Significance
Auxin is a simple small molecule, yet it elicits an enormous variety of responses. How a single molecule can encode such diverse and context-specific information is a long-standing question. One hypothesis is that variation in auxin response components generates diversity in local auxin responses. To test this, we transplanted the forward nuclear auxin signal transduction pathway from plants into yeast. We found that auxin circuit composition can tune the dynamic response, and we identified which properties of auxin response components had the largest impact on circuit performance. Reconstitution of eukaryotic pathways in novel contexts can inform our understanding of the pathways themselves and can generate tools for engineering novel cellular behaviors.
Keywords: synthetic biology, signaling dynamics
Abstract
Auxin influences nearly every aspect of plant biology through a simple signaling pathway; however, it remains unclear how much of the diversity in auxin effects is explained by variation in the core signaling components and which properties of these components may contribute to diversification in response dynamics. Here, we recapitulated the entire Arabidopsis thaliana forward nuclear auxin signal transduction pathway in Saccharomyces cerevisiae to test whether signaling module composition enables tuning of the dynamic response. Sensitivity analysis guided by a small mathematical model revealed the centrality of auxin/indole-3-acetic acid (Aux/IAA) transcriptional corepressors in controlling response dynamics and highlighted the strong influence of natural variation in Aux/IAA degradation rates on circuit performance. When the basic auxin response circuit was expanded to include multiple Aux/IAAs, we found that dominance relationships between coexpressed Aux/IAAs were sufficient to generate distinct response modules similar to those seen during plant development. Our work provides a new method for dissecting auxin signaling and demonstrates the key role of Aux/IAAs in tuning auxin response dynamics.
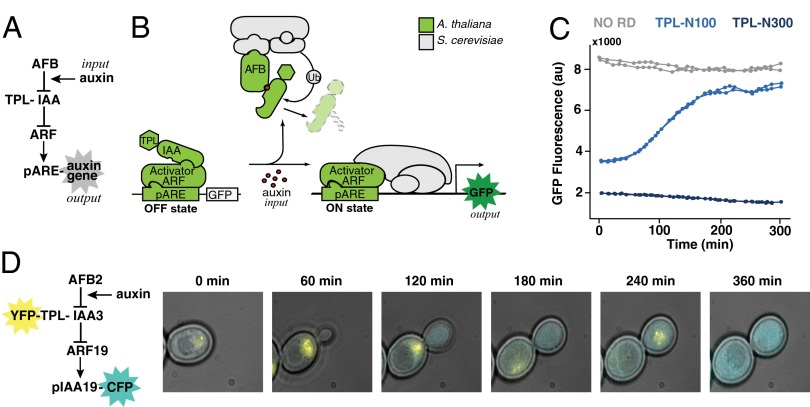
Evolution depends on the plasticity of existing signaling pathways. The small molecule auxin is linked to signaling modules that allowed plants to move to land, develop new organs, and respond to the environment (1, 2). Despite the wide range of auxin responses, the core auxin signal transduction pathway is quite simple, involving a small number of components from perception through transcription (Fig. 1A). Auxin triggers the rapid turnover of Aux/IAA (IAA) proteins, which repress the activity of auxin response factor (ARF) transcription factors through recruitment of TOPLESS (TPL) corepressors (3, 4). Auxin receptors, auxin signaling F-box (AFB) proteins, act as part of an E3 ubiquitin ligase to catalyze the ubiquitination and subsequent degradation of IAAs when auxin is present (5, 6). ARFs bound to auxin-responsive cis-regulatory elements (AuxREs) are then free to regulate the expression of auxin target genes, which include the IAAs themselves (7–9).
Fig. 1.
Auxin-induced transcription in yeast. (A) Network diagram of the forward auxin response pathway in yeast. An auxin input increases association of a member of the TIR1/AFB family of F-box proteins (AFB) and an IAA protein fused to the first 100 aa of the TPL corepressor (TPL-IAA). Auxin-induced association of an AFB and a TPL-IAA leads to degradation of the TPL-IAA, thereby freeing a transcriptional activator in the ARF family to induce expression of an output gene driven by a promoter containing an auxin response element (pARE). (B) The five A. thaliana components needed to recapitulate auxin response in S. cerevisiae are shown in light green. They were an AFB F-box receptor, an IAA, a TPL corepressor, an ARF transcription factor, and an auxin-responsive promoter. The remaining cellular machinery (gray) was supplied by yeast. Fluorescence from a GFP reporter was used as a quantitative output. (C) Synthetic auxin-reversible repression required fusion of a specific TPL truncation to the IAA protein. Flow cytometry was used to monitor the induction of a GFP reporter following auxin treatment in circuits containing either IAA14 with no repression domain (NO RD), shown in gray, or two different C-terminal TPL truncations fused to IAA14. Auxin was added at time 0. TPL-N300 (dark blue) includes the first 300 aa of TPL and excludes the multiple C-terminal WD repeats. Fusion of TPL-N300 to IAA proteins results in reporter repression that is largely auxin-insensitive. TPL-N100 (light blue) includes only the first 100 aa of TPL. When fused to an IAA, TPL-N100 provides auxin-reversible repression. Two replicate induction curves are shown for each circuit. (D) Auxin-induced IAA degradation and subsequent transcriptional activation could be simultaneously monitored in dual-labeled yeast strains. YFP-TPL-IAA3 and a Cerulean reporter driven by the auxin-responsive IAA19 promoter (pIAA19-CFP) were monitored following auxin treatment using time-lapse microscopy and a microfluidic chamber.
