Significance
Abscisic acid (ABA) is a key stress-responsive hormone. Subgroup III Snf1-related kinase 2s (SnRK2s) are crucial positive regulators in ABA signaling pathway, but it is still not clear how SnRK2s are activated. In addition, besides ABA, some abiotic stresses can also activate SnRK2s with unknown mechanisms. Here we provide several lines of evidence to strongly support that brassinosteroid insensitive 2, a glycogen synthase kinase 3 (GSK3)-like kinase, interacts with and phosphorylates SnRK2s on specific novel sites to activate SnRK2s, which provides significant insights into the function of GSK3-like kinases in ABA signaling and transactivation of SnRK2s. Moreover, many GSK3-like kinases are regulated at transcriptional and/or posttranslational levels by many abiotic stresses, implying the presence of direct regulation of ABA signaling by other abiotic stresses.
Keywords: signal transduction, phosphorylation cascades, kinase activation
Abstract
Arabidopsis glycogen synthase kinase 3 (GSK3)-like kinases have versatile functions in plant development and in responding to abiotic stresses. Although physiological evidence suggested a potential role of GSK3-like kinases in abscisic acid (ABA) signaling, the underlying molecular mechanism was largely unknown. Here we identified members of Snf1-related kinase 2s (SnRK2s), SnRK2.2 and SnRK2.3, that can interact with and be phosphorylated by a GSK3-like kinase, brassinosteroid insensitive 2 (BIN2). bin2-3 bil1 bil2, a loss-of-function mutant of BIN2 and its two closest homologs, BIN2 like 1 (BIL1) and BIN2 like 2 (BIL2), was hyposensitive to ABA in primary root inhibition, ABA-responsive gene expression, and phosphorylating ABA Response Element Binding Factor (ABF) 2 fragment by in-gel kinase assays, whereas bin2-1, a gain-of-function mutation of BIN2, was hypersensitive to ABA, suggesting that these GSK3-like kinases function as positive regulators in ABA signaling. Furthermore, BIN2 phosphorylated SnRK2.3 on T180, and SnRK2.3T180A had decreased kinase activity in both autophosphorylation and phosphorylating ABFs. Bikinin, a GSK3 kinase inhibitor, inhibited the SnRK2.3 kinase activity and its T180 phosphorylation in vivo. Our genetic analysis further demonstrated that BIN2 regulates ABA signaling downstream of the PYRABACTIN RESISTANCE1/PYR1-LIKE/REGULATORY COMPONENTS OF ABA RECEPTORS receptors and clade A protein phosphatase 2C but relies on SnRK2.2 and SnRK2.3. These findings provide significant insight into the modulation of ABA signaling by Arabidopsis GSK3-like kinases.
Abscisic acid (ABA) is a key phytohormone in responding to various abiotic stresses and in plant development, such as embryogenesis, seed dormancy and germination, and root elongation (1–4). Since the discovery of ABA receptors, PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS (RCAR) (5, 6), a core ABA signaling pathway has been proposed. Without ABA, clade A protein phosphatase 2Cs (PP2Cs) inhibit the activity of subgroup III Snf1-related kinase 2s (SnRK2s) by physical interaction and dephosphorylation (7, 8), leading to inhibition of downstream transcription factors required for ABA-responsive gene expression (9–11). Perception of ABA causes conformational changes of PYR/PYL/RCAR proteins, which facilitate their binding to PP2Cs to release their inhibition on SnRK2s (7, 11). The activated SnRK2s phosphorylate transcription factors, such as ABA Response Element Binding Factors (ABFs), to regulate ABA responsive gene expression (7, 11).
The Arabidopsis subgroup III SnRK2 family contains three members, SnRK2.2, SnRK2.3, and SnRK2.6 (12, 13). SnRK2.6 is specifically expressed in guard cells (12) to regulate ABA-mediated stomata movement. SnRK2.2 and SnRK2.3 are ubiquitously expressed and responsible for ABA-regulated seed germination and primary root elongation (13). Their triple knockout snrk2.2 snrk2.3 snrk2.6 displays a considerable resistance to ABA, whereas single or double mutants could not, suggesting their redundant role in mediating ABA signaling (14, 15). Besides ABA, osmotic stresses also activate SnRK2s, probably through a mechanism independent of ABA biosynthesis and clade A PP2Cs (3, 16–19). However, how SnRK2s are activated is not fully understood. It is reported that several members of SnRK2s can be regulated by upstream kinases (17, 20), and autophosphorylation activity of recombinant SnRK2.2 and SnRK2.3 is only one-tenth to one-fifth of that of SnRK2.6, suggesting that some SnRK2s may be activated by yet unknown kinases in vivo (21).
