Significance
In response to DNA damage, a proper balance between DNA repair and apoptosis is very important for genomic integrity. DNA-dependent protein kinase, catalytic subunit (DNA-PKcs) plays a key role in DNA repair. Many reports also indicate a prosurvival function for cytosolic DNA-PKcs. However, how this prosurvival activity of DNA-PKcs is regulated remains unknown. We identify Ring Finger protein 144A (RNF144A) as the first E3 ubiquitin ligase for cytosolic DNA-PKcs. RNF144A is induced in a p53-dependent manner during DNA damage and targets cytosolic DNA-PKcs for ubiquitination and degradation. The regulation of DNA-PKcs by RNF144A is important for proper apoptotic response during DNA damage, suggesting a tumor suppressor function for RNF144A.
Keywords: endosome, DDR, transmembrane domain
Abstract
Several ring between ring fingers (RBR) -domain proteins, such as Parkin and Parc, have been shown to be E3 ligases involved in important biological processes. Here, we identify a poorly characterized RBR protein, Ring Finger protein 144A (RNF144A), as the first, to our knowledge, mammalian E3 ubiquitin ligase for DNA-PKcs. We show that DNA damage induces RNF144A expression in a p53-dependent manner. RNF144A is mainly localized in the cytoplasmic vesicles and plasma membrane and interacts with cytoplasmic DNA-dependent protein kinase, catalytic subunit (DNA-PKcs). DNA-PKcs plays a critical role in the nonhomologous end-joining DNA repair pathway and provides prosurvival signaling during DNA damage. We show that RNF144A induces ubiquitination of DNA-PKcs in vitro and in vivo and promotes its degradation. Depletion of RNF144A leads to an increased level of DNA-PKcs and resistance to DNA damaging agents, which is reversed by a DNA-PK inhibitor. Taken together, our data suggest that RNF144A may be involved in p53-mediated apoptosis through down-regulation of DNA-PKcs when cells suffer from persistent or severe DNA damage insults.
Ring Finger protein 144A (RNF144A) and RNF144B belong to the RNF144 family and are very conserved in higher eukaryotes. Both proteins contain an RING1-in between rings (IBR) -RING2 [termed the ring between ring fingers (RBR)] domain in the N terminus and a potential single-transmembrane (TM) domain in the C terminus. RBR domain-containing proteins usually possess an E3 ubiquitin ligase activity and are involved in regulation of the cell cycle and apoptosis (1–3). These proteins function like RING (Really Interesting New Gene)/HECT (Homologous to the E6AP Carboxyl Terminus) hybrids (i.e., they use their RING1 to bind E2s) but then transfer ubiquitin onto a conserved cysteine residue in the RING2 domain through a thioester linkage similar to the E3 thioester-linked ubiquitin intermediates in HECT-type E3 ligases (4). It has been shown that DNA damage induces RNF144B expression to promote cell apoptosis through regulation of the stability of p21 (5), BAX (1), and p73 (6). RNF144B shares 71% homology of amino acids with RNF144A. RNF144A is able to interact with E2-conjugating enzymes UbcH7 and UbcH8 through its RING1 domain, and mutations on IBR or RING2 reduce this E2–E3 interaction (7). In addition, RNF144A is transcriptionally repressed by metastasis-associated protein 1 and inhibits migration and invasion in breast tumor cells (8). However, whether RNF144A is involved in apoptosis and DNA damage response (DDR) remains unexplored.
DDR includes cell-cycle arrest, DNA repair, apoptosis, and senescence (9). When cells suffer from severe or persistent DNA damage beyond repair, a shift from DNA repair to apoptosis will occur to avoid accumulation of unwanted mutations and potential tumor development. However, the detailed mechanisms for the shift remain unclear. Deregulation of DDR can lead to genome instability and ultimately, cancer development. In response to DNA double-strand breaks (DSBs), cells rely on two major pathways to repair DNA damage: homologous recombination and nonhomologous end-joining (NHEJ) (10). Homologous recombination requires a homologous template to accurately repair DSBs in S and G2 phases of the cell cycle (11). NHEJ does not need template DNA to repair DSBs; therefore, it often causes deletion or insertion mutations at sites of repair (12). NHEJ is the prevalent DSB repair pathway and can process the direct ligation of broken DNA ends throughout the whole cell cycle. This process can be regulated by assembly and disassembly of DNA-dependent protein kinase (DNA-PK) holoenzyme [comprising a catalytic subunit (DNA-PKcs) and Ku70/Ku80 heterodimer] and other DNA repair complexes on DSB sites (13). In addition, DNA-PKcs may also be inactivated by cleavage late in apoptosis to prevent DNA repair during DDR (14, 15).
DNA-PKcs also locates outside the nucleus and performs functions other than NHEJ. For example, DNA-PKcs can be found in the centrosomes, and it regulates microtubule spindles during DNA damage and normal mitosis (16–18). In addition, DNA-PKcs locates in the cytoplasm and plasma membrane and performs other functions (19–23). One of the functions of the cytoplasmic DNA-PKcs may be to phosphorylate protein kinase B (also known as Akt) at Ser-473 (20). DNA-PKcs in the cytoplasm has been suggested to recognize cytosine-phosphate-guanine (CpG)-oligodeoxynucleotides and play an important role in the induction of IL-10 production by macrophages (24). DNA-PKcs was shown to be responsible for Akt activation by CpG-oligodeoxynucleotide treatment in macrophages (19). A functional role of cytoplasmic DNA-PKcs is elucidated in another study (22), which showed that an endosome-resident receptor in natural killer (NK) cells, CD158d, associates with DNA-PKcs and recruits Akt to endosomes for phosphorylation by DNA-PKcs. These events subsequently lead to NF-κB activation and a proinflammatory response in NK cells. Recent study also showed that cytoplasmic DNA-PKcs can sense the presence of viral DNA to activate IFN regulatory factor 3-dependent innate immune response (25). Additional substrates of cytoplasmic DNA-PKcs have also been identified, such as HSP90α (23).
DNA-PKcs can be regulated by cellular shuttling by two cAMP signaling pathways (26) and an epidermal growth factor receptor endocytosis signal (27, 28). PKA activation induces DNA-PKcs nuclear entry, whereas exchange protein activated by cAMP facilitates DNA-PKcs nuclear exit through Rap2 (26). DNA-PKcs is highly ubiquitinated (29, 30), suggesting that ubiquitin may modulate its functions and turnover. However, no mammalian E3 ligase responsible for DNA-PKcs ubiquitination has been reported so far. More research is warranted to investigate the regulation of DNA-PK functions and turnover.
Our study shows that DNA damage can induce RNF144A in a p53-dependent manner. We also identify a previously unidentified role for RNF144A in DNA damage-induced apoptosis. We provide the first evidence, to our knowledge, for E3 ubiquitin ligase activity of RNF144A. We identify cytoplasmic DNA-PKcs as a substrate of RNF144A. RNF144A can interact with cytoplasmic DNA-PKcs and degrade it during normal growth and on DNA damage. Degradation of cytoplasmic DNA-PKcs reduces both cytoplasmic and nuclear DNA-PKcs. During DNA damage, an increased level of RNF144A leads to down-regulation of DNA-PKcs and promotes apoptosis. Our data suggest that p53 may promote apoptosis through RNF144A-mediated down-regulation of DNA-PKcs survival signaling.
Results
DNA Damage Induces a p53-Dependent Accumulation of RNF144A.