How such a simple pathway can orchestrate the large number of context-specific responses regulated by auxin is a long-standing question. Notably, each component in the auxin signaling pathway belongs to a large gene family (2, 10, 11). In Arabidopsis, there are 6 AFBs, 23 ARFs, and 29 IAAs. Functional divergence between component family members could provide variation in response to a generic auxin signal (12). Members of the ARF family can be classified as transcriptional activators or repressors (7, 8), and the role of repressor ARFs in regulating auxin signaling is still a matter of debate (13). In addition, expression studies of auxin-signaling gene families support the idea of an “auxin pre-pattern” where certain combinations of coexpressed components generate auxin circuits with distinct auxin response capabilities (14, 15). However, owing to the high level of redundancy and coexpression of auxin signaling component family members, feedback, and interference from other signaling pathways (16), the contributions of individual components to auxin circuit behavior have remained elusive.
Here, we ported several variations of the auxin transcriptional response pathway from Arabidopsis thaliana to Saccharomyces cerevisiae. In so doing, we defined a minimal auxin response circuit (ARC) sufficient to recapitulate auxin-induced transcription in a heterologous context. Because no feedback was engineered into these ARCs, they captured the dynamic potential of the simplest forward response architecture. The ARC consists of five plant components: an AFB, an IAA, TPL, an activating ARF transcription factor, and an auxin-responsive plant promoter (Fig. 1). The implementation of ARCs in yeast (ARCSc) relied on successful interfacing of the ARC with other essential elements of the yeast degradation and transcriptional machinery (Fig. 1B). This modular, plug-in nature of the ARC, similar to what was observed with auxin-induced degradation (17), highlights the likelihood that the ARC could be implemented in a variety of other eukaryotic contexts. The suite of ARC variants represented in the ARCSc collection presented an opportunity to quantitatively investigate the dynamic capabilities of the auxin response in isolation.
Auxin response pivots on relief of transcriptional repression, yet neither expression of an IAA alone nor coexpression with TPL repressed induction of the IAA19 promoter by activator ARFs (Fig. 1C and SI Appendix, Fig. S1). A fusion of full-length TPL with the ARF-interaction domain of IAA12 is able to fully recapitulate the stabilized IAA12 mutant phenotype (3). Following this logic, we identified two TPL truncations (TPL-N100 and TPL-N300) that conferred repression in yeast when fused to a full-length IAA. However, only repression by TPL-N100 (hereafter referred to as TPL) was relieved by auxin treatment (Fig. 1C). In a yeast strain where we could simultaneously monitor degradation of the TPL:IAA fusion and reporter activation, we observed reporter activation dynamics similar to those observed in cells where the TPL:IAA fusion was not labeled (Fig. 1D). Thus, performance of ARCSc is consistent with the established model of plant auxin signaling, with the exception that the TPL and IAA components are acting as a single protein.
