Significance
A major tumor suppressor, phosphatase and tensin homolog (PTEN), dephosphorylates the potent tumorigenic signaling lipid phosphatidylinositol (3,4,5)-trisphosphate (PIP3) at the plasma membrane. However, most PTEN is located in the cytosol and only transiently associated with the membrane to convert PIP3 to PIP2. Here, we developed a platform using a heterologous expression system, in which a library of randomly mutated human PTEN is expressed and localization of the protein is visually inspected in Dictyostelium. This unbiased approach revealed a membrane-binding regulatory interface that is negatively regulated by a phosphorylated C-terminal tail. Based on the mechanistic information, we created an enhanced PTEN that dramatically represses PIP3 signaling. Thus, PTEN activation readjusts PIP3 signaling in tumor cells and serves as a target for anticancer therapies.
Keywords: protein engineering, PI3 kinase signaling, membrane localization, chemotaxis, protein interaction
Abstract
The signaling lipid phosphatidylinositol (3,4,5)-trisphosphate (PIP3) is a key regulator of cell proliferation, survival, and migration and the enzyme that dephosphorylates it, phosphatase and tensin homolog (PTEN), is an important tumor suppressor. As excess PIP3 signaling is a hallmark of many cancers, its suppression through activation of PTEN is a potential cancer intervention. Using a heterologous expression system in which human PTEN-GFP is expressed in Dictyostelium cells, we identified mutations in the membrane-binding regulatory interface that increase the recruitment of PTEN to the plasma membrane due to enhanced association with PI(4,5)P2. We engineered these into an enhanced PTEN (ePTEN) with approximately eightfold increased ability to suppress PIP3 signaling. Upon expression in human cells, ePTEN decreases PIP3 levels in the plasma membrane; phosphorylation of AKT, a major downstream event in PIP3 signaling; and cell proliferation and migration. Thus, the activation of PTEN can readjust PIP3 signaling and may serve as a feasible target for anticancer therapies.
Phosphatidylinositol (3,4,5)-trisphosphate (PIP3) is a potent second messenger that drives many biological processes, such as cell growth, survival, and migration (1, 2). In many cancers, PIP3 levels are elevated due to mutations that either elevate the activity of phosphoinositide 3-kinases (PI3Ks) or decrease that of tumor suppressor phosphatase and tensin homolog (PTEN) (3–5). Although inhibition of PI3Ks has been extensively tried as a cancer drug target, activation of PTEN has been rarely studied (6). As PTEN is mainly located in the cytosol and its PIP3 phosphatase activity is suppressed at this location (7, 8), recruiting more PTEN to the plasma membrane and thereby stimulating its lipid phosphatase activity would seem to be an effective method to repress abnormal PIP3 levels in cancer cells.
PTEN comprises an N-terminal “PIP2-binding” motif, globular catalytic and C2 domains, and a C-terminal tail (8–10). Positively charged residues in the PIP2-binding and C2 domains have been proposed to recruit PTEN to the plasma membrane through associations with negatively charged head groups of membrane lipids (11–13). The C-terminal tail is thought to fold back and bind to the membrane-binding regions, maintaining the majority of PTEN in the cytoplasm (11, 14, 15). This intramolecular inhibition is controlled by phosphorylation of four serine/threonine residues in the tail domain. A PTEN mutant that carries an alanine substitution in the phosphorylation sites of the C-terminal tail (termed PTENA4) increases the membrane association of PTEN. However, most PTENA4 is still present in the cytoplasm, suggesting that the A4 mutations may not completely liberate the membrane-binding sites from inhibition by the tail.
To decipher the mechanisms underlying the membrane association of PTEN, we developed a visual screen for the localization of human PTEN expressed in Dictyostelium cells. PTEN is evolutionarily conserved, and human PTEN can functionally replace Dictyostelium PTEN (16–19). Using this heterologous expression system, we identified a membrane-binding regulatory interface in PTEN, consisting of regions of the catalytic domain and the CBR3 and Cα2 loops of the C2 domain (20). In the current study, we introduce multiple mutations in the membrane-binding regulatory interface that completely release the inhibitory effects of the tail, generating a synthetic enzyme, referred to as enhanced PTEN (ePTEN), with greatly increased membrane localization and PIP3 phosphatase activity. Our findings demonstrate that activation of PTEN is a feasible therapeutic strategy for cancers with increased PIP3 signaling.
Results
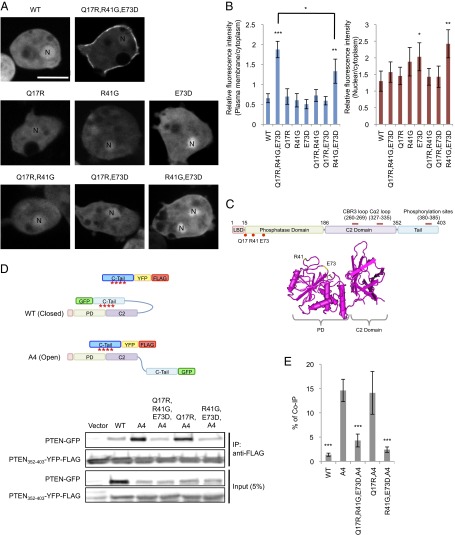
By expanding our visual screen for the membrane recruitment of human PTEN, we isolated another mutant, PTENQ17R, R41G, E73D, which showed more than a twofold increase in its association with the plasma membrane in Dictyostelium cells (Fig. 1 A and B). Q17 is located near the PIP2-binding domain, whereas R41 and E73 are located in the phosphatase domain and exposed on the surface of PTEN (Fig. 1C). All three substitutions are necessary for the robust membrane localization, although PTENR41G, E73D modestly promotes membrane association with additional nuclear accumulation. To determine whether the membrane localization of PTENQ17R, R41G, E73D is due to release of the core region (amino acid residues 1–352) from the inhibitory C-terminal tail domain (residues 352–403), we performed an “in trans” interaction assay. We assessed the binding of PTEN-GFP to a separate tail domain fused to FLAG after immunoprecipitation with anti-GFP antibodies (Fig. 1D). As previously reported, whereas PTEN-GFP did not interact with PTEN352–403-FLAG, PTENA4-GFP showed a strong interaction (Fig. 1 D and E). In contrast, PTENQ17R, R41G, E73D, A4 did not bind to PTEN352–403-FLAG (Fig. 1E). Further analyses showed that substitution of R41G and E73D was sufficient to block the core–tail interaction (Fig. 1E).
Fig. 1.
PTENQ17R, R41G and E73D-GFP is localized at the plasma membrane. (A) Dictyostelium cells expressing GFP fused to PTEN, PTENQ17R,R41G,E73D, PTENQ17R, PTENR41G, PTENE73D, PTENQ17R,R41G, PTENQ17RE73D, and PTENR41G,E73D were viewed by fluorescence microscopy. Nuclei (N) are indicated. (Scale bar, 10 µm.) (B) Intensity of GFP at the plasma membrane was quantified relative to that in the cytosol. Values represent the mean ± SD (n ≥ 15). (C) The domain structure of PTEN and the position of mutated residues Q17, R41, and E37 are shown. The 3D structure of PTEN suggests that R41 and E37 are at the surface of PTEN (34) (www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=11638). (D) Interaction of the C-terminal tail domain of PTEN with full-length PTEN was assessed by pull-down assay. PTEN maintains associations between the core and tail regions in the same molecule (close conformation) through phosphorylation of the tail (red asterisks). In contrast, the core region of PTENA4 dissociates from the tail (open conformation) and interacts with an exogenously added tail domain. Whole-cell lysates expressing PTEN-GFP, PTENA4-GFP, PTENQ17R,R41G,E73D,A4-GFP, PTENQ17R,A4-GFP, or PTENR41G,E73D,A4-GFP were incubated with PTEN352–403-YFP-FLAG. PTEN352–403-YFP-FLAG was immunoprecipitated with beads coupled to anti-FLAG antibodies. Bound fractions (immunoprecipitates, IP) were analyzed with antibodies to GFP and FLAG. (E) Band intensity was quantified (n = 3).
