Significance
Recent investigation of several mammalian hosts suggests that their intestinal bacterial communities display evidence of clusters characterized by differences in specific bacterial taxa, a concept referred to as enterotypes. By performing stable isotope analysis of environmental samples, monitoring communities during dietary shifts, and collecting functional metagenomic sequence data, we provide novel insight into the origins and dynamics of enterotype-like community clustering in wild house mice. Two clusters are present in wild mice, one associated with higher plant-derived and another with animal-derived food intake, which can shift within a period of 1 wk. Remarkably, these clusters display shared characteristics with those present in humans, chimpanzees, and laboratory mice, suggesting ancient shared traits among mammalian bacterial communities.
Abstract
Understanding the origins of gut microbial community structure is critical for the identification and interpretation of potential fitness-related traits for the host. The presence of community clusters characterized by differences in the abundance of signature taxa, referred to as enterotypes, is a debated concept first reported in humans and later extended to other mammalian hosts. In this study, we provide a thorough assessment of their existence in wild house mice using a panel of evaluation criteria. We identify support for two clusters that are compositionally similar to clusters identified in humans, chimpanzees, and laboratory mice, characterized by differences in Bacteroides, Robinsoniella, and unclassified genera belonging to the family Lachnospiraceae. To further evaluate these clusters, we (i) monitored community changes associated with moving mice from the natural to a laboratory environment, (ii) performed functional metagenomic sequencing, and (iii) subjected wild-caught samples to stable isotope analysis to reconstruct dietary patterns. This process reveals differences in the proportions of genes involved in carbohydrate versus protein metabolism in the functional metagenome, as well as differences in plant- versus meat-derived food sources between clusters. In conjunction with wild-caught mice quickly changing their enterotype classification upon transfer to a standard laboratory chow diet, these results provide strong evidence that dietary history contributes to the presence of enterotype-like clustering in wild mice.
First large-scale sequencing surveys of the human intestinal microbiome emphasized considerable differences between individuals (1). Numerous studies expanded upon these initial observations and identified factors, such as geography, host genetics, diet, and other environmental factors that contribute to interindividual variability in humans (2–4) and other animals (5–7). However, important questions pertaining to the nature and origins of gut microbial community structure remain to be answered. In particular, the presence of enterotypes, or distinct clusters characterized by the abundances of signature bacterial genera, is a debated concept first reported in humans (3). Wu et al. (8) soon after provided evidence that the proportion of protein and animal fat versus carbohydrates in long-term dietary habits contribute to determining and individual’s enterotype, whereas the intriguing existence of analogous enterotypes in chimpanzees suggests that they may reflect more ancient features of host-microbial physiology and homeostasis in the gut (9). On the other hand, a meta-analysis of enterotypes across human body sites found their identification to be sensitive to distance metrics and clustering methods used, in addition to a majority of gradients, rather than distinct clusters being present in signature taxa (10).
Most recently, Hildebrand et al. (11) provide a first assessment of the possibility of enterotypes in the house mouse, a critical and widely used model of gut microbiome research, whereby two enterotype-like clusters among the five laboratory strains studied are identified. Furthermore, differences in low-grade inflammation between these two groups suggest possible mechanisms driving clustering, for example, differences in inflammation-inducing taxa such as Enterobacteriaceae (12). However, which aspects of inflammation and differences in taxon abundance represent cause or consequence of each other remains unclear, as does the possibility of other contributing factors to community clustering. In addition, the natural state of bacterial communities is potentially misrepresented in laboratory settings because of practices, such as inoculating laboratory mice with limited mixtures of bacterial strains (e.g., altered Schaedler flora) and feeding standard diets.
In this study, we use the criteria outlined by Koren et al. (10) to first provide an assessment of enterotype-like clustering in a panel of wild-caught house mice previously included in a biogeographic survey of intestinal communities (7), and compare them to wild-caught mice housed in a laboratory environment. We find that two clusters similar to those identified in humans, chimpanzees, and laboratory mice are frequently present in the wild, but are nearly lost among mice housed in the laboratory for 1 y. As a follow-up, we caught additional mice, transferred them to the laboratory, and regularly monitored the dynamics of their fecal communities over a period of 12 wk beginning from the time of sampling. This process documents the rapid loss of one cluster abundant in the wild under laboratory conditions. Furthermore, deep shotgun metagenomic sequencing reveals significant differences in functional microbiomic categories, in particular pertaining to protein versus carbohydrate metabolism between the two groups. Finally, stable isotope analysis performed on wild-caught samples reveals significant differences in long-term diet between the two groups, consistent with the observations based on functional metagenomic data.
Results
Enterotype-Like Clusters in Wild Mice.