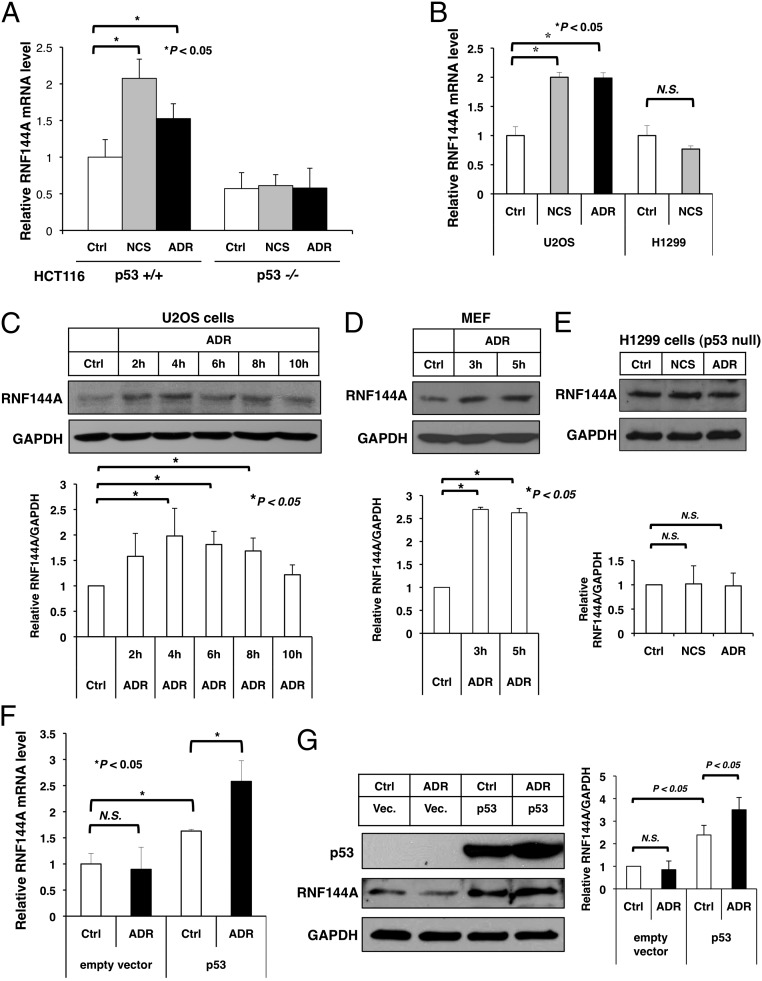
To investigate a potential role for RNF144A during DNA damage, we first examined the changes of RNF144A mRNA and protein level after adriamycin (ADR) and/or neocarzinostatin (NCS) treatment. Treatment with ADR and NCS induced a significant expression of RNF144A mRNA in human colon cancer HCT116 p53+/+ cells but not isogenic HCT116 p53−/− cells (Fig. 1A). Thus, RNF144A is induced by DNA damaging agents in a p53-dependent manner like RNF144B (5, 31). A similar increase of RNF144A mRNA level after ADR or NCS treatment was also found in human osteosarcoma U2OS cells but not p53-null human nonsmall cell lung carcinoma H1299 cells (Fig. 1B). Western blot analysis further confirmed induction of RNF144A protein levels on treatment with DNA-damaging agents and in U2OS cells and mouse embryonic fibroblasts but not H1299 cells (Fig. 1 C–E). To further investigate a role for p53 in RNF144A expression, we reconstituted the expression of p53 in H1299. As shown in Fig. 1 F and G, ADR treatment failed to induce RNF144A mRNA and protein in H1299 cells; however, transient overexpression of p53 increased RNF144A mRNA and protein levels in H1299 cells and rescued their induction by ADR treatment. These results suggest a role for p53 in the induction of RNF144A during DNA damage. In addition, a ChIP sequencing experiment performed in MCF7 cells shows binding of p53 on the RNF144A promoter (32). To see whether other forms of DNA damage could also induce RNF144A mRNA expression, we accessed the Gene Expression Omnibus database. Indeed, RNF144A mRNA was also induced by UV radiation in human skin fibroblasts (Fig. S1) (33). As such, it seems that the induction of RNF144A may be a general response to DNA damage. All together, these data show a p53-dependent accumulation of RNF144A mRNA and protein and imply a role for RNF144A in DNA damage response.
Fig. 1.
DNA-damaging agents induce the expression of RNF144A. (A and B) Real-time quantitative RT-PCR analysis shows that ADR and NCS induced the expression of RNF144A in (A) HCT116 p53+/+ and (B) U2OS cells but not (A) HCT116 p53−/− and (B) H1299 cells. Cells were treated with NCS (300 ng/mL) or ADR (10 μM) for 4 h. RNF144A mRNA level was determined by real-time RT-PCR in triplicate. Results were normalized to GAPDH levels and are expressed relative to the mock control (Ctrl) samples. (C–E) ADR induced RNF144A in (C) U2OS and (D) mouse embryonic fibroblast (MEF) but not (E) H1299 cells. Cells were treated with ADR (5 μM) for the indicated times, and RNF144A and GAPDH protein levels were determined by Western blot analysis. Quantitation of Western blots by ImageJ software from two to four independent experiments is shown in C–E, Lower. (F and G) Reexpression of p53 restored ADR-mediated induction of RNF144A mRNA in p53-null H1299 cells. An empty vector or p53 was transfected into H1299 cells. After 24 h, the cells were either mock-treated (Ctrl) or treated with ADR (10 μM) for 4 h. RNF144A mRNA level was determined by real-time RT-PCR in triplicate. (F) Results were normalized to GAPDH levels and are expressed relative to the empty vector mock-treated samples. (G) RNF144A and GAPDH protein levels were determined by Western blotting, and the signal intensity was quantified using ImageJ software. The relative RNF144A levels (normalized by GAPDH signals) from two independent experiments are shown as a graph in G, Right. Error bars represent SDs. *Significant difference with P < 0.05 (two-tailed t test). N.S., not significant.
Physiological Role for RNF144A in Apoptosis Induction on DNA Damage.
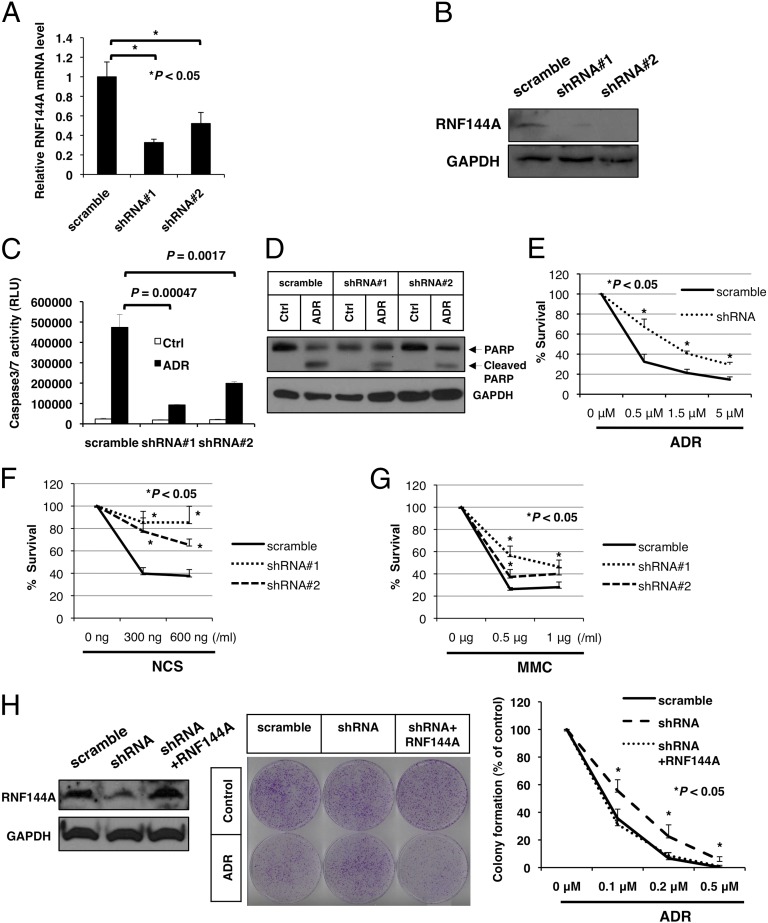
To determine the role of RNF144A in DNA damage response, we generated RNF144A-knockdown U2OS stable cell lines and examined caspase activation, poly(ADP-ribose) polymerase (PARP) cleavage, cell viability, and colony formation in these cells after DNA damage (Fig. 2). RNF144A mRNA and protein level were successfully depleted in U2OS cells using two different shRNA constructs (Fig. 2 A and B). We then used these three stable cell lines for subsequent studies in this work. As shown in Fig. 2C, ADR treatment resulted in a dramatic increase of caspase 3/7 activity in the scrambled shRNA control cells. RNF144A depletion significantly reduced the ADR-induced caspase 3/7 activities. We also measured the proteolytic cleavage of PARP, a substrate of active caspase 3. RNF144A depletion reduced ADR-induced cleavage of PARP (Fig. 2D). Knockdown of RNF144A also increased cell viability after treatment of ADR, NCS, or mitomycin C (Fig. 2 E–G). Finally, we rescued RNF144A expression in RNF144A-knockdown U2OS cells (Fig. 2H, Left) and performed colony formation assay after ADR treatment [Fig. 2H, Center (ADR = 20 nM) and Right (ADR = 0–0.5 μM)]. ADR treatment inhibited colony formation of U2OS cells, and the effect was attenuated by depletion of RNF144A. On rescue of RNF144A expression, ADR sensitivity was restored. Taken together, we conclude that RNF144A plays a proapoptotic role on DNA damage.
Fig. 2.