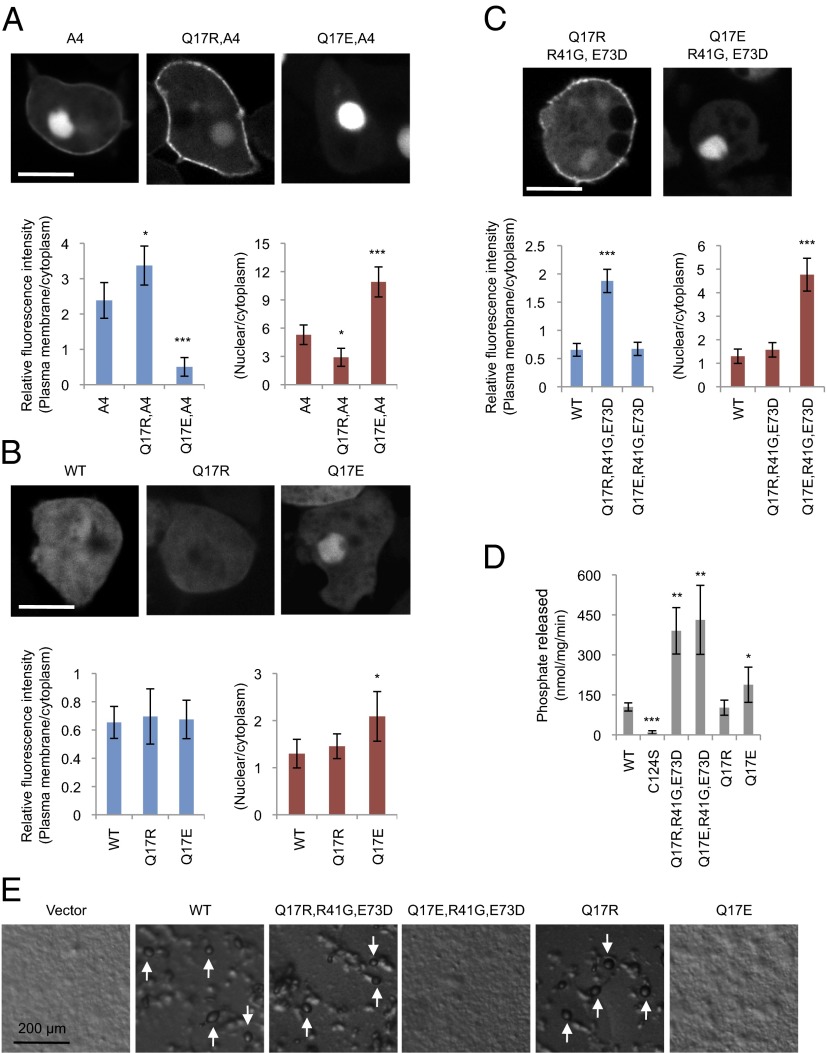
Q17 is located adjacent to a cluster of positively charged amino acids in the PIP2-binding domain (amino acid residues 6–15). It has been proposed that these cationic residues interact with anionic phospholipids, such as PIP2, and are masked by the inhibitory tail domain (10, 12, 16, 21). In addition, amino acid residues 13–15 are thought to function as a nuclear localization signal (13, 22, 23). As Q17R itself did not stimulate membrane recruitment, we combined Q17R with A4 and found increases in the membrane localization of PTENA4, Q17R (Fig. 2 A and B), suggesting that the effect of Q17R is seen only when the C-terminal tail is released. To test whether Q17R enhances the localization of PTEN at the plasma membrane by increasing positive charges in the PIP2-binding domain, we replaced Q17 with a negatively charged residue, glutamate (Q17E). PTENQ17E, R41G, E73D showed no accumulation at the plasma membrane but was highly localized in the nucleus (Fig. 2C). Q17E alone only modestly stimulated the nuclear localization of PTEN (Fig. 2B). A PTENA4, Q17E displayed a stronger nuclear localization, suggesting that the charge of residue 17 strongly influences the localization of PTEN between the plasma membrane and nucleus, when the tail is dissociated from the core region of PTEN. In contrast, substitution of Q17 with either R or E did not inhibit the PIP3 phosphatase activity of PTEN (Fig. 2D). Furthermore, PTENQ17R, R41G, E73D and PTENQ17R, but neither PTENQ17E, R41G, E73D nor PTENQ17E, rescued developmental defects in pten− Dictyostelium cells, showing the functional importance of the ability of PTEN to associate with the plasma membrane (Fig. 2E).
Fig. 2.
Q17R promotes PTEN membrane association in PTENA4 and PTENR41,E73D. (A–C) Dictyostelium cells expressing the indicated forms of PTEN-GFP were observed by fluorescence microscopy. (Scale bar, 10 µm.) Intensity of GFP at the plasma membrane was quantified relative to that in the cytosol. Values represent the mean ± SD (n ≥ 15). (D) PTEN-GFP, PTENC124S-GFP, PTENQ17R-GFP, and PTENQ17E-GFP were immunopurified from Dictyostelium cells, and phosphatase activities were measured (n ≥ 3). (E) PTEN-null Dictyostelium cells expressing different PTEN-GFP constructs were starved for 36 h to induce differentiation into fruiting bodies (white arrows).
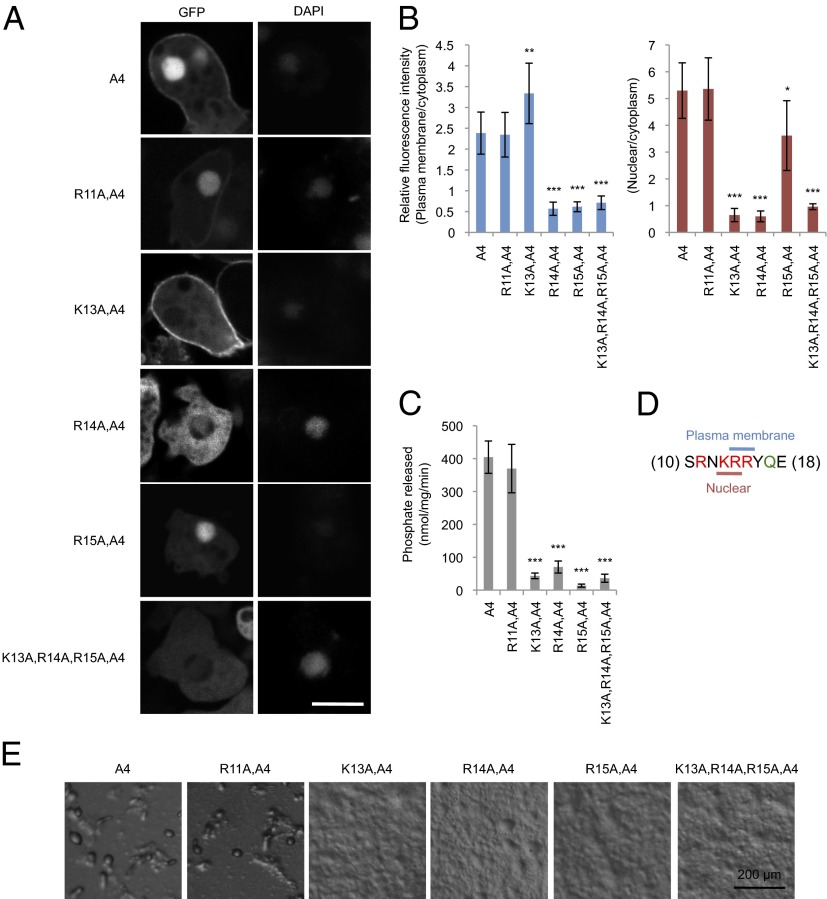
To examine the role of positively charged residues in the PIP2-binding domain for localization at the plasma membrane and nucleus, we individually substituted R11, K13, R14, and R15 with alanine in PTENA4. The effects of each mutation were distinct (Fig. 3 A and B): R11A showed no effect on the localization of PTENA4-GFP, K13A blocked nuclear localization but increased membrane association, R14A inhibited both nuclear and membrane localization, and R15A blocked only plasma membrane localization. These results suggest that K13 and R14 are part of the nuclear localization signal, whereas R14 and R15 target PTEN to the plasma membrane (Fig. 3D). Finally, K13A, R14A, and R15A abolished the ability of PTENA4 to rescue developmental defects in pten− Dictyostelium cells (Fig. 3E), likely due to decreases in PIP3 phosphatase activity (Fig. 3C).