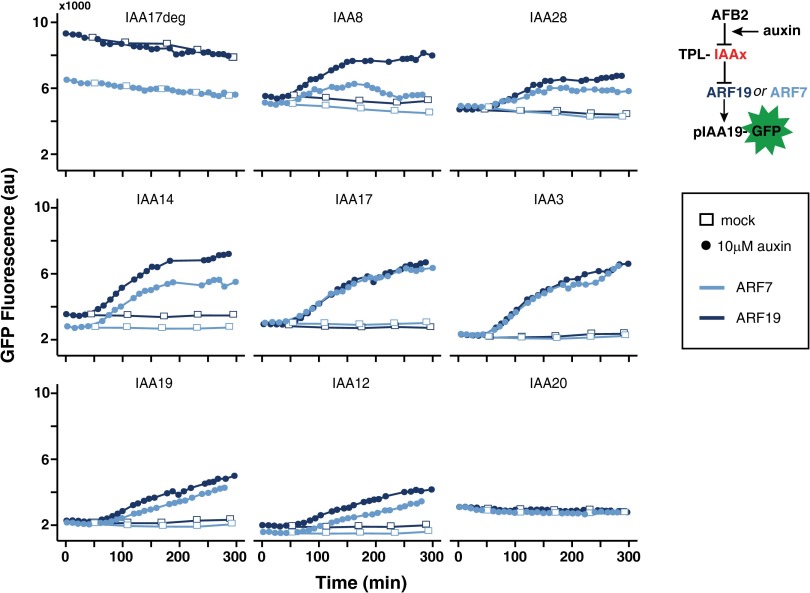
The expansion of the IAA gene family has been linked to the increase in auxin signal complexity of land plants (2), suggesting IAAs play an essential role in tuning the auxin response. To explore the range of behaviors possible in a basic ARC configuration, we therefore took advantage of the large number of naturally evolved variants in the IAA gene family. We tested the impact of different IAAs on response dynamics of ARCSc variants containing either ARF19 or ARF7 using time-lapse flow cytometry (Fig. 2 and SI Appendix, Fig. S2). Our analysis focused primarily on IAAs that display strong mutant phenotypes when stabilized by point mutations that disrupt formation of the AFB–auxin–IAA complex (18–23). Additional IAAs found in distinct subgroups in the IAA family tree were also included (10). Circuits containing an IAA missing the ARF interaction domain (IAA17deg) failed to repress ARF activity, and thus indicated the maximum potential for reporter activation (Fig. 2). Yeast expressing IAA20, an IAA with a substantially diverged degron sequence rendering it insensitive to auxin, showed no reporter activation. Circuits with other IAAs exhibited substantial differences in the initial repressed state, as well as in the magnitude and rate of GFP reporter induction. The induction curves for a given IAA with either ARF19 or ARF7 were qualitatively quite similar, suggesting that ARF7 and ARF19 have similar or overlapping functions. This interpretation is supported by genetic and phylogenetic analysis in plants (24–26). This does not exclude the possibility that other ARF–IAA combinations, or higher-order interactions between multiple components from the ARF and IAA families, could generate additional diversity.
Fig. 2.
IAAs drive auxin response dynamics. Representative auxin-induced reporter fluorescence curves are shown for ARCs with ARF7 (light blue) or ARF19 (dark blue) in the context of different IAAs. Auxin was added at time 0 in all graphs. ARF7 and ARF19 showed qualitatively similar patterns of auxin response, whereas the identity of the IAA had a dramatic effect on ARC dynamics. ARCSc recapitulated regulatory features of plant ARC function. Transcriptional repression required the known ARF–IAA interaction domain, because an IAA lacking this domain (IAA17deg) had no effect on ARF activity. In addition, auxin response was mediated by IAA degradation, because a naturally occurring IAA lacking a degron (IAA20) rendered the circuit insensitive to auxin treatment.
To quantify the differences in the behavior of ARCSc IAA variants, we developed a small model that expands upon the model used to quantify our degradation system (27). The present model captures the overall flow of information while avoiding reference to specific molecules and kinetic parameters that cannot be measured and limiting the potential for overfitting (28, 29). In our model the variable u represents the concentration of the applied auxin input and the variable g represents the GFP output of the reporter (Fig. 3A). A lumped internal state, which combines multiple reactions including the binding of auxin to the AFB receptor and related molecular machinery, is represented by x. IAA protein levels are represented by y. The model has eight parameters (k1–8) that intuitively correspond to the biological processes listed in Table 1. We tested the validity of this interpretation numerically by evaluating fits of the data using different assumptions about which parameters change when different ARFs or IAAs are used in the circuit (SI Appendix). Our analysis, and the interpretation in Table 1, suggests varying the IAA corresponds to tuning three parameters: expression level (k3), auxin-induced degradation (k5), and ARF affinity (k8) of each IAA. The rest of the parameters are independent of the changing components in the circuit except for k7, which depends on which ARF is present.
Fig. 3.
Model selection and sensitivity analysis of the auxin response pathway. (A) Gray-box model of the auxin response pathway. The auxin input is represented by the variable u and the GFP output of the reporter is represented by the variable g. The variable x represents a lumped internal state, which combines multiple reactions including the binding of auxin to the AFB receptor, and the variable y represents IAA protein levels. The model has eight parameters (k1–8) that intuitively correspond to the biological processes listed in Table 1. We focused on three parameters with strong effect on ARC dynamics: IAA expression level (k3), rate of auxin-induced IAA degradation (k5), and ARF-IAA affinity (k8). (B) A graphical representation of the two performance metrics used for sensitivity analysis: the preauxin steady state g0 and the activation time ΔT (t90–t10). Sample data from one ARC is shown as blue dots with a sample model fit in red. (C and D) Sensitivity analysis of preauxin steady state g0 (C) and activation time ΔT (D) to model parameter values of k3, k5, and k8 for each IAA. Each parameter was varied across its entire range of estimated values derived from our experimental dataset, and all other parameters were held constant. Each IAA is plotted as a single point, and the red line indicates the sensitivity curve computed from the model. The preauxin steady state g0 was accurately predicted by k3 (expression level) and to a lesser extent by k8 (ARF affinity), with k5 having little effect. In contrast, activation time ΔT was predicted with high accuracy by k5 (auxin-induced degradation) alone. Error bars represent SD (n = 2). More details can be found in SI Appendix.