RNF144A promotes apoptosis during DNA damage. (A and B) U2OS cells were transfected with two different RNF144A shRNA constructs containing a puromycin-resistant gene and then selected by puromycin for 3 wk. Stable cells were used in the subsequent experiments. (A) Real-time quantitative PCR and (B) Western blot were performed to confirm the establishment of RNF144A-knockdown U2OS stable cells. (C and D) Knockdown of RNF144A reduced ADR-mediated activation of caspase 3/7 and PARP cleavage. RNF144A-depleted U2OS stable cells were treated with ADR (5 μM) or vehicle control (Ctrl) for (C) 6 or (D) 10 h. (C) Caspase 3/7 activity assay and (D) Western blot analysis for cleavage of PARP were then performed. RLU, relative light units. (E–H) RNF144A promoted cell death during DNA damage response. Stable RNF144A knockdown cells were treated with indicated concentrations of (E) ADR, (F) NCS, and (G) mitomycin C (MMC) for 24 h. (H) RNF144A was transfected into RNF144A-knockdown U2OS cells to rescue its expression. (Left) After 2 wk of G418 selection for stable RNF144A transfection, cell lysates were harvested for immunoblotting to confirm rescue of RNF144A expression. The cells were then either mock-treated (control) or treated with ADR for 3 h and released for 10 d. Cell viability was determined by colony formation assay. (Center) Representative images of experiment after ADR (20 nM) treatment. (Right) Repeated experiments with ADR ranging from 0 to 0.5 μM. All results were derived from three independent experiments. Error bars represent SDs. *Significant difference with P < 0.05 (two-tailed t test).
RNF144A Interacts with DNA-PKcs Through Amino Acids 173–252.
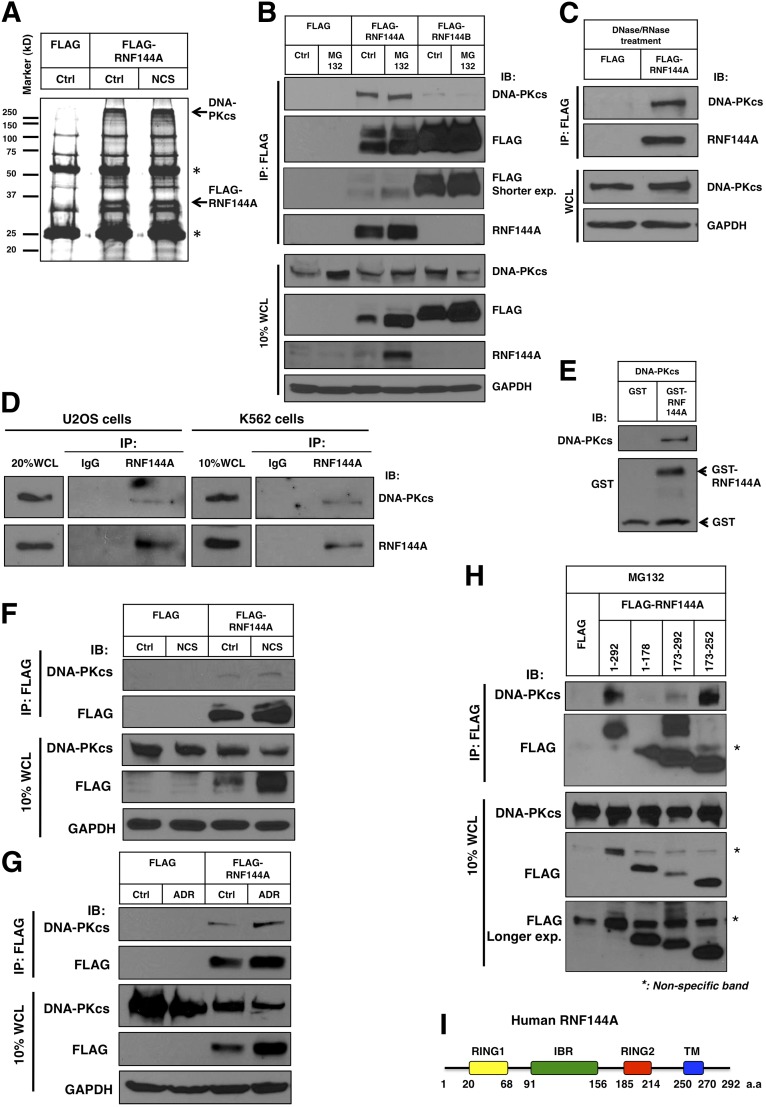
To understand the potential mechanisms for the proapoptotic function of RNF144A during the DNA damage response, we identified RNF144A substrates and/or its interacting proteins on DNA damage. We overexpressed FLAG-RNF144A in HEK293T cells and performed immunoprecipitation–MS (IP-MS). A SYPRO Ruby protein staining of the RNF144A-interacting proteins revealed predominant bands with molecular masses greater than 250 kDa (Fig. 3A). Interestingly, MS identified 66 peptide sequences matched to DNA-PKcs with a Mascot score of 3,156 (Table S1). Indeed, this RNF144A–DNA-PKcs interaction was further confirmed by subsequent IP/Western blot analysis (Fig. 3B). We also tested the interaction between RNF144B and DNA-PKcs in the same experiment. Despite that the expression level of FLAG-RNF144B was much higher than that of FLAG-RNF144A, the interaction between RNF144B and DNA-PKcs was very hard to detect, even in the presence of a proteasome inhibitor MG132 (Fig. 3B). Therefore, we only focused on the interaction between RNF144A and DNA-PKcs in this study. These data also suggest that the interaction between RNF144A and DNA-PKcs observed in our assays is specific. RNF144A interacted with Ku80 as well but not with Artemis, further supporting its binding with DNA-PKcs and suggesting its involvement with the DNA-PK complex (Fig. S2A). DNA-PK has DNA binding property (34). Thus, the observed interaction could indirectly result from protein–nucleic acid interactions. To exclude this possibility, we pretreated cell extracts with DNase/RNase and then performed IP. DNase/RNase treatment did not abolish the interaction between RNF144A and DNA-PKcs (Fig. 3C), suggesting that this interaction is independent of DNA and RNA. We also performed endogenous co-IP experiments in two different cell lines, U2OS and a human myelogenous leukemia cell line K562, and further showed that RNF144A physically interacted with DNA-PKcs under physiological conditions (Fig. 3D). This interaction was also seen in the GST pull-down assay, in which purified GST-RNF144A interacted with purified DNA-PKcs (active DNA-PK kinase purified from HeLa cells) in vitro (Fig. 3E). GST-RNF144A also pulled down DNA-PKcs in cell lysates prepared from untreated or ADR- or NCS-treated cells (Fig. S2 B and C). In addition, RNF144A–DNA-PKcs interaction was seen in both normal and DNA-damaging (NCS and ADR treatment) conditions (Fig. 3 F and G). DNA damage induced accumulation of RNF144A protein (Fig. 1); therefore, increased RNF144A protein might also increase RNF144A–DNA-PKcs complex formation on DNA damage (Fig. 3 F and G). To investigate further, we treated HEK293T cells with ADR for 2–24 h, prepared the cellular lysates, and incubated them with the same amount of FLAG-RNF144A (which was immunoprecipitated by anti-FLAG beads from other HEK293T cells transfected with an empty vector control plasmid or FLAG-RNF144A). The coimmunoprecipitated DNA-PKcs was relatively constant (Fig. S2D). We further used a series of RNF144A-truncated proteins and showed that the RNF144A amino acids 173–252 were responsible to interact with DNA-PKcs (Fig. 3H). The high-molecular weight smear seen above the band of FLAG-RNF144A(173–292) on IP:FLAG/IB:FLAG film in Fig. 3H is modified FLAG-RNF144A(173–292) protein, and it was also seen in a longer-exposed film of FLAG immunoblot of whole-cell lysates. The amino acids 173–252 contain RING2 and the intervening sequence between RING2 and the TM domain (Fig. 3I), suggesting that an RNF144A–DNA-PKcs–specific interaction does not require the TM domain.
Fig. 3.