Fig. 3.
Positively charged residues in the lipid-binding domain control membrane and nuclear localization of PTEN. (A–C) Dictyostelium cells expressing the indicated forms of PTEN-GFP were observed by fluorescence microscopy. (Scale bar, 10 µm.) (B) Intensity of GFP at the plasma membrane was quantified relative to that in the cytosol. Values represent the mean ± SD (n ≥ 15). (D) The indicated PTEN-GFP proteins were immunopurified from Dictyostelium cells, and phosphatase activities were measured (n ≥ 3). (E) PTEN-null Dictyostelium cells expressing different PTEN-GFP constructs were starved for 36 h to induce differentiation into fruiting bodies.
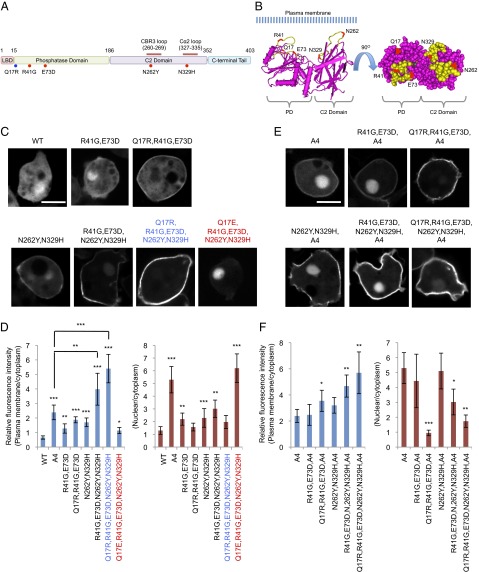
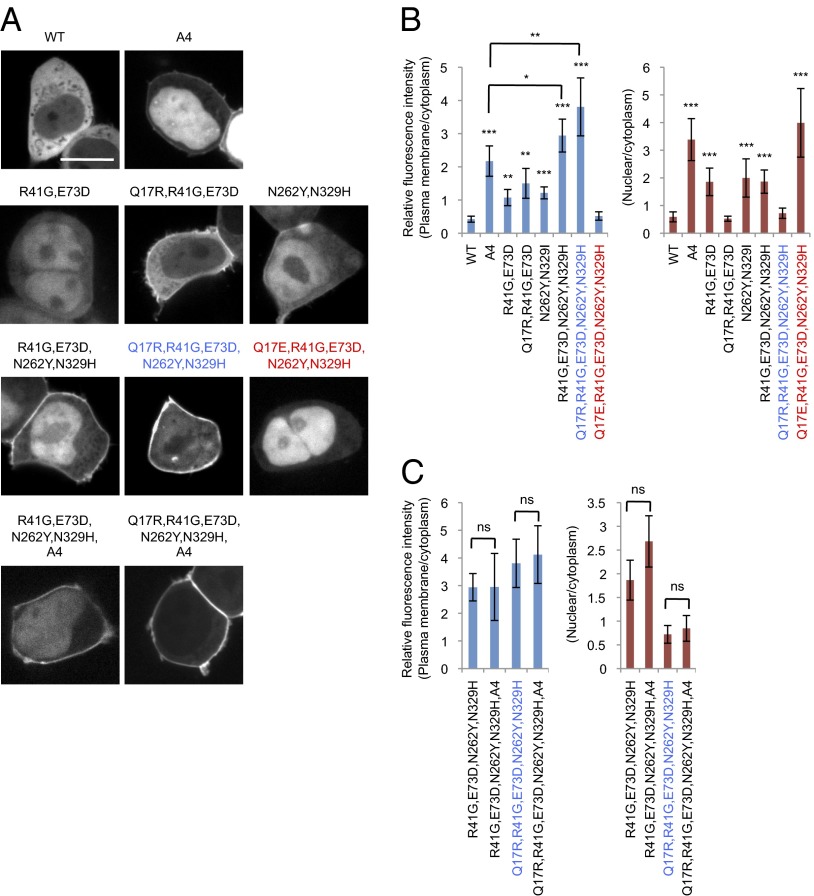
Next, we sought to engineer PTEN to increase its ability to associate with the plasma membrane. Because Q17R promotes the membrane localization of PTEN mutants, such as PTENR41G, E73D, which partially blocks the interaction with the C-terminal tail, we tested whether further release of the inhibition is possible with additional mutations. We have previously isolated a PTEN mutant, PTENN262Y, N329H, which also increases the membrane association of PTEN (20). These residues are located in the CBR3 loop and Cα2 loop of the C2 domain, respectively. Substitutions N262Y and N329H weaken the association of the core region with the tail in the in trans interaction assay but do not inhibit the PIP3 phosphatase activity (20). The structure of PTEN suggests that all four mutations (R41G, E73D, N262Y, and N329H) are located on the surface on the same side of the molecule (Fig. 4 A and B). Indeed, whereas PTENA4, PTENQ17R, R41G, E73D, and PTENN262Y, N329H showed twofold increases in membrane association compared with PTEN, PTENQ17R, R41G, E73D, N262Y, N329H showed an eightfold increase (Fig. 4 C and D). We designated PTENQ17R, R41G, E73D, N262Y, N329H as ePTEN. The addition of the A4 mutation to ePTEN did not further increase its membrane association (Fig. 4 E and F), suggesting that the core–tail interaction may be absent. Furthermore, ePTEN did not show nuclear localization. Remarkably, changing Q17R to Q17E (PTENQ17E, R41G, E73D, N262Y, N329H) almost completely blocked membrane recruitment and targeted the protein exclusively to the nucleus (nPTEN) (Fig. 4 C and D).
Fig. 4.
ePTEN-GFP (PTENQ17R, R41G, E73D, N262Y, N329H-GFP) is highly enriched at the plasma membrane. (A) The domain structure of PTEN and the position of mutations are shown. Mutations that affect membrane binding and tail interaction are colored in blue and red, respectively. (B) The 3D structure of PTEN suggests that Q17, R41, E37, N329, and N262Y form an interface that binds to the plasma membrane at the surface of PTEN (34) (www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=11638). (C–F) Dictyostelium cells expressing different forms of PTEN-GFP (C) and PTENA4-GFP (E) were observed by fluorescence microscopy. (Scale bar, 10 µm.) (D and F) Intensity of GFP at the plasma membrane was quantified relative to that in the cytosol. Values represent the mean ± SD (n ≥ 15).