Glycogen synthase kinase 3s (GSK3s) can phosphorylate a number of proteins to regulate their activity, stability, and subcellular localization in diverse systems (22, 23). In Arabidopsis, GSK3/Shaggy-like kinases (ASKs) contain at least 10 members and participate in many biological processes (23). BRASSINOSTEROID INSENSITIVE 2 (BIN2), a GSK3-like kinase, functions as a negative regulator in brassinosteroid (BR) signaling by phosphorylating transcription factors BRASSINAZOLE RESISTANT 1 and BRI1-EMS-SUPPRESSOR 1 (BES1) (24, 25). BIN2 also participates in many other biological processes, such as auxin signaling (26) and stomata development (27, 28). Plant GSK3-like kinases apparently play important roles in responding to many abiotic stresses (23). For example, BIL2, a close homolog of BIN2, can complement yeast salt insensitive mutants (29) and mediate salt tolerance in Arabidopsis (30), and another GSK3-like kinase, ASKα (AtSK11) participates in regulating redox stress response through phosphorylating glucose-6-phosphate dehydrogenase in Arabidopsis (31). In rice, knockout of OsGSK1, an AtBIN2 ortholog, showed an enhanced tolerance to cold, heat, high salt, and drought (32). Interestingly, bin2-1, a gain-of-function mutation of BIN2, was hypersensitive to ABA in primary root inhibition (33), implying its potential role in ABA signaling, but the underlying mechanism is unknown.
In this study, when we initially identified BIN2-interacting proteins with liquid chromatography tandem mass spectrometry (LC-MS/MS) using immunoprecipitated (IPed) proteins from BIN2-FLAG transgenic plants, we found that SnRK2.2 may interact with BIN2. We further confirmed that BIN2 physically interacts with all subgroup III SnRK2s both in vitro and in vivo and is able to phosphorylate SnRK2.2 and SnRK2.3 and enhances their kinase activity. We identified T180 as a novel phosphorylation site of SnRK2.3 by BIN2 kinase, which is important for SnRK2.3’s activation. Primary root inhibition assay, ABA-responsive gene expression, and phosphorylating ABF fragment by in-gel kinase assays using bin2-1 and bin2-3 bil1 bil2 (34) mutants indicated that BIN2 and its homologs act as positive regulators in ABA signaling. Immuno-kinase assay and quantitative MS results indicated that bikinin inhibited the T180 phosphorylation of SnRK2.3 and its kinase activity. We generated double and multiple mutants between BIN2-related mutants and ABA signaling mutants, and their phenotypic analysis indicated that BIN2 enhances ABA responses through SnRK2s, but not PYR/PYL/RCARs and PP2Cs. Therefore, we propose that BIN2 phosphorylates SnRK2s to promote their activity.
Results
BIN2 Interacts with Subgroup III SnRK2s and Phosphorylates SnRK2.2 and SnRK2.3.
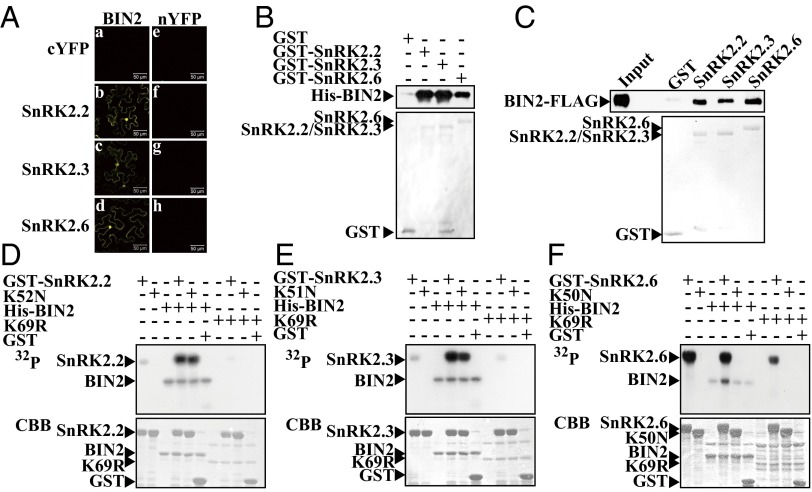
In an early study we found that ABA inhibits primary BR signaling outputs, likely through BIN2 or its upstream components (35). To identify proteins interacting with BIN2, we carried out an LC-MS/MS experiment with IPed BIN2-FLAG complex from 35S::BIN2-FLAG plants. Interestingly, we identified a peptide corresponding to SnRK2.2 (Fig. S1). We then tested physical interaction of BIN2 with SnRK2.2, SnRK2.3, and SnRK2.6 using a bimolecular fluorescence complementation (BiFC) assay, and we found that BIN2 interacts with all subgroup III SnRK2s in both cytoplasm and nucleus of Nicotiana benthamiana pavement cells (Fig. 1A). Pull-down assays demonstrated that GST-SnRK2s can directly interact with His-BIN2 in vitro (Fig. 1B). Using a semi-in vivo pull-down assay, we found that each SnRK2 protein was able to pull down BIN2-FLAG protein expressed in transgenic seedlings (Fig. 1C). We then conducted in vitro kinase assays to test whether BIN2 and SnRK2s can phosphorylate each other and found that BIN2 can phosphorylate both wild-type SnRK2.2 and SnRK2.3 and their kinase-dead forms, SnRK.2.2K52N (K52N) and SnRK2.3K51N (K51N) (Fig. 1 D and E), but not SnRK2.6 or SnRK2.6K50N (K50N) (Fig. 1F), and the SnRK2s cannot phosphorylate either BIN2 or its kinase dead form, BIN2K69R (K69R) (Fig. 1 D–F). Although SnRK2s failed to phosphorylate BIN2 in vitro, because interaction between SnRK2s and BIN2 may alter BIN2 activity, we conducted phosphorylation assays on BES1, a BIN2 substrate in the BR signaling pathway (25), by BIN2 kinase in vitro with or without SnRK2s. Additional SnRK2s did not affect BES1 phosphorylation by BIN2 kinase (Fig. S2), suggesting these SnRK2s may not directly influence BIN2 activity.