RNF144A physically interacts with DNA-PKcs. (A) An IP-MS experiment identified DNA-PKcs as an RNF144A-interacting protein. HEK293T cells were transfected with an empty FLAG vector or FLAG-RNF144A. After 24 h, some cells were treated with NCS (300 ng/mL) for an additional 5 h. Cell lysates were harvested for FLAG-IP. Shown is a Sypro Ruby protein stain of the gel. The indicated bands were verified to be DNA-PKcs and RNF144A by MS. *Anti-FLAG antibody heavy chain and light chain. MS identified 66 peptides derived from DNA-PKcs (sequences presented in Table S1). (B and C) IP analysis shows that (B) only FLAG-RNF144A but not FLAG-RNF144B interacted significantly with endogenous DNA-PKcs in both untreated and MG132-treated (for 6 h) conditions and (C) this interaction could still be seen after DNase/RNase treatment. IB, immunoblot; WCL, whole cell lysates. HEK293T cells were transfected with an FLAG vector or FLAG-RNF144A. After 24 h, cell lysates were harvested for FLAG IP and Western blot analysis as indicated. A shorter exposed film (labeled Shorter exp.) of FLAG immunoblot after FLAG IP is also shown. (D) Western blot analysis confirmed an interaction between endogenous RNF144A and DNA-PKcs in U2OS and K562 cells. Cell lysates were harvested for endogenous RNF144A IP followed by Western blot analysis. (E) GST pull-down experiment shows that RNF144A directly bound to DNA-PKcs. Purified DNA-PKcs proteins (Promega) were incubated with purified GST or GST-RNF144A protein followed by Glutathione Sepharose pull-down and SDS/PAGE and Western blot analyses. (F and G) RNF144A–DNA-PKcs complex exists in both NCS and ADR treatment conditions. HEK293T cells were transfected with FLAG or FLAG-RNF144A. After 24 h, cells were treated with NCS (300 ng/mL) or ADR (5 μM) for an additional 5 h. Cell lysates were then harvested for FLAG IP and immunoblotting. (H) Different truncated RNF144A fragments were transfected to HEK293T, and cells were treated with MG132 for 6 h; the FLAG IP was performed as above. This deletion analysis indicates that RNF144A (amino acids 173–252) interacted well with DNA-PKcs. (I) A scheme of human RNF144A and its domains. Ctrl, control.
RNF144A Has E3 Ubiquitin Ligase Activity and Promotes Ubiquitination of DNA-PKcs.
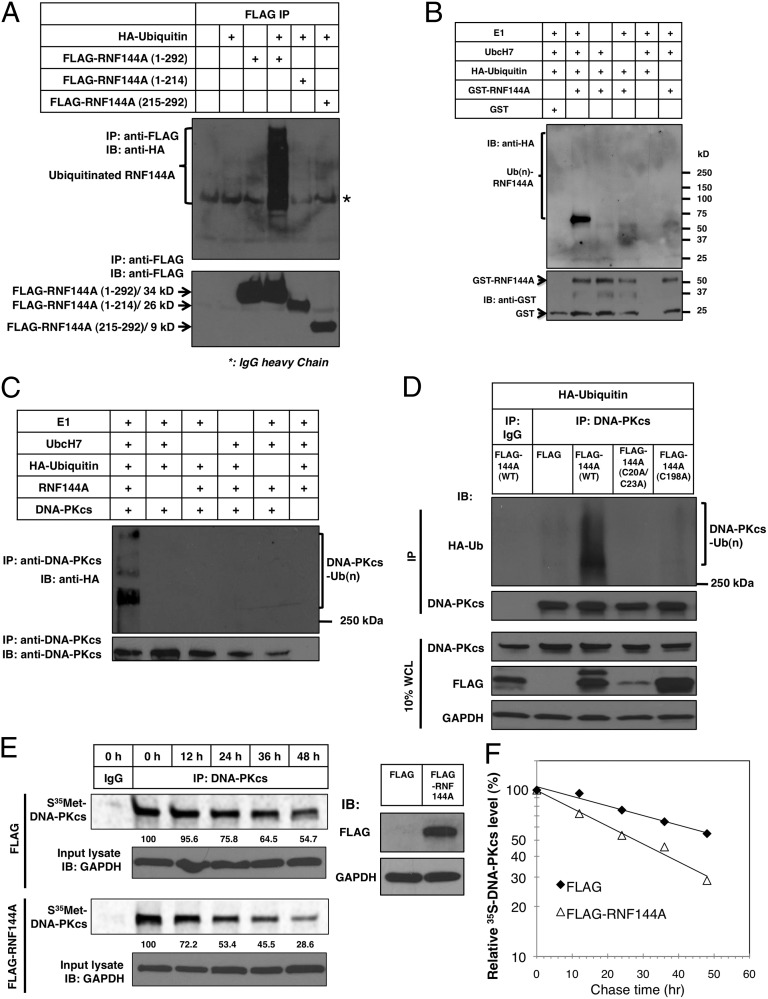
RNF144A contains the RING1-IBR-RING2 (RBR) domain and a potential single-TM domain, suggesting its potential E3 ubiquitin ligase activity (Fig. 3I). When RNF144A was overexpressed in HEK293T cells, it was readily ubiquitinated, which was revealed in the experiment of HA–ubiquitin cotransfection (Fig. 4A). This result suggested autoubiquitination of RNF144A when it is overexpressed inside the cells. The ubiquitinated smear disappeared when either the RBR domain or the TM domain was truncated (Fig. 4A), suggesting that its E3 ubiquitin ligase activity inside the cells may be highly regulated by the TM domain through unknown mechanisms. To show its bona fide E3 ligase activity, we purified GST and GST-RNF144A and performed in vitro ubiquitination assay. Indeed, GST-RNF144A could autoubiquitinate itself in vitro (Fig. 4B). These data show that RNF144A is a bona fide E3 ubiquitin ligase. Interestingly, under this in vitro condition, the autoubiquitination of RNF144A was mainly monoubiquitination. It was possible that additional factors for efficient polyubiquitination as seen in vivo were missing in this in vitro reaction. Taken together, these data show that RNF144A functions as an E3 ubiquitin ligase in vivo and in vitro.
Fig. 4.
RNF144A functions as an E3 ligase and promotes DNA-PKcs ubiquitination. (A) Different FLAG-tagged constructs were cotransfected to HEK293T cells with HA-ubiquitin. After 24 h, cell lysates were harvested for FLAG IP followed by HA immunoblotting to detect RNF144A ubiquitination in vivo. *IgG heavy chain. (B) Western blot analysis shows autoubiquitination of GST-RNF144A in vitro. The in vitro ubiquitination assay was performed as described in Materials and Methods. (C) In vitro ubiquitination assay of DNA-PKcs by RNF144A. GST-RNF144A was purified and treated with PreScission Protease to remove GST tag. The purified RNF144A protein was incubated with purified DNA-PKcs, E1, UbcH7, HA-Ubiquitin, and ATP reaction buffer at 30 °C for 1 h. Samples were separated by SDS/PAGE and immunoblotted. (D) RNF144A promoted ubiquitination of DNA-PKcs in vivo. WT RING1-dead mutant (C20A/C23A) and RING/HECT-inactive mutant of RNF144A (C198A) were cotransfected with HA-ubiquitin to HEK293T cells. After 24 h, cells were treated with MG132 for 6 h, and cell lysates were harvested and immunoprecipitated by anti–DNA-PKcs antibody followed by immunoblotting. (E and F) HEK293T cells were transfected with either an empty FLAG vector or FLAG-RNF144A; 24 h later, cells were labeled with [35S]methionine for 1 h and chased in media containing excess unlabeled methionine for the indicated times. (E, Left) DNA-PKcs was immunoprecipitated from equal cell numbers for each time point with DNA-PKcs antibody and analyzed by SDS/PAGE. The 35S-labeled DNA-PKcs was visualized by a PhosphorImager. The intensity at time 0 h was set as 100%. The relative intensity compared with that of the 0-h sample is shown below and plotted on a semilog scale graph. Control GAPDH immunoblots from the same input lysates for each IP are shown below each panel. Ub, ubiquitin.
To determine whether RNF144A regulates ubiquitination of DNA-PKcs, we also performed an in vitro ubiquitination assay using purified DNA-PKcs as a substrate. As shown in Fig. 4C, purified RNF144A(WT) promoted polyubiquitination of DNA-PKcs. These data indicate that RNF144A is capable of ubiquitinating DNA-PKcs in vitro. We also investigated the ubiquitination of DNA-PKcs in vivo. WT RING1-dead mutant (C20A/C23A) and RING/HECT-inactive mutant of RNF144A (C198A) were overexpressed in HEK293T cells, and ubiquitination of DNA-PKcs was analyzed. Only WT RNF144A was able to promote ubiquitination of DNA-PKcs, indicating that the E3 ubiquitin ligase activity of RNF144A was responsible for this increase of DNA-PKcs ubiquitination (Fig. 4D). These data show that RNF144A can bind to and ubiquitinate DNA-PKcs in vivo and in vitro. Combined with the observation that transient overexpression of RNF144A decreased DNA-PKcs protein level (Fig. 3 F and G), these data are consistent with the idea that RNF144A can promote DNA-PKcs degradation. To further obtain supporting evidence, we performed a 35S-methionine pulse-chase experiment to determine the half-life of DNA-PKcs in HEK293T cells that were transiently transfected with either a control empty vector or FLAG-tagged RNF144A (Fig. 4 E and F). Consistent with a previous study (35), phosphorimager analysis of these pulse-chase experiments showed that the half-life of DNA-PKcs was up to 48 h in control HEK293T cells (Fig. 4 E and F). In contrast, its half-life was reduced to around 24 h on expression of RNF144A. Taken together, these results indicate that RNF144A can ubiquitinate DNA-PKcs and promote its degradation.