To validate our findings in human cells, we expressed different PTEN-GFP constructs in HEK293T cells and examined their subcellular localization. Consistent with the behavior in Dictyostelium cells, ePTEN showed an eightfold increased association with the plasma membrane compared with PTEN and no nuclear accumulation (Fig. 5 A and B). Furthermore, nPTEN had no detectable membrane localization and showed strong nuclear accumulation (Fig. 5 A and B). Moreover, introduction of A4 into ePTEN did not show an additional increase in either localization. As was observed in Dictyostelium cells, PTENA4, PTENQ17R, R41G, E73D, and PTENN262Y, N329H showed two- to fourfold increases in membrane association (Fig. 5 A and B).
Fig. 5.
ePTEN-GFP (PTENQ17R, R41G, E73D, N262Y, N329H-GFP) is highly enriched at the plasma membrane in HEK293T cells. (A) HEK293T cells expressing the indicated forms of PTEN-GFP were observed by fluorescence microscopy. (Scale bar, 20 µm.) (B and C) Intensity of GFP at the plasma membrane was quantified relative to that in the cytosol. Values represent the mean ± SD (n ≥ 15). The values for PTENR41G, E73D, N262Y, N329H and PTENQ17R, R41G, E73D, N262Y, N329H are duplicated in B and C.
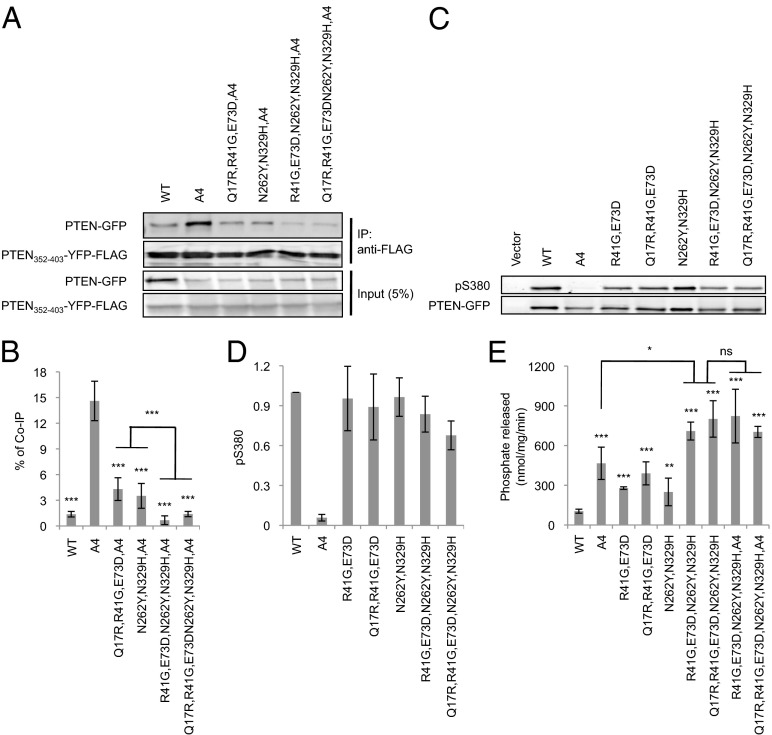
To further characterize ePTEN, we examined associations between the core and tail regions in in trans interaction assays (Fig. 1D), phosphorylation of the tail S/T cluster, and phosphatase activity. First, we found that the core–tail interaction in ePTEN was two- to threefold weaker than that in either PTENQ17R, R41G, E73D or PTENN262Y, N329H (Fig. 6 A and B). Second, the mutations in ePTEN did not act by interfering with the phosphorylation of the C-terminal region (Fig. 6 C and D). Third, it has been suggested that the C-terminal region may mask the catalytic site in addition to the membrane-binding regulatory interface because the reduced intramolecular interaction in PTENN262Y, N329H increased the PIP3 phosphatase activity (20). ePTEN provided further support for this idea because it had higher PIP3-C8 phosphatase activity than PTENA4, PTENQ17R, R41G, E73D, or PTENN262Y, N329H (Fig. 6E).
Fig. 6.
ePTEN-GFP (PTENQ17R, R41G, E73D, N262Y, N329H-GFP) does not bind to the inhibitory C-terminal tail. (A) Interaction of the C-terminal domain of PTEN with its N-terminal core region was assessed in pull-down assays. PTEN352–403-YFP-FLAG was added to whole-cell lysates expressing the indicated PTEN-GFP constructs and immunoprecipitated with beads coupled to anti-FLAG antibodies. Bound fractions (IP) were analyzed with antibodies to GFP and FLAG. (B) Band intensity was quantified (n = 3). (C) Whole-cell lysates expressing different PTEN-GFP constructs were analyzed by immunoblotting with antibodies against phospho-PTEN (pS380) and GFP (PTEN-GFP). (D) Band intensity was quantified (n ≥ 3). (E) The indicated PTEN-GFP proteins were immunopurified from Dictyostelium cells, and phosphatase activities were measured (n ≥ 3).
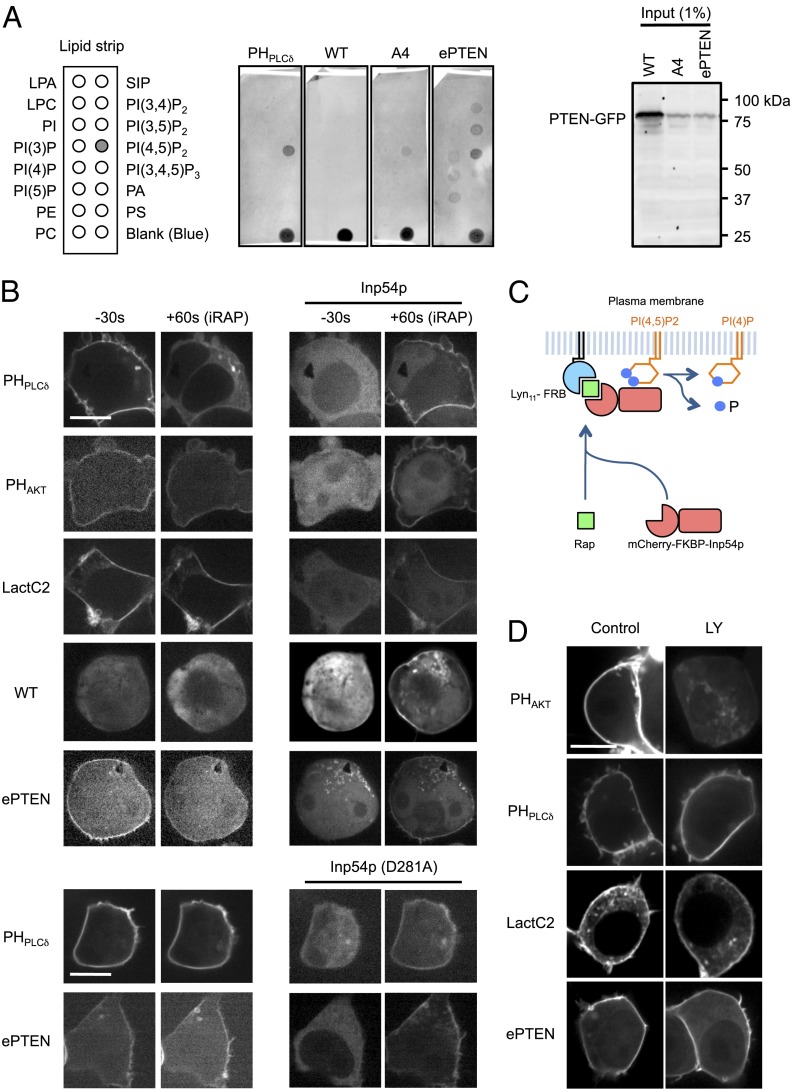
To determine mechanisms underlying the membrane association of ePTEN, we performed a lipid dot blot assay (Fig. 7A) (24, 25). Whole-cell lysates of Dictyostelium cells expressing GFP fused to PTEN, PTENA4, and ePTEN were incubated with nitrocellulose membranes carrying different phospholipids and their interactions were detected using anti-GFP antibodies. Whereas PTEN did not bind to any phospholipids, both PTENA4 and ePTEN showed increased association with PI(4,5)P2 (Fig. 7A). In addition to PI(4,5)P2, ePTEN was also found to interact with other PIP2 isomers such as PI(3,4)P2 and PI(3,5)P2. Immunoblotting of whole-cell lysates using anti-GFP antibodies confirmed full-length GFP fusion proteins (Fig. 7A). The positive control, GFP fused to the PH domain of PLCδ (GFP-PHPLCδ), bound selectively to PI(4,5)P2 (Fig. 7A). To further examine interactions of ePTEN with