Fig. 1.
BIN2 interacts with subgroup III SnRK2s and phosphorylates SnRK2.2 and SnRK2.3. (A) BIN2 interacts with SnRK2.2, SnRK2.3, and SnRK2.6 in BiFC assay. nYFP-BIN2 or nYFP was cotransformed into pavement cells of N. benthamiana with cYFP (a and e), SnRK2.2-cYFP (b and f), SnRK2.3-cYFP (c and g), or SnRK2.6-cYFP (d and h). (Scale bar, 50 μm.) (B) SnRK2.2, SnRK2.3, and SnRK2.6 interact with BIN2 in vitro. (C) SnRK2.2, SnRK2.3, and SnRK2.6 can pull down BIN2-FLAG in whole-protein extract from BIN2-FLAG transgenic plants. (D) BIN2 phosphorylates SnRK2.2 and SnRK2.2K52N (K52N) in vitro. (E) BIN2 phosphorylates SnRK2.3 and SnRK2.3K51N (K51N) in vitro. (F) BIN2 cannot phosphorylate SnRK2.6 or SnRK2.6K50N (K50N) in vitro. K69R is the abbreviation for BIN2K69R.
BIN2 and Its Homologs Play a Positive Role in Modulating ABA Signaling.
Therefore, we focused our investigation on the potential roles of BIN2 in ABA signaling. We discovered that bin2-3 bil1 bil2 was less sensitive to ABA in primary root inhibition than wild-type Ws-2 (Fig. 2 A and B). bin2-3 bil1 bil2 was hyposensitive to ABA in seed germination (Fig. S3 A–C). The expression levels of ABA-responsive genes determined by quantitative real-time RT-PCR (qRT-PCR) were also significantly compromised in bin2-3 bil1 bil2 (Fig. 2C and Fig. S3D). In contrast, bin2-1 was hypersensitive to ABA in both primary root inhibition (Fig. 2 D and E) and ABA-responsive gene expression compared with wild-type Col-0 (Fig. 2F). Basal levels of ABA-responsive gene expression were also significantly reduced in bin2-3 bil1 bil2 (Fig. 2G) and largely enhanced in bin2-1 (Fig. 2H). In addition, treatment of bikinin, an inhibitor of GSK3 kinases (36), significantly inhibited ABA-responsive gene expression (Fig. 2I). We further conducted an in-gel kinase assay on the ABF2 fragment (Gly73 to Gln120) by their protein extracts and found that the ABA-activated bands were significantly weaker in bin2-3 bil1 bil2 and largely stronger in bin2-1 than that in their corresponding wild types (Fig. 2 J and K and Fig. S3 E and F). Taken together, these data suggest that BIN2 and its homologs play a positive role in modulating ABA signaling.
Fig. 2.
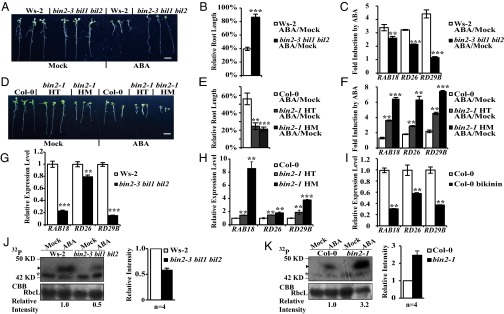
BIN2 plays positive roles in modulating ABA signaling outputs. (A) Primary root phenotype of wild-type (Ws-2) and bin2-3 bil1 bil2 grown on medium with or without (Mock) 10 μM ABA. (B) Statistic analysis of relative primary root length of Ws-2 (n = 25) and bin2-3 bil1 bil2 (n = 25). (C) Induction of RAB18, RD26, and RD29B expression by ABA in Ws-2 and bin2-3 bil1 bil2. Seedlings were treated with 50 μM ABA or solvent only (Mock) for 1 h. (D) Primary root phenotype of wild-type (Col-0) and bin2-1 grown on medium without (Mock) or with 10 μM ABA. (E) Statistic analysis of relative primary root length for Col-0 (n = 20), bin2-1 heterozygote (HT) (n = 35), and bin2-1 homozygote (HM) (n = 20). (F) Induction of RAB18, RD26, and RD29B expression by ABA in Col-0 and bin2-1. Seedlings were treated with 50 μM ABA or solvent only (Mock) for 1 h. (G) Expression levels of RAB18, RD26, and RD29B in bin2-3 bil1 bil2 and Ws-2. Expression level of each gene in Ws-2 was normalized to “1.” (H) Expression level of RAB18, RD26, and RD29B in bin2-1and Col-0. Expression level of each gene in Col-0 was normalized to “1.” (I) Bikinin inhibits expression of RAB18, RD26, and RD29B in Col-0. The expression level of each gene in Col-0 grown on medium with or without (Mock) 10 μM bikinin. Expression level of each gene in Col-0 grown on medium without bikinin was normalized to “1.” (J) Signal intensity of in-gel kinase assay on phosphorylating the ABF2 fragment after treatment with 50 μM ABA or solvent only (Mock) for 1 h. (K) Signal intensity of in-gel kinase assay on phosphorylating the ABF2 fragment after treatment with 50 μM ABA or solvent only (Mock) for 1 h. In J and K, the arrowheads indicate the ABA-induced bands representing the activated SnRK2s. “*” indicates unknown, none ABA-induced bands, and right graphs in J and K show relative radioactivity intensity of ABA-inducible bands (means ± SE, n = 4). The relative intensity of ABA-inducible bands in both Ws-2 and Col-0 after ABA treatment was normalized to “1.” Coomassie brilliant blue R250 staining (CBB) of the large subunit of Rubisco (RbcL) was used as the loading control. Also see replicates 2–4 in Fig. S3 E and F. (Scale bars, 1 cm in A and D.) Primary root length of each material in mock treatment was normalized to 100% in B and E. Expression level of each gene for each material under mock treatment was normalized to “1” in C and F. Values are means ± SE for B and C and E–I. Student's t test was used to determine the significance of the indicated comparisons. Significant levels: **P < 0.01; ***P < 0.001.