RNF144A Interacts with DNA-PKcs Mainly in the Cytosol.
The presence of a potential TM domain in RNF144A suggests its membrane localization (Fig. 3I). The DNA-PK complex has been reported to localize outside the nucleus, such as in lipid rafts (21), plasma membrane (20), and endosomes (22), etc., for signal transduction function, including activation of Akt (19, 20). Thus, we were interested in determining the localization of the RNF144A–DNA-PKcs complex.
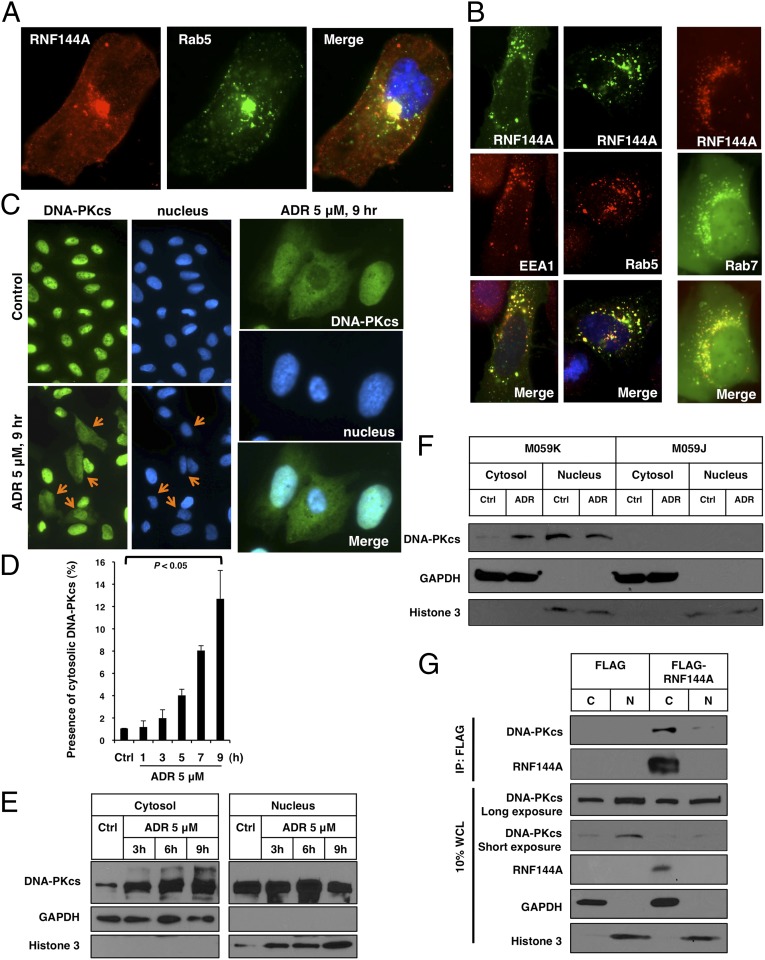
We characterized the subcellular localization of RNF144A. Limited by available RNF144A antibodies capable of detecting endogenous RNF144A by immunofluorescence, we used FLAG-tagged RNF144A to monitor its subcellular localization. RNF144B has been reported to mainly appear in the cytoplasm, plasma membrane, Golgi apparatus (5), and the mitochondria (1). However, unlike RNF144B, we observed that RNF144A partially localized with β-tubulin, calnexin (a marker for endoplasmic reticulum), and Rab5 (a marker for endosomes) (Fig. S3). Like Rab5, RNF144A is also distributed around plasma membrane and the perinuclear area (Fig. 5A). To gain additional evidence for the endosomal localization of RNF144A, we constructed EGFP- or mCherry-RNF144A and showed its colocalization with different endosomal markers: EEA1 and Rab5 (early endosome) and Rab7 (late endosome) (Fig. 5B and Fig. S4). Deletion of the TM domain (amino acids 250–270) caused a redistribution of RNF144A to a homogenous pattern throughout the cells, suggesting that the TM domain might restrict its subcellular localization (Fig. S5). The plasma membrane and endosomal localizations imply that RNF144A might participate in endocytosis, trafficking, activation, and degradation of target proteins in the cytosol.
Fig. 5.
RNF144A localizes at endosomes and interacts with cytoplasmic DNA-PKcs. (A and B) RNF144A localizes at endosomal organelles. (A) The immunofluorescence images (100×) show that RNF144A (red) partially overlapped with Rab5, an endosomal marker, and (green) positive dots. FLAG-tagged RNF144A was transfected into U2OS cells. On the next day, cells were fixed and stained with anti-FLAG antibody (red) and anti-Rab5 antibody (green). (B, Left and Center) EGFP-tagged RNF144A was transfected into U2OS cells for 1 d. Cells were fixed and stained with EEA1 or Rab5 antibody and a Texas Red-conjugated secondary antibody. (B, Right) mCherry-tagged RNF144A was cotransfected with EGFP-tagged Rab7 (48) into U2OS cells for 1 d. Immunofluorescence images (100×) were acquired by a Zeiss inverted microscope. Expanded image data are shown in Fig. S4. (C–F) ADR-induced redistribution of DNA-PKcs from the nucleus to the cytoplasm. U2OS cells were treated with ADR (5 μM) for the indicated time, and then, they were fixed and stained with anti–DNA-PKcs (green) and Hoechst 33258 (blue). (C) Orange arrow indicates the cells with cytoplasmic DNA-PKcs staining. High-magnification images of the ADR-treated cells are shown in Right. The specificity of anti–DNA-PKcs antibody for immunofluorescence study was verified using a pair of glioma cells: M059K (DNA-PKcs–proficient) and M059J (DNA-PKcs–deficient) cells (Fig. S6). (D) The percentages of cells with cytoplasmic DNA-PKcs staining on ADR treatment were counted (from 1,000 cells) and shown. Error bars represent SDs from three independent experiments. (E) Western blot analysis shows ADR-induced cytoplasmic DNA-PKcs in a time-dependent manner. (F) M059K (DNA-PKcs–proficient) and M059J (DNA-PKcs–deficient) cells were treated with ADR (5 μM) for 9 h, and then, they were harvested for fractionation and Western blot analysis. (G) FLAG-tagged RNF144A interacted with cytoplasmic DNA-PKcs. An empty FLAG vector or FLAG-tagged RNF144A was transfected to HEK293T cells. After 24 h, cell fractionation was performed. Cytosolic (C) or nuclear (N) extracts were then immunoprecipitated by FLAG M2 beads and immunoblotted with indicated antibodies. Ctrl, control.
We also checked the localization of DNA-PKcs after ADR treatment (Fig. 5 C–F and Fig. S6). Interestingly, although DNA-PKcs localizes mainly in the nucleus in untreated cells, ADR treatment caused a time-dependent increase of cytoplasmic DNA-PKcs in U2OS cells. This conclusion was supported by two independent assays, immunofluorescence staining (Fig. 5 C and D) and biochemical fractionation (Fig. 5E), suggesting a function of cytoplasmic DNA-PKcs during DDR. Also, a high-molecular weight smear in addition to a band corresponding to the anticipated 470-kDa protein was observed mainly in the cytosol fraction, suggesting that the cytoplasmic DNA-PKcs might be modified. To further confirm our finding in U2OS cells, we performed similar assays in a pair of glioma cell lines with different DNA-PKcs status, M059K (DNA-PKcs WT) and M059J (DNA-PKcs–deficient; as a control for DNA-PKcs staining and immunoblotting), and obtained similar results (Fig. 5F and Fig. S6). Because the nuclear/cytosolic shuttling of DNA-PKcs can be regulated (26), we suspect that ADR treatment might induce nuclear exit of DNA-PKcs. However, the exact mechanism needs additional investigation.
We then performed cell fractionation and co-IP to determine where the RNF144A–DNA-PKcs complex is located. Indeed, RNF144A interacted with DNA-PKcs mainly in the cytosol (Fig. 5G).
RNF144A Down-Regulates DNA-PKcs to Enhance Apoptosis During DNA Damage.