PI(4,5)P2 in cells, we depleted PI(4,5)P2 from the plasma membrane, using the chemically inducible dimerization system (Fig. 7 B and C) (26). In this experiment, an inositol polyphosphate 5-phosphatase (Inp54p) fused to mCherry and FKBP (mCherry-FKBP-Inp54p) was coexpressed with plasma membrane-located FRB (Lyn11-FRB) in HEK293T cells (Fig. 7C) (26). Upon addition of a chemical dimerizer (rapamycin) to the culture medium, FRB tightly binds to FKBP and recruits mCherry-FKBP-Inp54p to the plasma membrane. PI(4,5)P2 levels were decreased within 60 s after the addition of rapamycin, as revealed by the dissociation of GFP-PHPLCδ from the plasma membrane (Fig. 7B). In contrast, levels of PIP3 and phosphatidylserine in the plasma membrane were not decreased as shown by biosensors for PIP3 (GFP-PHAKT) and phosphatidylserine (GFP-LactC2) (Fig. 7B) (27). Supporting our in vitro binding data, depletion of PI(4,5)P2 resulted in dissociation of ePTEN-GFP from the plasma membrane (Fig. 7B). As a negative control, the recruitment of a catalytically inactive form of Inp54p (D281A) to the plasma membrane did not affect the localization of ePTEN-GFP and GFP-PHPLCδ (Fig. 7B). Unlike PI(4,5)P2, depletion of PIP3 by a PI3K inhibitor, LY294002, did not affect the localization of ePTEN-GFP (Fig. 7D).
Fig. 7.
ePTEN binds to PI(4,5)P2 in vitro and in cells. (A) Lipid dot blot assays. Dictyostelium cells expressing GFP-PHPLCδ, PTEN-GFP, PTENA4-GFP, and ePTEN-GFP were lysed and incubated with nitrocellulose membranes carrying the indicated phospholipids. Images are representative of more than three independent experiments. Protein–lipid interactions were detected by anti-GFP antibodies. Immunoblotting of whole-cell lysates using anti-GFP antibodies is shown. (B and C) HEK293T cells expressing mCherry-FKBP-Inp54p and Lyn11-FRB along with the indicated GFP fusion proteins were observed at 30 s before and 60 s after the addition of 2 nM rapamycin. (Scale bar, 20 µm.) Rapamycin dimerizes FKBP and FRB and brings mCherry-FKBP-Inp54p to the plasma membrane (C). (D) HEK293T cells carrying the indicated GFP constructs were treated with the PI3K inhibitor LY294002 for 5 min and observed by fluorescence microscopy.
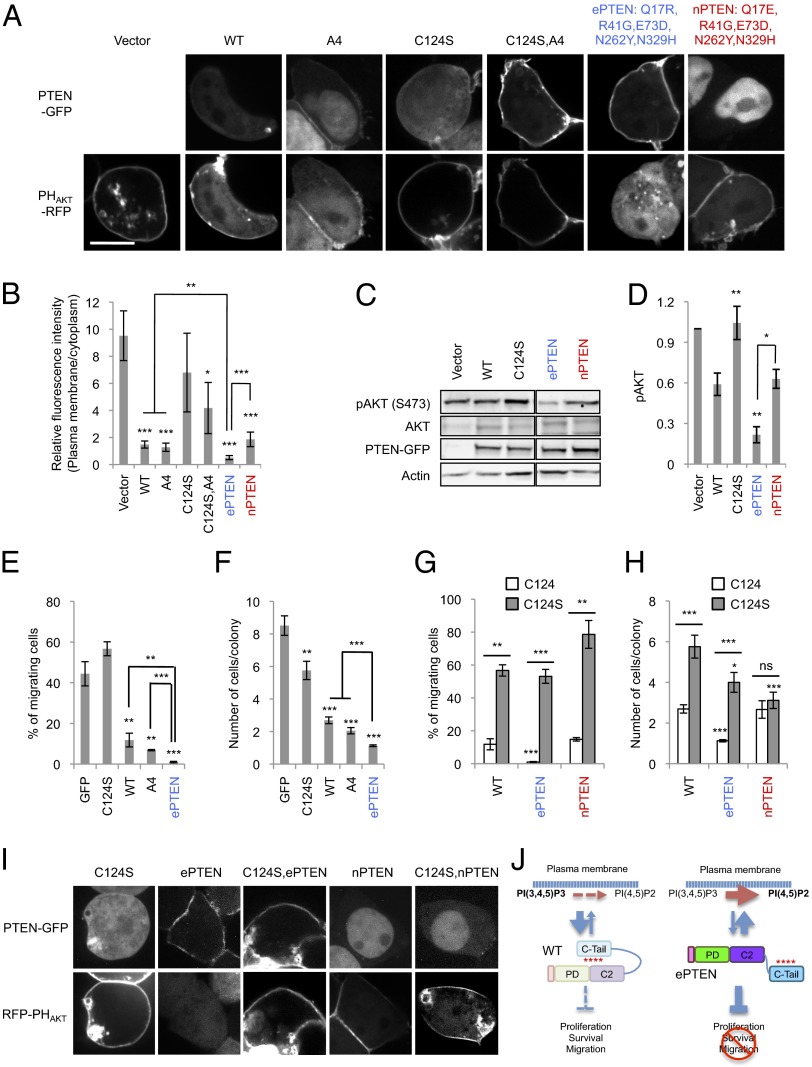
To analyze the function of ePTEN in PIP3 signaling, we coexpressed a PIP3 biosensor, PHAKT-RFP, along with ePTEN -GFP in HEK293T cells. Without the expression of PTEN, HEK293T cells showed high PIP3 levels at the plasma membrane, as observed by the accumulation of PHAKT-RFP (Fig. 8 A and B). Expression of PTEN-GFP and PTENA4-GFP decreased the amounts of PHAKT-RFP at the plasma membrane by approximately fivefold (Fig. 8 A and B). However, a negative control, PTENC124S-GFP, which is defective in its lipid phosphatase activity, showed increased membrane association but did not affect PHAKT-RFP localization (Fig. 8A and Fig. S1). Similarly, PTENC124S, A4, although mutated in the catalytic domain, significantly decreased PIP3 levels, suggesting that PTENC124S, A4 has a residual catalytic activity or helps recruit endogenous PTEN to the plasma membrane through dimerization (28). Remarkably, ePTEN-GFP eliminated detectable PHAKT-RFP at the plasma membrane (Fig. 8 A and B and Fig. S2), suggesting that PIP3 levels were decreased by over 95%. Consistent with the localization of PHAKT-RFP, phosphorylation of AKT at S473, which is stimulated by PIP3 (29, 30), was decreased approximately threefold in cells expressing ePTEN-GFP compared with PTEN-GFP (Fig. 8 C and D). Although most nPTEN accumulated in the nucleus, its ability to decrease levels of PHAKT-RFP at the plasma membrane or AKT phosphorylation was similar to that of PTEN (Fig. 8 A–D), suggesting that nPTEN can still interact with PIP3 in the plasma membrane. Supporting this notion, nPTEN rescued defects in the formation of fruiting bodies in PTEN-null Dictyostelium cells (Fig. S3).