BIN2 Phosphorylates SnRK2.3 on T180 to Promote Its Kinase Activity.
To investigate the biochemical mechanisms by which BIN2 phosphorylates SnRK2s and enhances ABA signaling, we used a mass spectrum approach and identified a number of potential phosphorylation sites of SnRK2.3K51N (K51N) by BIN2 kinase in vitro, including S172, S176, T177, and T180 (Fig. S4 A–C). We then mutated each site to Ala in combination with K51N mutation, and they were designated as K51N S172A, K51N S176A, K51N T177A, and K51N T180A, respectively. We also mutated S168, which is located in a typical GSK3 recognition motif S/TxxxS/T, to Ala (K51N S168A). In vitro phosphorylation assays with BIN2 kinase indicated that BIN2 can still strongly phosphorylate K51N S168A, K51N S172A, and K51N T177A but phosphorylates K51N S176A with a reduced activity and almost no phosphorylation on K51N T180A (Fig. 3A). The decreased phosphorylation of K51N T180A was not due to a reduced interaction between BIN2 and K51N T180A (Fig. S5A), suggesting that T180 of SnRK2.3 may be a key phosphorylation site by BIN2 kinase. Furthermore, SnRK2.3T180A (T180A) had extremely low autophosphorylation activity and decreased ability to phosphorylate ABF2 fragment, a widely used substrate of SnRK2s (7), demonstrating that T180 of SnRK2.3 is crucial for its activation (Fig. 3B). However, mutation of T180 to Asp (D) and Glu (E) failed to mimic a constitutively active SnRK2.3 in vitro (Fig. S5B). To investigate T180 function in vivo, we compared the responses of Col-0, SnRK2.3-OX (line 3), and SnRK2.3T180A-OX (line 7) to ABA by measuring expression levels of RAB18, RD26, and RD29B. SnRK2.3-OX showed hypersensitivity to ABA, whereas SnRK2.3T180A-OX had similar sensitivity to ABA compared with Col-0, implying that T180 is a key residue for transmitting ABA signaling in vivo (Fig. 3C). To test whether phosphorylation on T180 of SnRK2.3 by BIN2 can enhance its kinase activity, we then measured the activity of SnRK2.3 with additional BIN2 or BIN2K69R (kinase dead form) in vitro. We found that BIN2 greatly enhanced phosphorylation of SnRK2.3 and ABF2 fragment compared with BIN2K69R (Fig. 3D), although SnRK2.3 similarly interacted with both BIN2 and BIN2K69R (Fig. S5C), supporting that BIN2 phosphorylates SnRK2.3 at T180 to enhance its activation.
Fig. 3.