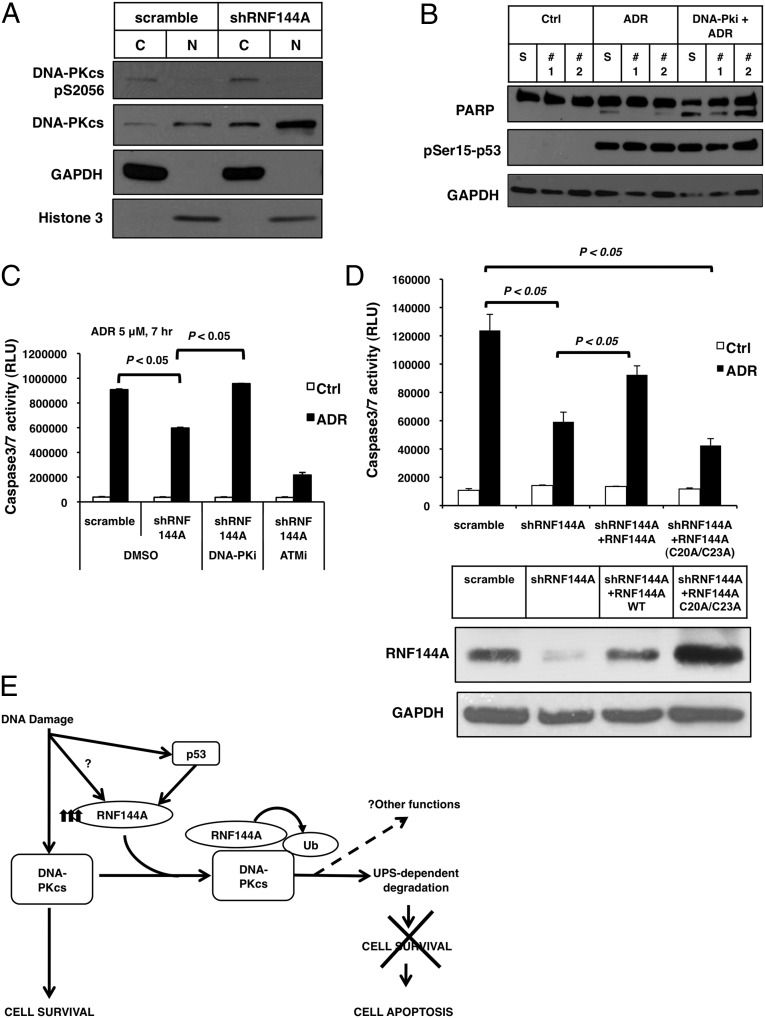
Consistent with the observations that RNF144A was able to ubiquitinate and degrade DNA-PKcs (Fig. 4), depletion of RNF144A in U2OS cells led to an increase in DNA-PKcs level (Fig. 6A). Interestingly, although RNF144A interacts with mainly cytoplasmic DNA-PKcs, knockdown of RNF144A caused an increase of DNA-PKcs in both cytosol and nucleus. It is quite likely that the DNA-PKcs accumulated in the cytosol of RNF144A-depleted cells can be translocated into the nucleus. Although there was more DNA-PKcs in the nucleus, the Ser-2056 autophosphorylated DNA-PKcs was found mainly in the cytosol, suggesting that it was highly active in the cytosol. To determine whether the apoptosis resistance by RNF144A knockdown on DNA damage is DNA-PKcs–dependent, we used a DNA-PKcs–specific inhibitor NU7441. Consistent with Fig. 2, RNF144A depletion significantly reduced the ADR-induced PARP cleavage and caspase 3/7 activities (Fig. 6 B and C). The DNA-PK inhibitor restored the effect of ADR-mediated cleavage of PARP and caspase 3/7 activities in RNF144A knockdown cells. However, an ataxia telangiectasia mutated (ATM)-specific inhibitor, KU55933, failed to rescue RNF144A depletion effect on caspase 3/7 activities (Fig. 6C). These results indicate that the apoptosis resistance in RNF144A-depleted cells can be reversed by inhibition of DNA-PK, thus supporting a role for DNA-PKcs in mediating the prosurvival phenotype in those RNF144A-depleted cells. We also investigated whether the effect on apoptosis by RNA144A depended on its E3 ligase activity. We rescued RNF144A-depleted cells with either WT or RING1-dead mutant (C20A/C23A) RNF144A. Expression of WT RNF144A could rescue ADR-induced apoptosis; however, the C20A/C23A mutant failed to do so, although the mutant was expressed at a higher level than WT (Fig. 6D). Thus, RNF144A requires its E3 ligase activity to promote apoptosis during DNA damage. Taken together, these data strongly suggest that RNF144A ubiquitinates and degrades DNA-PKcs to down-regulate its prosurvival functions and promote apoptosis after cells suffer from severe DNA damage.
Fig. 6.
RNF144A down-regulates DNA-PKcs to promote apoptosis during DNA damage. (A) Depletion of RNF144A increased pS2056–DNA-PKcs in the cytosol and total DNA-PKcs in both cytosol and nucleus. RNF144A-depleted stable U2OS cells were harvested for subcellular fractionation to separate cytoplasmic (C) and nuclear (N) fractions. Western blot analysis from each fraction was then performed. (B) A DNA-PK inhibitor restored ADR-induced cell apoptosis in RNF144A-depleted cells. S, scrambled shRNA control; #1, RNF144A shRNA#1; #2, shRNA2. RNF144A-depleted stable U2OS cells were treated with ADR (5 μM) for 4 h without or with pretreatment of DNA-PKi before ADR. Cell lysates were then harvested for Western blot analysis as indicated. (C) RNF144A-depleted stable U2OS cells were treated with ADR (5 μM) for 7 h with pretreatment of DNA-PKi or ATMi. Cells were then harvested for caspase 3/7 activity assay. (D) RNF144A (WT and C20A/C23A mutant) were transfected into RNF144A-knockdown U2OS cells to rescue its expression. Cells were treated with ADR (5 μM) for 10 h and then harvested for caspase 3/7 activity assay. (Lower) Because C20A/C23A mutant RNF144A sometimes expressed less than WT (Fig. 4D), to ensure that we had a least equal or more C20A/C23A mutant RNF144A expression than WT, we transfected more C20A/C23A mutant RNF144A plasmid than WT, which accounts for the higher expression of C20A/C23A mutant. (E) A proposed model for how RNF144A down-regulates the prosurvival signaling of DNA-PKcs and promotes cell death during DDR. A basal level of RNF144A may be responsible for other physiological functions or baseline down-regulation of DNA-PKcs. DNA damage stabilizes p53 and induces p53-dependent accumulation of RNF144A. RNF144A protein might also be stabilized by yet-to-be-determined mechanism(s). The accumulated RNF144A then interacts with and degrades more DNA-PKcs through the ubiquitin-proteasome system and inhibits the prosurvival function of DNA-PKcs. Through this mechanism, we propose that accumulation of RNF144A may shift the cell fates from cell survival to apoptosis on persistent or severe DNA damage.
Discussion
We have carried out the first, to our knowledge, biochemical and functional characterization of the RBR domain protein RNF144A. We found that RNF144A is induced on DNA damage and showed a role for p53 in the induction as well as DNA damage-induced apoptosis. Colocalization with Rab5-positive vesicles suggests that RNF144A may actively modulate trafficking and turnover of the plasma membrane and endosomal residents. Using the IP-MS method, we identified DNA-PKcs as an RNF144A-interacting protein and showed their interaction in the cytosol. RNF144A can ubiquitinate cytoplasmic DNA-PKcs and promote its degradation. This regulation leads to inhibition of DNA-PKcs prosurvival function during DDR. These p53–RNF144A–DNA-PKcs mechanisms may shift cell fates from the survival mode to programmed cell death when the cells encounter severe or persistent DNA damage beyond repair (Fig. 6E).
During evolution, cells develop many conserved pathways to adapt to DNA damage and maintain genome integrity. The signals for initiating cell-cycle arrest, DNA repair, apoptosis, and senescence can be sequentially triggered while genome surveillance proteins detect DNA damage (9). Competition between DNA repair and apoptosis will decide the final cell fate. Cells are able to use the NHEJ pathway to repair DSBs throughout the whole cell cycle. Although the NHEJ pathway can repair DSBs, it can generate mutations and potentially, tumor transformation (12, 13). Induction of p53-dependent apoptosis is important to abort the process (36, 37). We now provide a previously unidentified link between the p53 signaling pathway and DNA-PKcs regulation through an endosomal E3 ligase RNF144A to induce apoptosis and eliminate the damaged cells when the damage is beyond repair.