Fig. 8.
ePTEN-GFP (PTENQ17R, R41G, E73D, N262Y N329H-GFP) decreases PIP3 levels at the plasma membrane and suppresses cell migration and proliferation. (A) HEK293T cells expressing different PTEN-GFP constructs along with PHAKT-RFP were observed by fluorescence microscopy. (Scale bar, 10 µm.) (B) Intensity of RFP at the plasma membrane was quantified relative to that in the cytosol. Values represent the mean ± SD (n ≥ 8). (C) Whole-cell lysates expressing different PTEN-GFP constructs were analyzed by immunoblotting with antibodies against phospho-AKT, AKT, GFP (PTEN-GFP), and actin. (D) Band intensity was quantified (n ≥ 3). (E and G) MCF-10A PIK3CA cells were infected with lentiviruses carrying the indicated PTEN constructs and examined for cell migration, using transwell plates with pore size of 8 µm. Values represent the mean ± SD (n ≥ 3). (F and H) The number of cells in individual colonies was determined. Values represent the mean ± SD (n ≥ 20). (I) HEK293T cells expressing different PTEN constructs along with PHAKT-RFP were observed by fluorescence microscopy. (Scale bar, 10 µm.) (J) A model for ePTEN function.
To determine the effect of ePTEN on cancer cell migration, we expressed PTEN, PTENC124S, PTENA4, or ePTEN in the MCF-10A PIK3CA cell line, which carries a constitutive active form of PI3Kα (31, 32). Cells were plated in transwell chambers and their EGF-stimulated transmigration behaviors were quantitatively examined (Fig. 8E). Forty percent of cells expressing GFP or PTENC124S-GFP migrated in 16 h. This migration behavior was suppressed by PTEN-GFP and PTENA4 to 10%. Remarkably, ePTEN-GFP almost completely blocked migratory activity (Fig. 8F). Similarly, ePTEN showed the strongest suppression of cell proliferation (Fig. 8F). MCF-10A PIK3CA cells were plated and their numbers in individual colonies were determined after being cultured for 9 d. We found that PTEN and PTENA4 significantly suppressed cell proliferation and decreased the number of cells in each colony by approximately threefold (Fig. 8F). ePTEN further inhibited cell proliferation with a sixfold decrease (Fig. 8F).
Recent studies have shown that, in addition to its role as a lipid phosphatase at the plasma membrane, PTEN also controls cell proliferation in a phosphatase-independent manner in the nucleus (8, 33). We used ePTEN and nPTEN to assess the relative importance of the membrane or nuclear localizations for cell migration and proliferation. Clearly, ePTEN showed stronger inhibition for both cell migration (Fig. 8G) and cell proliferation (Fig. 8H) compared with PTEN and nPTEN. The inhibitory activity of PTEN, ePTEN, and nPTEN for cell migration depended on their lipid phosphatase activity as the mutation C124S abolished their suppression activity. However, nPTEN was able to block cell proliferation independently of its lipid phosphatase activity (Fig. 8H), supporting a role of nuclear PTEN independent of its phosphatase activity. As a control, we confirmed that the C124S mutation does not affect the localization of ePTEN and nPTEN (Fig. 8I).
Discussion
We have previously identified the membrane-binding regulatory interface in PTEN, consisting of the catalytic domain, the CBR3 loop, and the Cα2 loop (20). The regulatory interface provides a binding site for the C-terminal tail, which masks the membrane-binding site located in the same interface and regulates the plasma membrane localization of PTEN. The CBR3 loop of the C2 domain protrudes from the surface of PTEN and likely presents positively charged residues including K260 and K263 to negatively charged head groups of phospholipids in the plasma membrane. Importantly, residues N262 and K269 in the same loop are not required for membrane binding but are needed for interaction with the inhibitory tail, providing a competitive mechanism for membrane association. In addition, the Cα2 loop that extends near the CBR3 loop also supports interactions with the tail domain. Individual mutations in either the CBR3 or the Cα2 loop do not appear to disrupt interactions of the C2 domain with the tail, but the combination eliminates these interactions, leading to the recruitment of PTEN to the plasma membrane. Our model supports and extends the earlier hypothesis that the CBR3 loop of the C2 domain is a membrane-binding site (34).
In the current study, we found that two residues, R41 and E73, which are located in the phosphatase domain, also play important roles in the interaction with the tail domain. Both residues are located in the loop extending from the regulatory interface toward the plasma membrane. Unlike previously identified mutations in the catalytic center, such as C124R/S and H93R, R41G and E73D do not inhibit the phosphatase activity of PTEN, separating the residues necessary for catalytic activity from those for membrane association. Combined mutations in the C2 and phosphatase domains have a stronger effect on membrane association than do the A4 mutations, which substitute four phosphorylation sites in the C-terminal tail. Although the A4 mutations have been thought to completely open the conformation of PTEN (e.g., completely dissociate the tail from the core region), our findings suggest that PTENA4 only partially releases the tail inhibition.
The PIP2-binding domain is located in the N terminus (amino acid residues 6–15) and has been suggested to be important for membrane association and nuclear localization (10, 13, 22). By individually mutating four positively charged residues, we separated the residues necessary for membrane association from nuclear localization. These two functions of the PIP2-binding domain are mediated by distinct, and partially overlapping, residues. Whereas R14 and R15 likely interact with membrane lipids in cells, K13 and R14 serve as part of the nuclear localization signal. Because K13 and R14 are involved in PIP2 binding (12, 16, 19, 21, 35), it is conceivable that PIP2 might play a role in the localization in the nucleus as well as at the plasma membrane. Immediately adjacent to this region, the mutation Q17R promotes membrane recruitment when inhibition by the tail is removed. Q17R also blocks nuclear localization, thereby enriching PTEN on the plasma membrane. The levels of positive charges in the PIP2-binding domain appear to be a key mechanism of localization to the plasma membrane.
Based on the mechanistic information, we genetically engineered ePTEN, PTENQ17R, R41G, E73D, N262Y, N329H, a version of PTEN with significantly increased membrane association and PIP3 phosphatase activity (Fig. 8J). The four mutations, R41G, E73D, N262Y, and N329H, synergistically dissociate the C-terminal tail from the core region of PTEN and therefore expose the membrane-binding site. However, it is also possible that these mutations can directly enhance interactions with the plasma membrane. Furthermore, we speculate that increases in the phosphatase activity of ePTEN result from opening the conformation, rather than bringing ePTEN to the plasma membrane, because increased activity was observed in our in vitro assay in which a monodisperse substrate, C8-PIP3, was used. The C-terminal tail likely masks the catalytic domain as well as the membrane-binding site. Releasing C-terminal inhibition allows ePTEN to more efficiently bind to its substrates.
ePTEN was found to be a potent repressor of PIP3 production and an inhibitor of AKT phosphorylation. In addition to the release of C-terminal inhibition, the substitution of a residue in the PIP2-binding domain (Q17R) was a critical component in the creation of ePTEN. If similar activations can be achieved pharmacologically, it may represent an excellent strategy to enhance the tumor suppressor activity of PTEN as a therapeutic target. In many cancers, PIP3 signaling is overactivated by abnormal activation of PI3Ks or RasGTPases, leaving intact PTEN available for activation. In addition, heterozygous defects of PTEN are associated with cancers, and therefore the activation of the remaining PTEN could be useful in such cases. Finally, by introducing ePTEN into cancer cells, it may be possible to achieve gene therapy for certain types of cancers in the future.