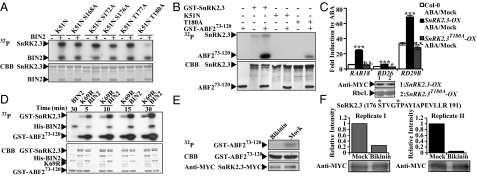
BIN2 phosphorylates SnRK2.3 and enhances its kinase activity. (A) In vitro kinase assays of various mutant forms of SnRK2.3 with BIN2 kinase. Potentially phosphorylated Ser or Thr residues of SnRK2.3 were mutated to Ala, and each mutation was combined with a kinase dead mutation K51N. SnRK2.3K51N S168A is abbreviated for K51N S168A, and the same rule also applied for other mutant forms. (B) In vitro phosphorylation assays of SnRK2.3 and SnRK2.3T180A on the ABF2 fragment. (C) Relative expression levels of RAB18, RD26, and RD29B in Col-0, SnRK2.3-OX, and SnRK2.3T180A-OX after treatment with 50 μM ABA or solvent only (Mock) for 1 h. Expression level of each gene for each material under mock treatment was normalized to “1.” Values are means ± SE. Student's t test was used to determine the significance of the indicated comparisons. Significant levels: *P < 0.05; **P < 0.01; ***P < 0.001; and n.s. (not significant, P > 0.05). (D) In vitro phosphorylation assays of SnRK2.3 on ABF2 fragment in the presence of BIN2 or BIN2K69R (K69R). (E) Immuno-kinase assays of SnRK2.3-MYC on ABF2 fragment. The SnRK2.3-MYC-OX plants were grown on 1/2 MS without (Mock) or with 5 μM bikinin for 10 d and treated with 50 μM ABA for 1 h. The IPed SnRK2.3-MYC was used to phosphorylate ABF2 fragment. (F) Quantitative mass spectrum analysis on phosphorylation of the IPed SnRK2.3-MYC. SnRK2.3-MYC was IPed from seedling as described in Fig. 3E and digested by trypsin. Phospho-peptides were enriched with TiO2 beads. The sequence of phospho-peptide for quantification is shown. Phosphorylation sites are marked with “*.” Signal intensity of the target phospho-peptide from the mock treatment was normalized to “1.” Two independent experiments were conducted.
To determine the effect of GSK3s on SnRK2.3 activation in vivo, we IPed SnRK2.3-MYC from transgenic seedlings grown on 1/2 MS medium with or without (Mock) 5 μM bikinin after 50 μM ABA treatment for 1 h, and we found that bikinin significantly inhibited the activity of SnRK2.3-MYC on phosphorylating ABF fragment (Fig. 3E), and bikinin did not directly inhibit SnRK2.3 in vitro (Fig. S5D). We quantified the T180 phosphorylation level of the IPed SnRK2.3-MYC from seedlings after ABA treatment. We found that T180s were significantly less phosphorylated in seedlings grown on bikinin than the mock (Fig. 3F and Fig. S5 E and F).
BIN2-Promoted ABA Signaling Is Dependent on SnRK2s.
To investigate whether BIN2-enhanced ABA signaling is mediated through SnRK2.2 and SnRK2.3 or other components of ABA signaling, we conducted genetic analysis by generating a set of double or multiple mutants of BIN2-related mutants with several known ABA signaling mutants. We first generated a multiple mutant of bin2-1 with ABA receptor quadruple mutant pyr1 pyl1 pyl2 pyl4. Because pyl2 is linked with erecta locus (5), we crossed bin2-1 heterozygote with er105 to obtain Col-0:er and bin2-1:er plants, which were used as controls. We found that pyr1 pyl1 pyl2 pyl4 quadruple mutant was insensitive to ABA in inhibiting primary root elongation, but bin2-1 pyr1 pyl1 pyl2 pyl4 showed an enhanced sensitivity to ABA, which was similar to the bin2-1:er single mutant (Fig. 4 A and B), and induction of RAB18, RD26, and RD29B expression by ABA in bin2-1 pyr1 pyl1 pyl2 pyl4 was much higher than that in pyr1 pyl1 pyl2 pyl4 (Fig. 4C). The intensity of ABA-induced bands was also largely higher in bin2-1 pyr1 pyl1 pyl2 pyl4 than in pyr1 pyl1 pyl2 pyl4 by an in-gel kinase assay (Fig. S6 A and B), implying that BIN2 can modulate ABA signaling downstream of ABA receptors. We obtained an abi2G168D mutant in Col-0 background through a bri1-301 suppressor screening in seed germination, which was designated as abi2-3. A double mutant of bin2-1 and abi2-3 also partially suppressed the insensitive phenotype of abi2-3 to ABA in primary root inhibition (Fig. 4 D and E). The induction of RAB18, RD26, and RD29B expression by ABA was higher in abi2-3 bin2-1 than in abi2-3 (Fig. 4F).
Fig. 4.