Table 1.
Parameter interpretations
| Parameter | Biological interpretation |
| k1 | Assembly of auxin-primed AFB receptor |
| k2 | Basal degradation/dissociation of auxin-primed AFB receptor |
| k3 | IAA expression |
| k4 | Basal degradation/dilution of IAA |
| k5 | Auxin-induced degradation of IAA |
| k6 | Basal degradation/dilution of GFP |
| k7 | GFP expression |
| k8 | ARF affinity of IAA |
To identify how IAA features influence the observed differences in response behavior, we defined two performance metrics: the preauxin steady state g0 and the activation time ΔT (Fig. 3B). The metrics were plotted against the estimated IAA dependent parameter values for each variant to quantify the sensitivity of the pathway to these parameters. The preauxin steady state g0 was accurately predicted by k3 (expression level) and to a lesser extent by k8 (ARF affinity), whereas, as the model suggests, k5 did not reliably affect g0 (Fig. 3C and SI Appendix). In other words, the “OFF” state of the circuit before auxin treatment is largely determined by the amount of IAA present and only modestly affected by the affinity of the IAA for the ARF. Experimentally, we also confirmed that ARCs containing the high-expressing IAA1.1, encoded by a yeast codon-optimized IAA1 (27), had a much lower preauxin steady state than those with IAA1 (SI Appendix, Fig. S3). In contrast, activation time ΔT was predicted with high accuracy by k5 alone (Fig. 3D). This result indicates that natural variation in IAA degradation rates—and not similar levels of variation in the other parameters tested here—is sufficient to dramatically alter the dynamics of auxin response.
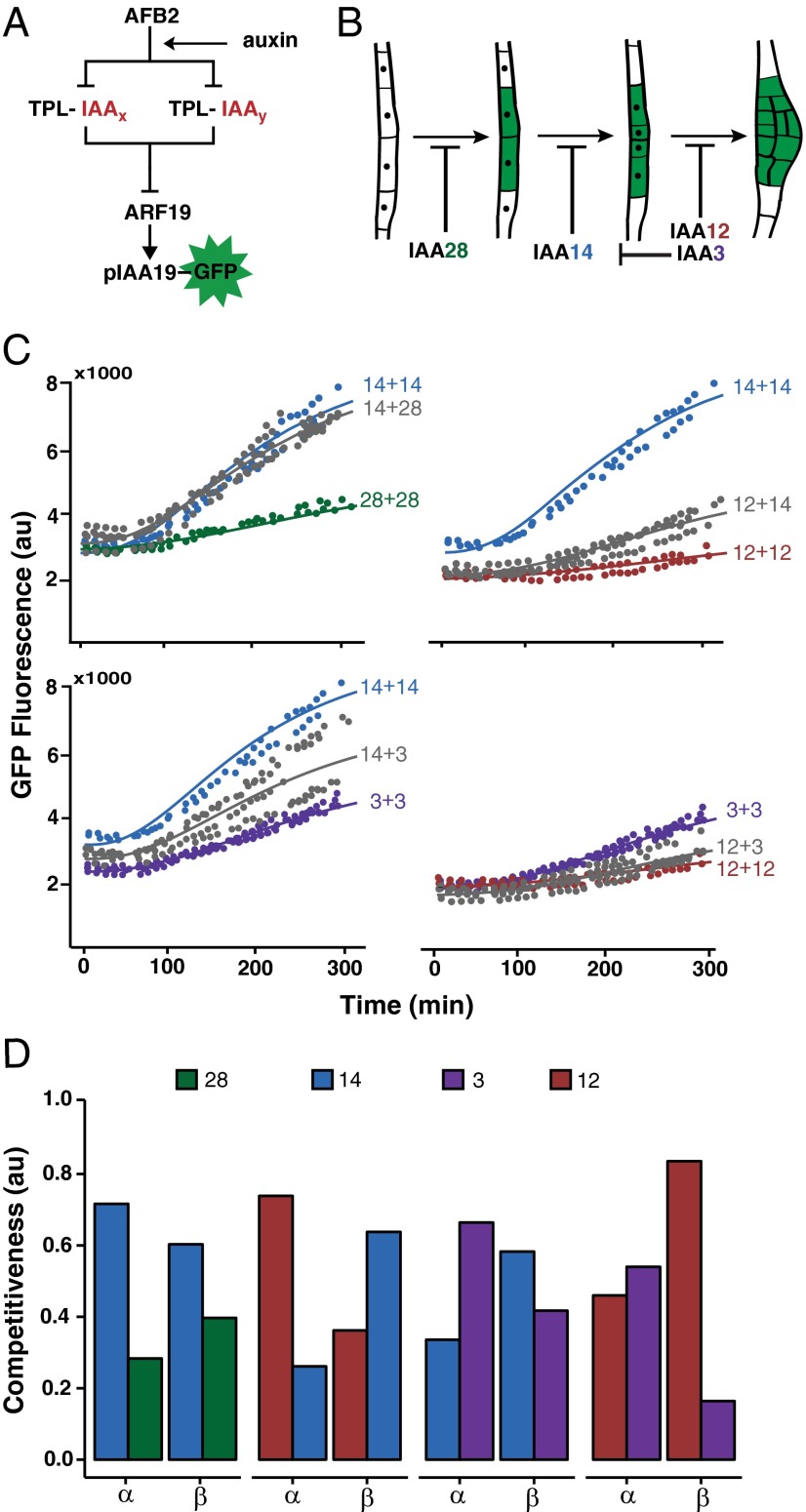
We next engineered ARCSc variants with pairs of IAAs to test the impact of competition between IAAs on auxin response (Fig. 4A). This architecture was inspired by work suggesting that sequential modules of ARF–IAA partners drive discrete stages in the development of lateral roots (30–32). IAA28, IAA14, IAA12, and IAA3 have all been implicated in lateral root development (Fig. 4B). IAA28 regulates founder-cell specification (33) and IAA14 subsequently drives the initial cell divisions (22). IAA12 is involved in later divisions and outgrowth (30), and IAA3 has been shown to be involved in emergence as well as negative regulation of the IAA14 module (34). Limited expression studies available for IAA and ARF family members indicate that multiple family members are often coexpressed in plant cells, raising the question of how specificity or ordering is encoded (14, 15). Our data and those of others suggest that multiple IAAs can interact with and repress the same activator ARF (15). One attractive hypothesis is that a hierarchy of ARC function is encoded by a hierarchy of function within the IAAs themselves.
Fig. 4.