Experimental Procedures
Cell Culture and Plasmids.
All Dictyostelium cells were cultured in HL5 medium at 22 °C. Cells expressing PTEN-GFP were selected by G418 (20 µg/mL). The primers and plasmids used in this study are listed in Tables S1 and S2. Human PTEN was mutagenized using overlap extension PCR as previously described (20, 36) and cloned into pKF3, a Dictyostelium-expressing plasmid carrying GFP. All constructs were confirmed by DNA sequencing. HEK293T cells were maintained in DMEM (Invitrogen) supplemented with 10% (vol/vol) FBS (Invitrogen). Cells were transiently transfected with 1 µg of DNA plasmids on eight-well chambered coverglass (Lab-TekII; Nunc), using 3 µL of GeneJuice (Novagen), following the manufacturer’s protocol. Cells were then incubated for 24 h before observation.
Isolation of PTEN Mutants.
We generated a library containing human PTEN randomly mutagenized as described previously (20). Briefly, the cDNA of human PTEN was mutagenized using a Diversity PCR random mutagenesis kit (Clontech) and cloned into the Dictyostelium expression vector pKF3 plasmid. The PTEN library was electroporated into Dictyostelium cells (25) and screened for increased membrane localization of PTEN-GFP. To identify mutations that resulted in increased membrane localization, the PTEN gene isolated from individual clones was PCR amplified and sequenced.
PTEN-GFP Localization.
Cells expressing different PTEN-GFP constructs were washed, resuspended in development buffer (DB) containing 20 µM MG132 to block proteosomal degradation of PTEN-GFP, and placed on eight-well chambered coverglass (Lab-TekII; Nunc), as described in ref. 20. Fluorescent images were acquired under a Leica DMI 6000 inverted microscope equipped with a 63× objective and captured by a CoolSNAP EZ camera. All images were analyzed using ImageJ software. To quantify fluorescence intensity of PTEN-GFP at the plasma membrane and nucleus relative to the cytosol, we measured and averaged fluorescence intensity in a 1-pixel area from three different positions in each compartment. Background fluorescence intensity was subtracted from each measurement. The nucleus was identified by DAPI staining. To integrate fluorescence intensity of PTEN-GFP at the plasma membrane, the total intensity at the cell periphery was measured and normalized relative to that in the cytosol.
Development.
PTEN-null Dictyostelium cells were grown exponentially (4 × 105 cells/mL), washed twice in DB (10 mM phosphate buffer, 2 mM MgSO4, 0.2 mM CaCl2), and plated on 1% nonnutrient DB agar for 36 h (37, 38). Cells were observed under an Olympus SZ-PT dissecting microscope equipped with a 6× objective and their images were taken by a Nikon 4500 camera.
Interactions Between PTEN and Its C-Terminal Tail.
Interactions between PTEN and its C-terminal tail (PTEN352–403-YFP-FLAG) were examined, as described in ref. 11. HEK293T cells were cultured in DMEM supplemented with 10% (vol/vol) FBS on a 100-mm dish, transiently transfected with 8 µg of the plasmid carrying PTEN352–403-YFP-FLAG using GeneJuice (Novagen), and then cultured for 24 h in DMEM with 10% (vol/vol) FBS. HEK293T cells were lysed in 1 mL of lysis buffer containing 1% Nonidet P-40, 50 mM NaCl, 20 mM Tris⋅HCl (pH 7.5), 10% (vol/vol) glycerol, 0.1 mM EDTA, phosphatase inhibitor mixture (Sigma), and protease inhibitor mixture (Roche). The lysates were then cleared by centrifugation at 16,000 × g for 20 min at 4 °C. Dictyostelium cells expressing WT and mutant versions of PTEN-GFP were lysed in 1% Nonidet P-40, 300 mM NaCl, 10 mM Tris⋅HCl (pH 7.5), 2 mM EDTA, phosphatase inhibitor mixture (Sigma) and protease inhibitor mixture (Roche). The lysates were then cleared by centrifugation at 13,000 rpm for 20 min at 4 °C. One hundred microliters of the HEK293T lysates was mixed with 500 µL of the Dictyostelium lysates. Beads coupled to anti-FLAG antibodies (Sigma) were added to the mixtures and incubated for 2 h. The beads were washed twice in the lysis buffer and the bound fractions were analyzed by SDS/PAGE and immunoblotting, using antibodies to FLAG and GFP.
Immunoblotting.
Proteins were resolved by SDS/PAGE and transferred onto a PVDF membrane. Antibodies against PTEN (138G6; Cell Signaling); phosphorylated PTEN at residues S380, T382, and T383 (44A7; Cell Signaling); AKT (9272; Cell Signaling); phosphorylated AKT (Serine 473; Cell Signaling 4058); GFP (11E5; Molecular Probes); and actin (C-11; Santa Cruz Biotechnology) were used. Immunocomplexes were visualized by fluorophore-conjugated secondary antibodies including anti-rabbit Alexa Fluoro 488 (Invitrogen), anti-goat Aelxa Fluoro 647 (Invitrogen), and anti-mouse Dylight 649 (Jackson ImmunoResearch Laboratories) and detected using a PharosFX Plus molecular imager (Bio-Rad).
Phosphatase Activity.
The phosphatase activity of PTEN was measured, as described previously (19). WT and mutant forms of PTEN fused to GFP were expressed in Dictyostelium cells and immunopurified using GFP-Trap agarose beads (Allele Biotech). The phosphatase activity was determined by measuring release of phosphates from PIP3 diC8, using a Malachite Green Phosphatase assay kit (Echelon). The activity was normalized to amounts of purified PTEN-GFP proteins.
Lipid Dot Blot Assay.
The lipid dot blot assay was performed as described previously (24, 25, 36). Briefly, Dictyostelium cells expressing different PTEN-GFP constructs were starved for 2 h in 10 mM phosphate buffer (pH 7.0), 2 mM MgSO4, and 0.2 mM CaCl2 and then washed twice in 20 mL of ice-cold 10 mM sodium phosphate (pH 7.0). Then, cells were lysed at 5 × 108 cells/mL in 10 mM sodium phosphate (pH 7.0), 0.5% Nonidet P-40, 150 mM NaCl, and 1% protein inhibitor mixture (Sigma) for 10 min on ice. Cell lysates were cleared twice at 10,000 × g for 10 min at 4 °C. The supernatants were mixed with equal volumes of 10 mM sodium phosphate (pH 7.0) and 150 mM NaCl. Nitrocellulose membranes spotted with different phospholipids (PIP membrane P-6001; Echelon) were incubated in PBS containing 0.05% Tween 20 and 3% (wt/vol) fatty acid-free BSA to block nonspecific binding and then mixed with the lysates for at least 3 h. After extensive washing, the membranes were probed with anti-GFP antibodies followed by secondary antibodies conjugated with Alexa 647 (Invitrogen) and scanned using a PharosFX Plus molecular imager.
Cell Proliferation Assay.
MCF-10A cells that carry a PIK3CA knock-in mutation (E545K) were cultured in DMEM/F12 medium supplemented with 1% FBS, 10 µg/mL insulin, 0.5 µg/mL hydrocortisone, and 0.1 µg/mL cholera toxin in the absence of EGF (31, 32). Cells were infected with lentiviruses expressing different PTEN-GFP constructs and cultured for 5 d to allow the expression of PTEN constructs. Cells were plated at the density of 100 cells per well on 96-well plates (Nunc) and cultured for 9 d. The numbers of cells were determined in individual colonies that expressed PTEN-GFP constructs, using a Zeiss Axio Observer inverted microscope equipped with an AxioCam.