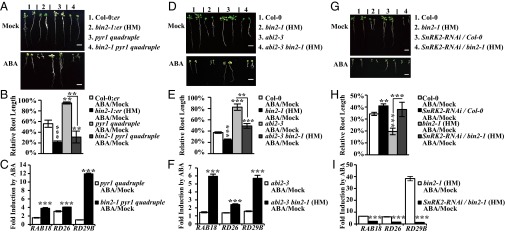
BIN2 promotes ABA signaling outputs through subgroup III SnRK2s. (A) Root phenotype of Col-0:er, bin2-1:er, pyr1 pyl1 pyl2 pyl4, and bin2-1 pyr1 pyl1 pyl2 pyl4 grown on medium with or without (Mock) 10 μM ABA. (B) Relative primary root length in Col-0:er (n = 25), bin2-1:er (n = 15), pyr1 pyl1 pyl2 pyl4 (n = 25), and bin2-1 pyr1 pyl1 pyl2 pyl4 (n = 20). (C) Induction of RAB18, RD26, and RD29B expression by ABA in pyr1 pyl1 pyl2 pyl4 and bin2-1 pyr1 pyl1 pyl2 pyl4. Seedlings were treated with 50 μM ABA or solvent only (Mock) for 1 h. (D) Root phenotype of Col-0, abi2-3, bin2-1, and abi2-3 bin2-1. (E) Relative primary root length in Col-0 (n = 25), bin2-1 (n = 15), abi2-3 (n = 25), and abi2-3 bin2-1 (n = 20). (F) Induction of RAB18, RD26, and RD29B expression by ABA in abi2-3 and abi2-3 bin2-1. Seedlings were treated with 50 μM ABA or solvent only (Mock) for 1 h. (G) Primary root phenotype of Col-0, bin2-1, SnRK2-RNAi/Col-0, and SnRK2-RNAi/bin2-1. (H) Relative primary root length in Col-0 (n = 20), SnRK2-RNAi/Col-0 (n = 20), bin2-1 (HM) (n = 20), and SnRK2-RNAi/bin2-1 (HM) (n = 20). (I) Induction of RAB18, RD26, and RD29B expression by ABA in bin2-1 and SnRK2-RNAi/bin2-1. Seedlings were treated with 50 μM ABA or solvent only (Mock) for 1 h. (Scale bars, 1 cm in A, D, and G). Primary root length of each material in mock treatment was normalized to 100%. Expression level of each gene for each material under mock treatment was normalized to “1” in C, F, and I. Values are means ± SE in B, C, E, F, H, and I. Student's t test was used to evaluate the significance of indicated comparisons. Significant levels: *P < 0.05; **P < 0.01; and ***P < 0.001.
To test whether BIN2-modulated ABA responses are mediated by SnRK2s, we created knockdown lines of SnRK2s using an RNAi approach in wild-type and bin2-1, designated as SnRK2-RNAi/Col-0 and SnRK2-RNAi/bin2-1, respectively. In both RNAi lines, the expression level of SnRK2.2 was reduced to 1% of wild-type, and the SnRK2.3 expression level was reduced by 40–60% of wild-type, but the expression level of SnRK2.6 was only slightly reduced (Fig. S6C). In primary root inhibition assays, SnRK2-RNAi/bin2-1 and SnRK2-RNAi/Col-0 showed a similar sensitivity to ABA (Fig. 4 G and H), and the induction of RAB18, RD26, and RD29B expression by ABA in SnRK2s RNAi/bin2-1 was much lower than in bin2-1 (Fig. 4I), suggesting that BIN2 enhances ABA signaling outputs through these SnRK2s.
Discussion
This study provides several lines of evidence to strongly support that Arabidopsis GSK3-like kinases play positive roles in modulating ABA signaling outputs. First, bin2-3 bil1 bil2 was less sensitive to ABA in primary root inhibition, ABA-induced gene expression, and phosphorylating ABF fragment. Second, gain-of-function mutation of BIN2, bin2-1 was hypersensitive to ABA in both assays. Third, a GSK3 kinase inhibitor, bikinin, strongly inhibited ABA signaling outputs. Fourth, BIN2 can directly interact with SnRK2s and activate their kinase activity. Fifth, bikinin inhibited T180 phosphorylation of SnRK2.3 and its kinase activity in vivo. Finally, genetic analysis also indicated that BIN2 promoted ABA signaling through SnRK2s. Phosphorylation of SnRK2s by GSK3-like kinases is apparently important for their full activation. It has been speculated that activation of SnRK2.2 and SnRK2.3 may require other components (3, 17–19). GSK3-like kinases acting as positive modulators to promote SnRK2.2 and SnRK2.3 activity is providing a direct example.
Arabidopsis GSK3-like kinases may play important roles in integrating environmental cues with ABA signaling. It was reported that both ABA and NaCl can induce expression of BIL2, and plants overexpressing BIL2 exhibited an elevated tolerance to salt stress (29, 30). Furthermore, high salinity can activate ASKα, and its overexpression also enhanced seedlings’ tolerance to salt (31), and most Arabidopsis GSK3-like kinases are up-regulated by NaCl and osmotic stress (37). In addition, low humidity, NaCl, and sorbitol can rapidly activate SnRK2s, which is independent of Clade A PP2C and ABA biosynthesis (16, 17). Therefore, many abiotic stresses may activate SnRK2s by regulating certain GSK3-like kinases at transcriptional or posttranslational levels.
Although BR and ABA have antagonistic effects on seed germination (35, 38), the effect of BRs on ABA signaling outputs is apparently complicated. It was known that BR-deficient mutant de-etiolated-2 mutant (det2) (38) and BR-perception mutant bri1-301 (35) were hypersensitive to ABA in seed germination. BR-deficient mutants constitutive photomorphogenesis and dwarfism (cpd) and det2 (39) and BR-perception mutant bri1 were hypersensitive to ABA in primary root inhibition (40) (Fig. S6 D and E). A knockout mutant of Brassinosteroid signaling kinase 5 (BSK5), a positive regulator in BR signaling upstream of BIN2, was also hypersensitive to ABA in seed germination and primary root inhibition (41). However, bes1-D and bzr1-D, dominant mutants with enhanced BR signaling outputs and acting downstream of BIN2, were also hypersensitive to ABA in primary root inhibition (39) (Fig. S6 F and G). Therefore, BR signaling may not only play a role in inhibiting BIN2 activity to inhibit ABA signaling but also promote ABA signaling through downstream transcription factors.