IAA coexpression reveals dominance relationships between IAAs. (A) Modified ARF19 circuits were built that contained pairs of IAAs (IAAx and IAAy). (B) Schematic showing the simplified sequence of lateral root development: founder cell specification, first cell division and patterning/outgrowth. Auxin response modules are proposed to be sequentially triggered by degradation of the indicated IAAs. (C) One IAA could dictate the auxin response dynamics of yeast cells expressing multiple IAAs. Comparisons are shown between circuits containing two copies of the same IAA (colored points) or one copy of each IAA (gray points). Model fits are shown as solid lines. (D) α (AFB binding competiveness) or β (ARF binding competitiveness) can affect the contribution of an IAA to ARC dynamics. Estimated α and β values from the modified model are shown for each of the IAAs tested here. More details can be found in SI Appendix.
Although it was not possible to experimentally determine a mechanism for this potential for hierarchical action, we were able to test whether such a pattern can be simulated in a minimal synthetic context. Toward that end, we generated ARCSc variants expressing pairwise combinations of IAA3, IAA12, IAA14, and IAA28. These competition strains made it possible to characterize the impact of an additional IAA on the dynamics of an ARC. Induction curves of ARCSc competition strains indicated that transcriptional dynamics were largely biased toward the IAA that acts in later developmental modules (Fig. 4C). The 12+14 and 12+3 circuits exhibited response dynamics similar to that of a 12+12 circuit, whereas transcriptional dynamics of 14+28 circuits were nearly identical to a 14+14 circuit. The 14+3 transcriptional dynamics showed a high degree of variation from one experiment to another, following response dynamics of either 14+14 or 3+3 circuits. This leads to a dominance hierarchy of 28 < 14, 3 < 12, which matches the proposed sequence of roles in lateral root development.
The preferential response to one of the two coexpressed IAAs suggests dominance relationships between IAAs could play a role in the generation of sequential auxin responses. Novel dynamics created by heterodimers may also be a factor in some cases, although the strong dominance of one IAA over another makes this less likely for most of the examples tested here. The dominance relationships uncovered here could explain how transcriptional dynamics could be switched from one regime to another over time. IAAs are among the earliest auxin response genes (9), suggesting that differences in synthesis and degradation rates among family members could lead to a dramatic change in the cellular complement of IAAs after exposure to auxin. In this way, IAAs may tune the intensity and duration of downstream transcriptional outputs similar to IκB repressors in the mammalian NF-κB pathway (35, 36).