RNF144A is an RBR-containing protein, and it is able to interact with E2-conjugating enzymes, UbcH7 and UbcH8, through its RING1 domain (7). To our knowledge, our study is the first one to show its E3 ubiquitin ligase activity and show that RNF144A is able to promote its autoubiquitination and ubiquitination of DNA-PKcs both in vivo and in vitro (Fig. 4 A–D). RNF144A is unstable and degraded through the proteasome-dependent pathway (Fig. 3B). Intriguingly, the TM domain seems to play an important role in regulation of RNF144A E3 ligase activity and physiological function(s). On one hand, RNF144A might require its TM for full activity (Fig. 4A). On the other hand, the TM domain might also restrict the interaction between RNF144A and its targets in specific subcellular regions (Fig. S5). Thus, RNF144A seems to be tightly regulated at multiple levels: (i) transcription (induction on DNA damage), (ii) posttranslation (protein stability), and (iii) regulation of activity and possible substrate selectivity (through TM). Recent studies suggest that the ligase activities or the RBR proteins are tightly regulated. For example, the E3 ligase activity of Parkin or HOIP is inhibited by its N terminus in the absence of a binding partner. Binding of the ubiquitin-interacting motifs of Eps15 or the SH3 domain from endophilin A1 to the Parkin N-terminal ubiquitin-like domain releases its autoinhibition (38). The association with HOIL-1L or SHARPIN unblocks the inhibition and activates HOIP E3 ligase activity (39, 40). The requirement of the TM domain for its full E3 ligase activity of RNF144A might suggest a yet-to-be identified mechanism to regulate its activity.
We also looked at the expression profile of RNF144A in the gene expression BioGPS database, which profiled a panel of 79 human and 61 mouse tissues (41). In human tissues/cell lines, expression of RNF144A is differentially expressed in the human CNS, particularly the cerebellum, retina, adrenal gland, uterus, pancreatic islet cells, CD34+ and CD4+ T cells, CD56+ NK cells, and a chronic myelogenous leukemia cell line K562. According to SAGE Geni analysis in the SAGE Anatomic Viewer server from the National Cancer Institute, its expression is also found in human brain, retina, spinal cord, muscle, and some breast tissues (CD24+ luminal epithelium and possibly, CD10+ myoepithelium). At least 36 RNF144A somatic mutations (F98I, A137V, R139C, C203F, D221N, and more) are discovered in renal carcinoma, prostate cancer, large intestine cancer, lung cancer, and others (42–44). It will be very interesting to further determine whether these mutations affect the function of the RNF144A–DNA-PKcs complex to cause tumors.
Although RNF144A also shares high homology sequence with RNF144B in amino acids 173–252, we did not observe a strong interaction between RNF144B and DNA-PKcs (Fig. 3B). This observation might be because of a different subcellular localization or protein orientation on the plasma membrane between RNF144A and RNF144B or other unknown mechanisms. Another possibility is that the motif (216ESLDDDFLL224) in RNF144A, but not RNF144B, might contain a similar conserved human Ku80 binding motif (720EEGGDVDDLL729) for DNA-PKcs (45). This difference also implies that RNF144A and RNF144B may regulate different subsets of proteins. Additional experiments will be needed to address these interesting questions. During our manuscript preparation, Jiang et al. (46) showed that DNA-PKcs is ubiquitinated and bound to valosin-containing protein (VCP), possibly for regulation of proteasome-dependent degradation. VCP is a member of the AAA class of ATPases, found predominantly in the cytoplasm, and involved in diverse biological processes, including endoplasmic reticulum-associated degradation and ubiquitin fusion degradation (47). Knockdown of VCP in glioblastoma cells led to accumulation of DNA-PKcs and resistance to ionizing irradiation, ultimately resulting in shorter survival times in an orthotopic xenograft mouse model (46). This study supports the importance of regulation of DNA-PKcs ubiquitination in the cytoplasm. However, the E3 ligase for DNA-PKcs was not identified. Here, we show that RNF144A is the first, to our knowledge, mammalian E3 ligase for DNA-PKcs. It will be very interesting to determine whether VCP is also involved in RNF144A–DNA-PKcs regulation in the future. A previous study suggested that RNF144A suppresses tumor migration and invasion (8). Our study identifies a previously unidentified role for RNF144A in apoptosis induction through suppression of the prosurvival signaling function of DNA-PKcs. This regulation may provide a mechanism for the organisms to remove the cells suffering from severe DNA damage and prevent future cancer development. Therefore, these studies suggest a tumor suppressor function for RNF144A.
Materials and Methods
Cell Culture and Transfection.
HEK293T and H1299 cells were maintained in DMEM supplemented with 10% (vol/vol) FBS, penicillin [50 international unit (IU)/mL], and streptomycin (50 μg/mL). M059K and M059J cells were maintained in DMEM:F12 at 1:1 supplemented with 10% (vol/vol) FBS, penicillin, streptomycin, 1% nonessential amino acid, and 0.5 mM sodium pyruvate. HCT116 and U2OS cells were grown in McCoy’s 5A medium supplemented with 10% FBS, penicillin, and streptomycin. Mouse embryonic fibroblast cells were cultured in DMEM supplemented with heat-inactivated 10% FBS, penicillin (100 IU/mL), and streptomycin (100 μg/mL). All cells were grown in a humidified incubator at 37 °C with 5% CO and 95% air. A standard Lipofectamine 2000 (Life Technologies) method was used for transfection of HEK293T, U2OS, and H1299 cells.
In Vivo and in Vitro Ubiquitination Assay.
For in vivo ubiquitination assay, HEK293T cells were transfected with HA-ubiquitin, FLAG-tagged RNF144A (WTs or mutants), or a control FLAG vector as indicated. One day later, cells were treated with 20 μM MG132 for 6 h. Cells were then lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.5% sodium deoxycholate, 1% Nonidet P-40, 0.1% SDS) or SDS lysis buffer followed by boiling for 5 min at 95 °C. The cell lysates were reconstituted to 0.1% SDS by 1:10 dilution in TNN (Tris, NaCl, NP40) buffer supplemented with a mixture of protease inhibitors. The lysates were then sonicated and clarified by 10 min of centrifugation at 16,000 × g in a microfuge. Equivalent amounts of lysates were incubated overnight with a DNA-PKcs antibody/Protein G agarose or FLAG-M2 agarose beads (SIGMA) at 4 °C. Beads were washed five times with RIPA buffer. The beads were then boiled in Laemmli buffer and analyzed by SDS/PAGE followed by Western blot analysis using HA antibody. The in vitro ubiquitination assay was performed in 50 μL reaction buffer (50 mM Tris, pH 8, 1 mM DTT, 5 mM MgCl2, 100 mM NaCl, 5 mM ATP or 5 μL 10× energy regeneration solution; Boston Biochem). In each reaction, 15 nM human recombinant E1 (Boston Biochem), 0.5 mM UbcH7 (Boston Biochem), and 5 mM HA-ubiquitin (Boston Biochem) were mixed with 0.77–1 mM GST-RNF144A or GST-RNF144A mutant and DNA-PKcs (Promega) in the reaction buffer and incubated at 37 °C for 1 h. Samples then were subjected to IP with the indicated antibody and then immunoblotting with HA antibody.
Measurement of Protein Stability.
HEK293T cells transiently expressing FLAG or FLAG-tagged RNF144A for 24 h were prestarved by replacing the culture media with DMEM without l-methionine and l-cysteine (Gibco by Life Technologies) for 30 min. Cells were labeled in vivo with [35S]methionine/Cysteine EasyTagTMEXPRESS35S Protein Labeling Mix (PerkinElmer) using 300 μCi/mL for 1 h. After labeling, cells were washed one time with DMEM containing 5 mM l-methionine and 3 mM l-cysteine and then incubated in the same media for the indicated chase times. Equal cell numbers from each time point were harvested, and labeled DNA-PKcs protein was immunoprecipitated with DNA-PKcs antibody and resolved by SDS/PAGE. Labeled DNA-PKcs was visualized by exposure to a phosphorimager screen, scanned using a PhosphorImager (Molecular Dynamics), and quantitated by ImageJ software (National Institutes of Health).
Real-time RT-PCR; plasmid and shRNA construction; reagents and antibodies; IP, Western blot analysis, and immunofluorescence; cell survival assay; caspase 3/7 activity assay; protein purification; subcellular fractionation assay; colony formation assay; and statistical analysis are discussed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. David H. Hawke (MD Anderson Cancer Center) for help with the RNF144A MS analysis and Dr. Richard Pagano for the EGFP-rab7 WT plasmid. We thank members of the laboratories of W.-C.L. and Dr. Fang-Tsyr (Fannie) Lin for discussion. This work was supported by National Institutes of Health Grants R01CA100857, R01CA138641, and ARRA 3 P30CA125123-03S and Department of Defense Breast Cancer Research Program Grant W81XWH-09-1-0338. S.-R.H. was supported by T32 Fellowship T32DK60445. C.S.M. was supported by Cancer Prevention and Research Institute of Texas Predoctoral Fellowship CPRIT RP101499. Y.-J.L. is in the Interdepartmental Program in Translational Biology and Molecular Medicine, Baylor College of Medicine, which was supported, in part, by a grant from the Howard Hughes Medical Institute through the Med into Grad Initiative.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323107111/-/DCSupplemental.