We identified T180 as a novel transphosphorylation site of SnRK2.3 by BIN2, which was not among these well-known autophosphorylation sites of SnRK2s (21). Addition of bikinin to 1/2 MS medium inhibited the T180 phosphorylation in vivo (Fig. 3F). The subgroup III SnRK2s share high similarity in amino acid sequence with an identical activation loop (Fig. S7). It is likely that BIN2 can phosphorylate the conserved T181 of SnRK2.2 to enhance its activity. Interestingly, in vivo phosphorylation of SnRK2.3 on T180 and SnRK2.2 on T181 had been found in a previous study after ABA treatment (42). In addition, the fact that SnRK2.6K50N cannot be phosphorylated by BIN2 indicated that BIN2 may not activate SnRK2.6 in vitro. Because SnRK2.6 may also be activated through a PP2C-independent pathway (16, 17), it could be regulated by other GSK3-like kinases or other proteins in vivo. On the basis of these current and previous studies, we propose that certain GSK3-like kinases may be induced or activated by various abiotic stresses, including NaCl, drought, and wounding at transcription or protein levels (30, 31, 37). The activated GSK3-like kinases promote SnRK2.2 and SnRK2.3 by phosphorylating T181 and T180, probably also S177 and S176, respectively, to enhance ABA responses (Fig. S8). In addition, ABA early signaling may also regulate GSK3-like kinases, such as induction of BIL2 expression, to form a positive feedback loop to enhance ABA signaling. Further studies are needed to fully understand the mechanisms of how abiotic stresses, such as NaCl, activate GSK3-like kinases.
Materials and Methods
Plant Materials and Growth Condition.
Arabidopsis materials, including various mutants, transgenic plants, and their corresponding wild types used in this study, are summarized in Table S1. Plants were grown in a growth room at 23 °C under long-day conditions (16 h light/8 h dark). For primary root inhibition assay and RNA samples for marker gene expression, seedlings were grown on 1/2 MS medium in a growth chamber (Percival) at 23 °C under long-day conditions (16 h light/8 h dark). SI Materials and Methods describes in detail abi2-3 and the construction of SnRK2 RNAi lines (Table S2).
Primary Root Inhibition Assay.
Seeds were sterilized and sown on 1/2 MS medium containing 0.4% gellan gum and 1% (m/vol) sucrose. After stratification at 4 °C for 4 d, plates were put into a growth chamber under long-day conditions (16 h light/8 h dark) at 23 °C. After 4 d, seedlings were transferred to 1/2 MS medium containing 1% (m/vol) sucrose with or without 10 μM ABA under long-day conditions at 23 °C. Seedlings were allowed to grow for another 10 d and then collected for phenotypic analysis.
Semi-in Vivo Pull-Down.
Details in SI Materials and Methods.
Gene Expression Analysis by Quantitative Real-Time PCR.
Seeds were sterilized and then sown to 1/2 MS medium containing 0.4% gellan gum, with or without 10 μM bikinin (Calbiochem). The growth condition was the same as that in Primary Root Inhibition Assay. Ten-day-old seedlings were collected for indicated treatments and then were ground to fine powder in liquid nitrogen. Total RNA was extracted using the Tiangen RNApre Plant Kit. Procedures described previously (43) were used for cDNA synthesis and qRT-PCR. Primers for qRT-PCR are listed in Table S3. PCRs were performed with an Eppendorf iCycler.
Kinase Assay with the IPed SnRK2.3-MYC.
Details in SI Materials and Methods.
In-Gel Kinase Assay.
Details in SI Materials and Methods.
In Vitro Kinase Assay.
Details in SI Materials and Methods.
Transient Transformation and BiFC Assay.
Agrobacterium strain LBA4404 was transformed with SnRK2-cYFP, nYFP-BIN2, or control vectors and then infiltrated into young leaves of N. benthamiana. Plants were put in the dark for 1 d and then allowed to grow under long-day conditions (16 h light/8 h dark). Fluorescence signals in pavement cells were visualized by confocal microscopy (Zeiss) after 24–36 h.
Quantitative Mass Spectrum Analysis on the in Vivo Phosphorylation of SnRK2.3.
Details in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank S. Cutler for providing seeds of the quadruple mutant pyr1 pyl1 pyl2 pyl4, and Y. Li and X. Zhang for facilitating quantitative mass spectrum analysis. This work was supported by Grant 2012CB114300 from the National Basic Research Program of China (to X.W.), Grants 30925020 and 91117005 from the National Natural Science Foundation of China (to X.W.), Grant 10JC1400800 from the Key Project of Shanghai Science and Technology Committee (to X.W.), Grant 11XD1400700 from the Program of Shanghai Subject Chief Scientist (to X.W.), and by the Academy of Finland [Project Nos. 259169, 263853, and 271832 (to H.F.)].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316717111/-/DCSupplemental.