We hypothesized that dominance between IAAs could reflect competition for two key interactions in the transmission of an auxin signal: binding to the AFB and binding to the ARF (Fig. 4A). These interactions are approximately captured by the parameters k5 (auxin-induced degradation rate) and k8 (ARF affinity). In plants, where IAAs are strongly and differentially auxin-induced, IAA expression level is also likely to play a critical role in determining which IAA drives transcriptional dynamics. However, because both IAAs in the yeast strains tested here are expressed from the same strong constitutive yeast promoters, we predicted that k3 (expression level) was unlikely to have a significant role in determining dominance in our assays. Indeed, we found only minor differences in expression levels between coexpressed IAAs, and these small differences had no correlation to dominance (SI Appendix, Fig. S4). Because all of our rate estimates were obtained from experiments where only a single IAA is expressed, these estimates cannot be directly applied to predict competition outcomes in the more complex setting of multiple coexpressed IAAs. Instead, we modified our model to introduce two new parameters, α and β (SI Appendix), which represent the relative contributions of the two competing IAAs to the parameters k5 and k8, respectively. Thus, α and β describe which IAA is dominant in terms of its affinity to either the AFB (α) or the ARF (β). IAAs that demonstrated clear dominance in the behavior of the induction curve in mixed IAA circuits had a higher α and/or β than the other coexpressed IAA (Fig. 4D). It is worth noting that each of the original parameters, as well as the derived α and β, encompass multiple rates controlled by distinct biochemical mechanisms. Given the current limits for experimental measurement of each of the constituent rates within these composite parameters, it is impossible to predict the dominance relationship of any IAA pair or to explain observed dominance by an explicit molecular mechanism. Instead, these competition experiments serve to underline the striking complexity possible with even relatively simple circuits and generate a new hypothesis for sequential auxin circuit function during development.
By transplanting auxin response into yeast and defining a small mathematical model able to explain ARC dynamics, we were able to rigorously test how auxin response is controlled and tuned using only the core ARC components. When combined with feedback and interactions with other pathways, such tuning knobs likely have even more dramatic effects. By finding the ARC tuning knobs, this approach generated hypotheses about the evolution of natural ARC variants in plants, as well as showing a path to engineer novel circuit behaviors in synthetic contexts. The inherent modularity of the ARC lends itself to adding additional layers of complexity and quantitative analysis of other challenging features of auxin response, such as feedback or higher-order complexes between ARFs and IAAs. Indeed, recent structural studies indicate the potential for higher-order ARF–IAA complexes than previously suspected (37–39). ARCSc may be a useful tool for quantifying the impact of this structural complexity on response dynamics. In addition, incorporation of additional auxin-responsive promoters could expand the functionality of our system and allow incorporation of combinatorial control by multiple transcription factors. More generally, ARCSc demonstrates how the reconstitution of eukaryotic pathways can serve as a powerful companion to systems biology-driven studies that generate predictions about functional signaling modules.
Materials and Methods
Strain Construction.
A library of IAAs was cloned downstream of a GPD promoter in TRP1 integration vectors using a standard Gateway LR reaction (LRClonase II; Life Technologies) and transformed into W814-29B MATa strains (YKL381) containing pGP5G-AFB2 [AFB2 integration at LEU2, expression from the GPD promoter (27)]. ARFs were amplified from Arabidopsis cDNA and subcloned into the Gateway pDONR221 plasmid using a standard Gateway BP reaction (BP Clonase II; Life Technologies). Each ARF was then subsequently cloned into pGP8A-ccdB (integration at HIS3, expression from the ADH1 promoter) and transformed into a W303-1A MATa strain containing the pIAA19-GFP reporter integrated at URA3 (YKL12). Strains containing AFB2 and an IAA were then mated to strains containing the reporter and an ARF to generate the library of ARCSc variants. See SI Appendix, Table S1 for strains used in this study.
Plasmid Construction.
Arabidopsis genes were inserted into single integrating derivatives of pAG Gateway vectors (27, 40) (SI Appendix, Fig. S5 and Table S2). TRP1 targeting single integrating Gateway vectors (27) were further modified to contain an N-terminal TPL repression domain (RD) for auxin response assays, or an N-terminal YFP-RD tag to monitor the degradation of RD–IAA fusions. Repression domains TPL-N100 and TPL-N300 were amplified from A. thaliana Columbia ecotype cDNA. For the GFP auxin induction reporter plasmid, 300 bp immediately upstream of the IAA19 start codon was amplified from Arabidopsis gDNA and inserted upstream of GFP in the pAG306 (URA3, multiply inserting) targeting vector.
