fusion in primate genomes. Proc Natl Acad Sci USA. 2013;110(7):E583–E592. doi: 10.1073/pnas.1216542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yap MW, Mortuza GB, Taylor IA, Stoye JP. The design of artificial retroviral restriction factors. Virology. 2007;365(2):302–314. doi: 10.1016/j.virol.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006;103(14):5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortuza GB, et al. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature. 2004;431(7007):481–485. doi: 10.1038/nature02915. [DOI] [PubMed] [Google Scholar]
- 23.Ohkura S, et al. Novel escape mutants suggest an extensive TRIM5α binding site spanning the entire outer surface of the murine leukemia virus capsid protein. PLoS Pathog. 2011;7(3):e1002011. doi: 10.1371/journal.ppat.1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarthy KR, et al. Gain-of-sensitivity mutations in a Trim5-resistant primary isolate of pathogenic SIV identify two independent conserved determinants of Trim5α specificity. PLoS Pathog. 2013;9(5):e1003352. doi: 10.1371/journal.ppat.1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dodding MP, Bock M, Yap MW, Stoye JP. Capsid processing requirements for abrogation of Fv1 and Ref1 restriction. J Virol. 2005;79(16):10571–10577. doi: 10.1128/JVI.79.16.10571-10577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forshey BM, Shi J, Aiken C. Structural requirements for recognition of the human immunodeficiency virus type 1 core during host restriction in owl monkey cells. J Virol. 2005;79(2):869–875. doi: 10.1128/JVI.79.2.869-875.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilditch L, et al. Ordered assembly of murine leukemia virus capsid protein on lipid nanotubes directs specific binding by the restriction factor, Fv1. Proc Natl Acad Sci USA. 2011;108(14):5771–5776. doi: 10.1073/pnas.1100118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biris N, Tomashevski A, Bhattacharya A, Diaz-Griffero F, Ivanov DN. Rhesus monkey TRIM5α SPRY domain recognizes multiple epitopes that span several capsid monomers on the surface of the HIV-1 mature viral core. J Mol Biol. 2013;425(24):5032–5044. doi: 10.1016/j.jmb.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langelier CR, et al. Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication. J Virol. 2008;82(23):11682–11694. doi: 10.1128/JVI.01562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Sodroski J. The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J Virol. 2008;82(23):11495–11502. doi: 10.1128/JVI.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, et al. Virus-specific effects of TRIM5α(rh) RING domain functions on restriction of retroviruses. J Virol. 2013;87(13):7234–7245. doi: 10.1128/JVI.00620-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diaz-Griffero F, et al. A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction. J Virol. 2009;83(20):10737–10751. doi: 10.1128/JVI.01307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mylonas E, Svergun DI. Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering. J Appl Cryst. 2007;40(s1):s245–s249. [Google Scholar]
- 34.Svergun DI, Koch MHJ. Small-angle scattering studies of biological macromolecules in solution. Rep Prog Phys. 2003;66(10):1735–1782. [Google Scholar]
- 35.Goldstone DC, et al. Structural and functional analysis of prehistoric lentiviruses uncovers an ancient molecular interface. Cell Host Microbe. 2010;8(3):248–259. doi: 10.1016/j.chom.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 36.Price AJ, et al. Active site remodeling switches HIV specificity of antiretroviral TRIMCyp. Nat Struct Mol Biol. 2009;16(10):1036–1042. doi: 10.1038/nsmb.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ganser-Pornillos BK, et al. Hexagonal assembly of a restricting TRIM5alpha protein. Proc Natl Acad Sci USA. 2011;108(2):534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao G, et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497(7451):643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430(6999):569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 40.Shi J, Friedman DB, Aiken C. Retrovirus restriction by TRIM5 proteins requires recognition of only a small fraction of viral capsid subunits. J Virol. 2013;87(16):9271–9278. doi: 10.1128/JVI.00713-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez JG, et al. The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer. Proc Natl Acad Sci USA. 2014;111(7):2494–2499. doi: 10.1073/pnas.1318962111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haller O. Dynamins are forever: MxB inhibits HIV-1. Cell Host Microbe. 2013;14(4):371–373. doi: 10.1016/j.chom.2013.10.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.