References
- 1.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 2.Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15(7):395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev. 2010;24(16):1695–1708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Umezawa T, et al. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010;51(11):1821–1839. doi: 10.1093/pcp/pcq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324(5930):1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324(5930):1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 7.Fujii H, et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462(7273):660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umezawa T, et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA. 2009;106(41):17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi Y, et al. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005;44(6):939–949. doi: 10.1111/j.1365-313X.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- 10.Furihata T, et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA. 2006;103(6):1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soon FF, et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science. 2012;335(6064):85–88. doi: 10.1126/science.1215106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14(12):3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19(2):485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakashima K, et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50(7):1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- 15.Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA. 2009;106(20):8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida R, et al. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem. 2006;281(8):5310–5318. doi: 10.1074/jbc.M509820200. [DOI] [PubMed] [Google Scholar]
- 17.Boudsocq M, Droillard MJ, Barbier-Brygoo H, Laurière C. Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol Biol. 2007;63(4):491–503. doi: 10.1007/s11103-006-9103-1. [DOI] [PubMed] [Google Scholar]
- 18.Fujii H, Zhu JK. Osmotic stress signaling via protein kinases. Cell Mol Life Sci. 2012;69(19):3165–3173. doi: 10.1007/s00018-012-1087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujita Y, Yoshida T, Yamaguchi-Shinozaki K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol Plant. 2013;147(1):15–27. doi: 10.1111/j.1399-3054.2012.01635.x. [DOI] [PubMed] [Google Scholar]
- 20.Burza AM, et al. Nicotiana tabacum osmotic stress-activated kinase is regulated by phosphorylation on Ser-154 and Ser-158 in the kinase activation loop. J Biol Chem. 2006;281(45):34299–34311. doi: 10.1074/jbc.M601977200. [DOI] [PubMed] [Google Scholar]
- 21.Ng LM, et al. Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases. Proc Natl Acad Sci USA. 2011;108(52):21259–21264. doi: 10.1073/pnas.1118651109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimes CA, Jope RS. The multifaceted roles of glycogen synthase kinase 3β in cellular signaling. Prog Neurobiol. 2001;65(4):391–426. doi: 10.1016/s0301-0082(01)00011-9. [DOI] [PubMed] [Google Scholar]
- 23.Saidi Y, Hearn TJ, Coates JC. Function and evolution of ‘green’ GSK3/Shaggy-like kinases. Trends Plant Sci. 2012;17(1):39–46. doi: 10.1016/j.tplants.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 24.He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA. 2002;99(15):10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109(2):181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 26.Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA. 2008;105(28):9829–9834. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim TW, Michniewicz M, Bergmann DC, Wang ZY. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature. 2012;482(7385):419–422. doi: 10.1038/nature10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gudesblat GE, et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat Cell Biol. 2012;14(5):548–554. doi: 10.1038/ncb2471. [DOI] [PubMed] [Google Scholar]
- 29.Piao HL, et al. An Arabidopsis GSK3/shaggy-like gene that complements yeast salt stress-sensitive mutants is induced by NaCl and abscisic acid. Plant Physiol. 1999;119(4):1527–1534. doi: 10.1104/pp.119.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piao HL, Lim JH, Kim SJ, Cheong GW, Hwang I. Constitutive over-expression of AtGSK1 induces NaCl stress responses in the absence of NaCl stress and results in enhanced NaCl tolerance in Arabidopsis. Plant J. 2001;27(4):305–314. doi: 10.1046/j.1365-313x.2001.01099.x. [DOI] [PubMed] [Google Scholar]
- 31.Dal Santo S, et al. Stress-induced GSK3 regulates the redox stress response by phosphorylating glucose-6-phosphate dehydrogenase in Arabidopsis. Plant Cell. 2012;24(8):3380–3392. doi: 10.1105/tpc.112.101279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koh S, et al. T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol Biol. 2007;65(4):453–466. doi: 10.1007/s11103-007-9213-4. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127(1):14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan Z, Zhao J, Peng P, Chihara RK, Li J. BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 2009;150(2):710–721. doi: 10.1104/pp.109.138099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S, Cai Z, Wang X. The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc Natl Acad Sci USA. 2009;106(11):4543–4548. doi: 10.1073/pnas.0900349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Rybel B, et al. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem Biol. 2009;16(6):594–604. doi: 10.1016/j.chembiol.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charrier B, Champion A, Henry Y, Kreis M. Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol. 2002;130(2):577–590. doi: 10.1104/pp.009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steber CM, McCourt P. A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 2001;125(2):763–769. doi: 10.1104/pp.125.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodrigues A, et al. The short-rooted phenotype of the brevis radix mutant partly reflects root abscisic acid hypersensitivity. Plant Physiol. 2009;149(4):1917–1928. doi: 10.1104/pp.108.133819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111(3):671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li ZY, et al. A mutation in Arabidopsis BSK5 encoding a brassinosteroid-signaling kinase protein affects responses to salinity and abscisic acid. Biochem Biophys Res Commun. 2012;426(4):522–527. doi: 10.1016/j.bbrc.2012.08.118. [DOI] [PubMed] [Google Scholar]
- 42.Wang P, et al. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc Natl Acad Sci USA. 2013;110(27):11205–11210. doi: 10.1073/pnas.1308974110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, et al. Dual role of BKI1 and 14-3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev Cell. 2011;21(5):825–834. doi: 10.1016/j.devcel.2011.08.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.