Yeast Methods.
Standard yeast drop-out and yeast extract–peptone–dextrose plus adenine (YPAD) media were used, with care taken to use the same batch of synthetic complete (SC) media for related experiments. A standard lithium acetate protocol (41) was used for transformations of digested plasmids. All cultures were grown at 30 °C with shaking at 220 rpm.
Flow Cytometry.
Fluorescence measurements were taken with a BD Accuri C6 flow cytometer and CSampler plate adapter using an excitation wavelength of 488 and an emission detection filter at 533 nm. A total of 10,000 events above a 400,000 FSC-H threshold (to exclude debris) were measured for each sample and data exported as FCS 3.0 files for processing using the flowCore R software package and custom R scripts (27).
Induction Assays.
A freshly grown colony for each strain was inoculated in SC and the cell density (in events per microliter) was estimated using cytometry data gated for yeast by a custom R script (27). Each culture was then diluted to 0.25 events per microliter in 12 mL of SC. This dilution was split into duplicate 5-mL aliquots and incubated for 16 h. At 16 h, duplicate aliquots were combined, mixed, and resplit into two tubes and initial measurements taken in the cytometer (starting density was ∼200 events per microliter). For each strain, one replicate was mock-treated [95% (vol/vol) ethanol] and one replicate was treated with 10 μM indole-3-acetic acid. Immediately after auxin addition, fluorescence for the 0-min time point was recorded and subsequent measurements acquired at 10-min intervals for 5 h. Up to 12 strains were measured in parallel.
Steady-State Expression Measurements.
A freshly grown colony was inoculated into 1 mL of SC in a 96-well plate and grown to stationary overnight. The next morning each culture was diluted into fresh SC and fluorescence measurements taken ∼6 h later.
Microscopy.
Yeast cells grown overnight in SC at 30 °C were diluted 1:100 in SC, incubated for 4–5 h, and then diluted 1:20 before loading onto a Y04D plate (CellASIC). Using the CellASIC-ONIX microfluidic system and associated software, cells were treated with 10 μM auxin in SC medium. An inverted Nikon Eclipse Ti microscope with a 60×, numerical aperture and 1.4 oil objective was used to image the yeast cells at 1-h intervals using a YFP-HYQ and CFP bandpass filter (Nikon) with a CoolSNAP HQ2 14-bit camera.
Expression Analysis.
Yeast cells were grown in YPAD and collected at an OD of 0.9. RNA was extracted using the RiboPure RNA purification kit (Life Technologies). One microgram of RNA was used for cDNA synthesis using the iScript cDNA Synthesis kit (Bio-Rad). Samples were analyzed using the iQ SYBR Green Supermix (Biorad) reactions in a C100 Thermal Cycler fitted with a CFX96 Real-Time Detection System (Biorad). Relative expression levels were calculated using the formula (Etarget)−CPtarget/(Eref)−CPref (42) and normalized to the ACT1 reference gene.
Quantitative Analysis.
A gray-box modeling approach was used to identify a minimal mathematical model for the auxin response characterized by ARCSc variants. A more detailed description of the modeling and quantitative analysis is found in SI Appendix. The annotated code and supporting data are available for download at http://klavinslab.org/data.html.
Supplementary Material
Acknowledgments
We thank Takato Imaizumi, Ben Kerr, Georg Seelig, and Tom Daniel for careful reading of our manuscript; Nick Bolten for work on optimizing plasmid construction; Miles Gander for technical assistance; and members of the J.L.N. and E.K. groups for helpful discussions. This work was supported by the Paul G. Allen Family Foundation (J.L.N. and E.K.) and National Science Foundation (NSF) Grant CISE-0832773 (to E.K.). E.P.J. was supported by an NSF Graduate Research Fellowship and the Seattle Chapter of the Achievement Rewards for College Scientists Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1324147111/-/DCSupplemental.