Statistical Analysis.
P values were determined using Student’s t test: *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
Acknowledgments
We thank Dr. Todd Waldman (Georgetown University School of Medicine) for lentiviral constructs. We thank M. Rahdar, K. F. Swaney, and T. Inoue for providing plasmids. This work was supported by National Institutes of Health Grants GM084015 (to M.I.), GM28007 and GM34933 (to P.N.D.), and GM089853 and NS084154 (to H.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409433111/-/DCSupplemental.
References
- 1.Vanhaesebroeck B, Stephens L, Hawkins P. PI3K signalling: The path to discovery and understanding. Nat Rev Mol Cell Biol. 2012;13(3):195–203. doi: 10.1038/nrm3290. [DOI] [PubMed] [Google Scholar]
- 2.Sulis ML, Parsons R. PTEN: from pathology to biology. Trends Cell Biol. 2003;13(9):478–483. doi: 10.1016/s0962-8924(03)00175-2. [DOI] [PubMed] [Google Scholar]
- 3.Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer. 2011;11(4):289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carracedo A, Alimonti A, Pandolfi PP. PTEN level in tumor suppression: How much is too little? Cancer Res. 2011;71(3):629–633. doi: 10.1158/0008-5472.CAN-10-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leslie NR, Dixon MJ, Schenning M, Gray A, Batty IH. Distinct inactivation of PI3K signalling by PTEN and 5-phosphatases. Adv Biol Regul. 2012;52(1):205–213. doi: 10.1016/j.advenzreg.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: Lessons learned from early clinical trials. Nat Rev Clin Oncol. 2013;10(3):143–153. doi: 10.1038/nrclinonc.2013.10. [DOI] [PubMed] [Google Scholar]
- 7.Baker SJ. PTEN enters the nuclear age. Cell. 2007;128(1):25–28. doi: 10.1016/j.cell.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 8.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13(5):283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 9.Tamguney T, Stokoe D. New insights into PTEN. J Cell Sci. 2007;120(Pt 23):4071–4079. doi: 10.1242/jcs.015230. [DOI] [PubMed] [Google Scholar]
- 10.Gericke A, Leslie NR, Lösche M, Ross AH. PtdIns(4,5)P2-mediated cell signaling: Emerging principles and PTEN as a paradigm for regulatory mechanism. Adv Exp Med Biol. 2013;991:85–104. doi: 10.1007/978-94-007-6331-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahdar M, et al. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc Natl Acad Sci USA. 2009;106(2):480–485. doi: 10.1073/pnas.0811212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker SM, Leslie NR, Perera NM, Batty IH, Downes CP. The tumour-suppressor function of PTEN requires an N-terminal lipid-binding motif. Biochem J. 2004;379(Pt 2):301–307. doi: 10.1042/BJ20031839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denning G, Jean-Joseph B, Prince C, Durden DL, Vogt PK. A short N-terminal sequence of PTEN controls cytoplasmic localization and is required for suppression of cell growth. Oncogene. 2007;26(27):3930–3940. doi: 10.1038/sj.onc.1210175. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Shi Y, Wang J, Huang G, Jiang X. Crucial role of the C-terminus of PTEN in antagonizing NEDD4-1-mediated PTEN ubiquitination and degradation. Biochem J. 2008;414(2):221–229. doi: 10.1042/BJ20080674. [DOI] [PubMed] [Google Scholar]
- 15.Odriozola L, Singh G, Hoang T, Chan AM. Regulation of PTEN activity by its carboxyl-terminal autoinhibitory domain. J Biol Chem. 2007;282(32):23306–23315. doi: 10.1074/jbc.M611240200. [DOI] [PubMed] [Google Scholar]
- 16.Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109(5):599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 17.Vazquez F, et al. Tumor suppressor PTEN acts through dynamic interaction with the plasma membrane. Proc Natl Acad Sci USA. 2006;103(10):3633–3638. doi: 10.1073/pnas.0510570103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iijima M, Huang YE, Devreotes P. Temporal and spatial regulation of chemotaxis. Dev Cell. 2002;3(4):469–478. doi: 10.1016/s1534-5807(02)00292-7. [DOI] [PubMed] [Google Scholar]
- 19.Iijima M, Huang YE, Luo HR, Vazquez F, Devreotes PN. Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J Biol Chem. 2004;279(16):16606–16613. doi: 10.1074/jbc.M312098200. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen HN, et al. 2013. Mechanism of human PTEN localization revealed by heterologous expression in Dictyostelium. Oncogene, 10.1038/onc.2013.507.
- 21.Campbell RB, Liu F, Ross AH. Allosteric activation of PTEN phosphatase by phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 2003;278(36):33617–33620. doi: 10.1074/jbc.C300296200. [DOI] [PubMed] [Google Scholar]
- 22.Gil A, Andrés-Pons A, Pulido R. Nuclear PTEN: A tale of many tails. Cell Death Differ. 2007;14(3):395–399. doi: 10.1038/sj.cdd.4402073. [DOI] [PubMed] [Google Scholar]
- 23.Liu F, et al. PTEN enters the nucleus by diffusion. J Cell Biochem. 2005;96(2):221–234. doi: 10.1002/jcb.20525. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Senoo H, Sesaki H, Iijima M. Rho GTPases orient directional sensing in chemotaxis. Proc Natl Acad Sci USA. 2013;110(49):E4723–E4732. doi: 10.1073/pnas.1312540110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CL, Wang Y, Sesaki H, Iijima M. Myosin I links PIP3 signaling to remodeling of the actin cytoskeleton in chemotaxis. Sci Signal. 2012;5(209):ra10. doi: 10.1126/scisignal.2002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heo WD, et al. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314(5804):1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeung T, et al. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319(5860):210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 28.Papa A, et al. Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell. 2014;157(3):595–610. doi: 10.1016/j.cell.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawkins PT, Anderson KE, Davidson K, Stephens LR. Signalling through class I PI3Ks in mammalian cells. Biochem Soc Trans. 2006;34(Pt 5):647–662. doi: 10.1042/BST0340647. [DOI] [PubMed] [Google Scholar]
- 30.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4(12):988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 31.Gustin JP, et al. Knockin of mutant PIK3CA activates multiple oncogenic pathways. Proc Natl Acad Sci USA. 2009;106(8):2835–2840. doi: 10.1073/pnas.0813351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang GM, et al. Single copies of mutant KRAS and mutant PIK3CA cooperate in immortalized human epithelial cells to induce tumor formation. Cancer Res. 2013;73(11):3248–3261. doi: 10.1158/0008-5472.CAN-12-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song MS, et al. Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell. 2011;144(2):187–199. doi: 10.1016/j.cell.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JO, et al. Crystal structure of the PTEN tumor suppressor: Implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99(3):323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 35.Das S, Dixon JE, Cho W. Membrane-binding and activation mechanism of PTEN. Proc Natl Acad Sci USA. 2003;100(13):7491–7496. doi: 10.1073/pnas.0932835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang P, Wang Y, Sesaki H, Iijima M. 2010. Proteomic identification of phosphatidylinositol (3,4,5) triphosphate-binding proteins in Dictyostelium discoideum. Proc Natl Acad Sci USA 107(26):11829–11834.
- 37.Cai H, Huang CH, Devreotes PN, Iijima M. Analysis of chemotaxis in Dictyostelium. Methods Mol Biol. 2012;757:451–468. doi: 10.1007/978-1-61779-166-6_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, et al. Dictyostelium huntingtin controls chemotaxis and cytokinesis through the regulation of myosin II phosphorylation. Mol Biol Cell. 2011;22(13):2270–2281. doi: 10.1091/mbc.E10-11-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.