References
- 1.Benard G, et al. IBRDC2, an IBR-type E3 ubiquitin ligase, is a regulatory factor for Bax and apoptosis activation. EMBO J. 2010;29(8):1458–1471. doi: 10.1038/emboj.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen D, Li X, Zhai Z, Shu HB. A novel zinc finger protein interacts with receptor-interacting protein (RIP) and inhibits tumor necrosis factor (TNF)- and IL1-induced NF-kappa B activation. J Biol Chem. 2002;277(18):15985–15991. doi: 10.1074/jbc.M108675200. [DOI] [PubMed] [Google Scholar]
- 3.Nikolaev AY, Li M, Puskas N, Qin J, Gu W. Parc: A cytoplasmic anchor for p53. Cell. 2003;112(1):29–40. doi: 10.1016/s0092-8674(02)01255-2. [DOI] [PubMed] [Google Scholar]
- 4.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474(7349):105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng CC, Arakawa H, Fukuda S, Kondoh H, Nakamura Y. p53RFP, a p53-inducible RING-finger protein, regulates the stability of p21WAF1. Oncogene. 2003;22(28):4449–4458. doi: 10.1038/sj.onc.1206586. [DOI] [PubMed] [Google Scholar]
- 6.Sayan BS, et al. Differential control of TAp73 and DeltaNp73 protein stability by the ring finger ubiquitin ligase PIR2. Proc Natl Acad Sci USA. 2010;107(29):12877–12882. doi: 10.1073/pnas.0911828107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez-Noel G, Müller U, Harbers K. Identification of molecular determinants required for interaction of ubiquitin-conjugating enzymes and RING finger proteins. Eur J Biochem. 2001;268(22):5912–5919. doi: 10.1046/j.0014-2956.2001.02541.x. [DOI] [PubMed] [Google Scholar]
- 8.Marzook H, et al. Metastasis-associated protein 1 drives tumor cell migration and invasion through transcriptional repression of RING finger protein 144A. J Biol Chem. 2012;287(8):5615–5626. doi: 10.1074/jbc.M111.314088. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Zhou BB, Bartek J. Targeting the checkpoint kinases: Chemosensitization versus chemoprotection. Nat Rev Cancer. 2004;4(3):216–225. doi: 10.1038/nrc1296. [DOI] [PubMed] [Google Scholar]
- 10.Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol. 2009;10(4):243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11(3):196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao Z, Bozzella M, Seluanov A, Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008;7(18):2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song Q, et al. DNA-dependent protein kinase catalytic subunit: A target for an ICE-like protease in apoptosis. EMBO J. 1996;15(13):3238–3246. [PMC free article] [PubMed] [Google Scholar]
- 15.Han Z, et al. DNA-dependent protein kinase is a target for a CPP32-like apoptotic protease. J Biol Chem. 1996;271(40):25035–25040. doi: 10.1074/jbc.271.40.25035. [DOI] [PubMed] [Google Scholar]
- 16.Lee KJ, et al. Involvement of DNA-dependent protein kinase in normal cell cycle progression through mitosis. J Biol Chem. 2011;286(14):12796–12802. doi: 10.1074/jbc.M110.212969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shang ZF, et al. Inactivation of DNA-dependent protein kinase leads to spindle disruption and mitotic catastrophe with attenuated checkpoint protein 2 Phosphorylation in response to DNA damage. Cancer Res. 2010;70(9):3657–3666. doi: 10.1158/0008-5472.CAN-09-3362. [DOI] [PubMed] [Google Scholar]
- 18.Zhang S, Hemmerich P, Grosse F. Centrosomal localization of DNA damage checkpoint proteins. J Cell Biochem. 2007;101(2):451–465. doi: 10.1002/jcb.21195. [DOI] [PubMed] [Google Scholar]
- 19.Dragoi AM, et al. DNA-PKcs, but not TLR9, is required for activation of Akt by CpG-DNA. EMBO J. 2005;24(4):779–789. doi: 10.1038/sj.emboj.7600539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng J, Park J, Cron P, Hess D, Hemmings BA. Identification of a PKB/Akt hydrophobic motif Ser-473 kinase as DNA-dependent protein kinase. J Biol Chem. 2004;279(39):41189–41196. doi: 10.1074/jbc.M406731200. [DOI] [PubMed] [Google Scholar]
- 21.Lucero H, Gae D, Taccioli GE. Novel localization of the DNA-PK complex in lipid rafts: A putative role in the signal transduction pathway of the ionizing radiation response. J Biol Chem. 2003;278(24):22136–22143. doi: 10.1074/jbc.M301579200. [DOI] [PubMed] [Google Scholar]
- 22.Rajagopalan S, Moyle MW, Joosten I, Long EO. DNA-PKcs controls an endosomal signaling pathway for a proinflammatory response by natural killer cells. Sci Signal. 2010;3(110):ra14. doi: 10.1126/scisignal.2000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quanz M, et al. Heat shock protein 90α (Hsp90α) is phosphorylated in response to DNA damage and accumulates in repair foci. J Biol Chem. 2012;287(12):8803–8815. doi: 10.1074/jbc.M111.320887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yotsumoto S, Saegusa K, Aramaki Y. Endosomal translocation of CpG-oligodeoxynucleotides inhibits DNA-PKcs-dependent IL-10 production in macrophages. J Immunol. 2008;180(2):809–816. doi: 10.4049/jimmunol.180.2.809. [DOI] [PubMed] [Google Scholar]
- 25.Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. eLife. 2012;1:e00047. doi: 10.7554/eLife.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huston E, et al. EPAC and PKA allow cAMP dual control over DNA-PK nuclear translocation. Proc Natl Acad Sci USA. 2008;105(35):12791–12796. doi: 10.1073/pnas.0805167105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bandyopadhyay D, Mandal M, Adam L, Mendelsohn J, Kumar R. Physical interaction between epidermal growth factor receptor and DNA-dependent protein kinase in mammalian cells. J Biol Chem. 1998;273(3):1568–1573. doi: 10.1074/jbc.273.3.1568. [DOI] [PubMed] [Google Scholar]
- 28.Liccardi G, Hartley JA, Hochhauser D. EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res. 2011;71(3):1103–1114. doi: 10.1158/0008-5472.CAN-10-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44(2):325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner SA, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10(10):013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J, Xu LG, Liu T, Zhai Z, Shu HB. The p53-inducible E3 ubiquitin ligase p53RFP induces p53-dependent apoptosis. FEBS Lett. 2006;580(3):940–947. doi: 10.1016/j.febslet.2005.09.105. [DOI] [PubMed] [Google Scholar]
- 32.Nikulenkov F, et al. Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ. 2012;19(12):1992–2002. doi: 10.1038/cdd.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gentile M, Latonen L, Laiho M. Cell cycle arrest and apoptosis provoked by UV radiation-induced DNA damage are transcriptionally highly divergent responses. Nucleic Acids Res. 2003;31(16):4779–4790. doi: 10.1093/nar/gkg675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turchi JJ, Henkels K. Human Ku autoantigen binds cisplatin-damaged DNA but fails to stimulate human DNA-activated protein kinase. J Biol Chem. 1996;271(23):13861–13867. doi: 10.1074/jbc.271.23.13861. [DOI] [PubMed] [Google Scholar]
- 35.Lees-Miller SP, et al. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol. 1996;70(11):7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meek DW. Tumour suppression by p53: A role for the DNA damage response? Nat Rev Cancer. 2009;9(10):714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 37.Reinhardt HC, Schumacher B. The p53 network: Cellular and systemic DNA damage responses in aging and cancer. Trends Genet. 2012;28(3):128–136. doi: 10.1016/j.tig.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaugule VK, et al. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 2011;30(14):2853–2867. doi: 10.1038/emboj.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smit JJ, et al. The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension. EMBO J. 2012;31(19):3833–3844. doi: 10.1038/emboj.2012.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stieglitz B, Morris-Davies AC, Koliopoulos MG, Christodoulou E, Rittinger K. LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep. 2012;13(9):840–846. doi: 10.1038/embor.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su AI, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. 2002;99(7):4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463(7279):360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peifer M, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44(10):1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434(7033):605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 46.Jiang N, et al. Valosin-containing protein regulates the proteasome-mediated degradation of DNA-PKcs in glioma cells. Cell Death Dis. 2013;4:e647. doi: 10.1038/cddis.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye Y. Diverse functions with a common regulator: Ubiquitin takes command of an AAA ATPase. J Struct Biol. 2006;156(1):29–40. doi: 10.1016/j.jsb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 48.Choudhury A, et al. Rab proteins mediate Golgi transport of caveola-internalized glycosphingolipids and correct lipid trafficking in Niemann-Pick C cells. J Clin Invest. 2002;109(12):1541–1550. doi: 10.1172/JCI15420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.