References
- 1.Finet C, Berne-Dedieu A, Scutt CP, Marlétaz F. Evolution of the ARF gene family in land plants: Old domains, new tricks. Mol Biol Evol. 2013;30(1):45–56. doi: 10.1093/molbev/mss220. [DOI] [PubMed] [Google Scholar]
- 2.Paponov IA, et al. The evolution of nuclear auxin signalling. BMC Evol Biol. 2009;9:126. doi: 10.1186/1471-2148-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319(5868):1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- 4.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9(11):1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 6.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 7.Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA. 1999;96(10):5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15(2):533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abel S, Nguyen MD, Theologis A. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol. 1995;251(4):533–549. doi: 10.1006/jmbi.1995.0454. [DOI] [PubMed] [Google Scholar]
- 10.Remington DL, Vision TJ, Guilfoyle TJ, Reed JW. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 2004;135(3):1738–1752. doi: 10.1104/pp.104.039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9(1):109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Lokerse AS, Weijers D. Auxin enters the matrix—assembly of response machineries for specific outputs. Curr Opin Plant Biol. 2009;12(5):520–526. doi: 10.1016/j.pbi.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Pierre-Jerome E, Moss BL, Nemhauser JL. Tuning the auxin transcriptional response. J Exp Bot. 2013;64(9):2557–2563. doi: 10.1093/jxb/ert100. [DOI] [PubMed] [Google Scholar]
- 14.Rademacher EH, et al. Different auxin response machineries control distinct cell fates in the early plant embryo. Dev Cell. 2012;22(1):211–222. doi: 10.1016/j.devcel.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 15.Vernoux T, et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol. 2011;7:508. doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Bianco M, Kepinski S. Context, specificity, and self-organization in auxin response. Cold Spring Harb Perspect Biol. 2011;3(1):a001578. doi: 10.1101/cshperspect.a001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6(12):917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 18.Tian Q, Reed JW. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development. 1999;126(4):711–721. doi: 10.1242/dev.126.4.711. [DOI] [PubMed] [Google Scholar]
- 19.Nagpal P, et al. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 2000;123(2):563–574. doi: 10.1104/pp.123.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouellet F, Overvoorde PJ, Theologis A. IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell. 2001;13(4):829–841. doi: 10.1105/tpc.13.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogg LE, Lasswell J, Bartel B. A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell. 2001;13(3):465–480. doi: 10.1105/tpc.13.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29(2):153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- 23.Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16(13):1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukaki H, Taniguchi N, Tasaka M. PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant J. 2006;48(3):380–389. doi: 10.1111/j.1365-313X.2006.02882.x. [DOI] [PubMed] [Google Scholar]
- 25.Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19(1):118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilmoth JC, et al. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005;43(1):118–130. doi: 10.1111/j.1365-313X.2005.02432.x. [DOI] [PubMed] [Google Scholar]
- 27.Havens KA, et al. A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol. 2012;160(1):135–142. doi: 10.1104/pp.112.202184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ljung L. System Identification: Theory for the User. 2nd Ed. Upper Saddle River, NJ: Prentice Hall; 1999. [Google Scholar]
- 29.Neuert G, et al. Systematic identification of signal-activated stochastic gene regulation. Science. 2013;339(6119):584–587. doi: 10.1126/science.1231456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Smet I, et al. Bimodular auxin response controls organogenesis in Arabidopsis. Proc Natl Acad Sci USA. 2010;107(6):2705–2710. doi: 10.1073/pnas.0915001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Smet I. Lateral root initiation: One step at a time. New Phytol. 2012;193(4):867–873. doi: 10.1111/j.1469-8137.2011.03996.x. [DOI] [PubMed] [Google Scholar]
- 32.Lavenus J, et al. Lateral root development in Arabidopsis: Fifty shades of auxin. Trends Plant Sci. 2013;18(8):450–458. doi: 10.1016/j.tplants.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 33.De Rybel B, et al. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol. 2010;20(19):1697–1706. doi: 10.1016/j.cub.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 34.Goh T, Kasahara H, Mimura T, Kamiya Y, Fukaki H. Multiple AUX/IAA-ARF modules regulate lateral root formation: The role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philos Trans R Soc Lond B Biol Sci. 2012;367(1595):1461–1468. doi: 10.1098/rstb.2011.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kearns JD, Basak S, Werner SL, Huang CS, Hoffmann A. IkappaBepsilon provides negative feedback to control NF-kappaB oscillations, signaling dynamics, and inflammatory gene expression. J Cell Biol. 2006;173(5):659–664. doi: 10.1083/jcb.200510155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian B, Nowak DE, Brasier AR. A TNF-induced gene expression program under oscillatory NF-kappaB control. BMC Genomics. 2005;6:137. doi: 10.1186/1471-2164-6-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boer DR, et al. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell. 2014;156(3):577–589. doi: 10.1016/j.cell.2013.12.027. [DOI] [PubMed] [Google Scholar]
- 38.Korasick DA, et al. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci USA. 2014;111(14):5427–5432. doi: 10.1073/pnas.1400074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nanao MH, et al. Structural basis for oligomerization of auxin transcriptional regulators. Nat Commun. 2014;5:3617. doi: 10.1038/ncomms4617. [DOI] [PubMed] [Google Scholar]
- 40.Alberti S, Gitler AD, Lindquist S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007;24(10):913–919. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 42.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.