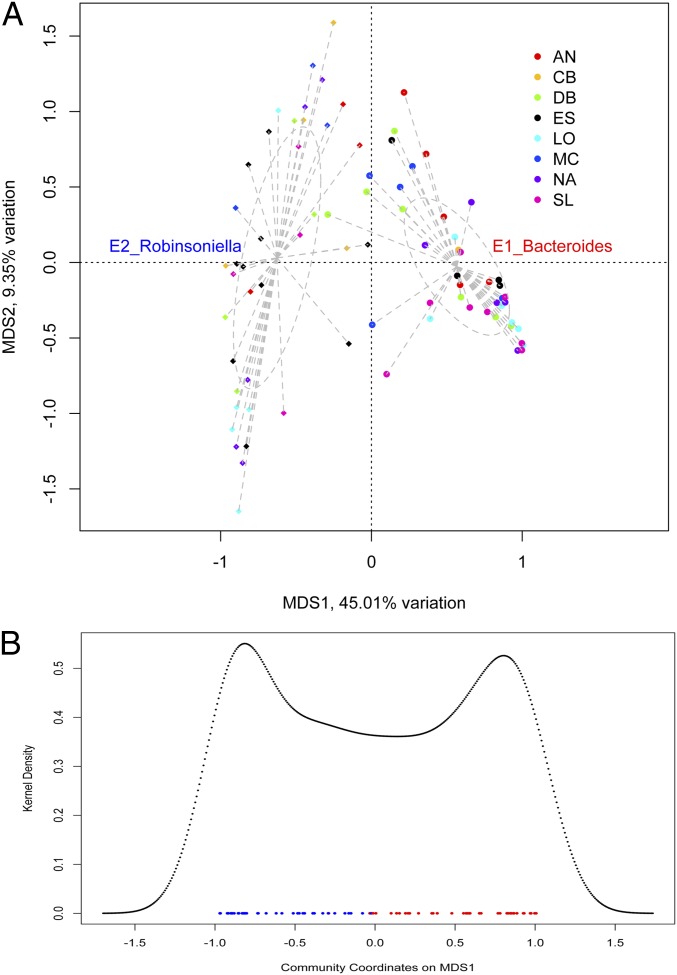
To evaluate the presence of enterotypes in wild mice, we first analyzed the cecal contents of 80 samples collected and dissected on-site at eight geographic locations in Western Europe [on average 530-km apart, across Germany and France (7)] following previous studies of enterotypes (10, 11). In total, optimal clustering was evaluated for five β-diversity metrics (i.e., measures of interindividual variability in community structure), six clustering methods, and three judging criteria (Methods and Table S1). However, to describe and compare our results to previous studies of humans (3, 8), chimpanzees (9), and laboratory mice (11), we focus mainly on the Bray–Curtis (BC) distance applied to genus-level abundances, evaluated by the partitioning around means (pam) clustering method. The BC distance is an abundance-based measure and thus particularly suited to describe changes in the abundance of indicator taxa, such as those associated with enterotypes. Furthermore, BC is highly correlated to the Jensen–Shannon (JS) distance (Mantel test, r = 0.9632, P = 0.001) applied in previous studies (3), but offers the advantage of being used in similarity percentage analysis (SIMPER) (13), a well-established method to identify taxa contributing to similarity within- and dissimilarity between groups (see below). Using BC and pam, an optimum of two clusters is revealed using all three judging criteria (Table S1). These two clusters, which we define as “E1” and “E2,” are present in all eight geographic regions [80 sites distributed across eight geographic locations on average 530 km apart (7)] (Fig. 1A), explain 39.45% of the variation in BC distances (as determined by the multivariate ANOVA implemented in analysis of dissimilarity, “adonis”) (Methods), and display a clear bimodal distribution along the first multidimensional scaling (MDS) axis (Fig. 1B). The support for these clusters (prediction strength >0.8) is comparable to that found for fecal samples included in the Human Microbiome Project (10).
Fig. 1.
Principle coordinate analysis of wild captured mice based on BC dissimilarity. (A) Colors denote each of eight unique sampling locations across Germany and France that are on average 530 km apart (7) (n = 80). Dots and diamonds represent the E1 enterotype (Bacteroides-dominant) and E2 enterotype (Robinsoniella-dominant) classification, respectively. (B) Kernel density of samples’ distribution along the first axis (MDS1) display a bimodal pattern, with red and blue dots denoting coordinates of samples belonging to E1 and E2, respectively.
To assess the influence of sequence binning methodology, we also carried out analyses on operational taxonomic units (OTUs) at 97% and 95% similarity thresholds, which yields less support for distinct clusters (Table S2). Interestingly, further examination indicates that this difference is at least partly a result of differences in the taxonomic levels at which geography influences community structure (7). At the 97% similarity threshold, geographic location has a significant influence on BC distance (adonis, 9.91% variation explained, P = 0.039), which is also observed at the 95% threshold (adonis, 10.13% variation explained, P = 0.041). By comparison, the two optimal clusters defined based on 97% and 95% OTUs explain only half the variation in BC distance compared with geography (adonis, 5.34% and 5.48% variation explained, P = 0.001 and P = 0.001, respectively). In contrast, BC distance based on genus-level classification is not significantly influenced by geographic location (adonis, 10.11% variation explained, P = 0.269), but displays nearly eightfold more variation explained by optimal clustering than that applied to OTUs (adonis, 39.45% variation explained, P = 0.001). Thus, OTU-based community profiling appears to reveal finer-scale variation in community structure that is shaped by geography (sampling region), whereas broader-scale patterns across geographical regions, such as enterotypes, are more prominent at the genus level. Accordingly, our subsequent analyses are based on the E1 and E2 clusters identified based on genus-level abundances described above.
Compositional Analysis of Enterotype-Like Clusters.
To identify signature taxa, we first applied the SIMPER method, which identifies taxa that contribute to differences between groups of samples (13). This analysis identifies Bacteroides as the largest contributor (31.68%) to dissimilarity between groups and similarity within E1 (77.78%). The second largest contributor (8.59%) to dissimilarity between groups is Robinsoniella, which contributes 14.04% to similarity within E2 and is also the most abundant genus in this group (average 13.6%). In addition, we identified an unclassified genus belonging to Lachnospiraceae (unclassified_Lachnospiraceae) that contributes 17.15% of the similarity within E2, but is second in terms of its abundance in E2 (average 12.45%). Second, we tested for significant differences in abundance among all taxa displaying ≥1% abundance in the whole dataset. Ten of these 16 genera displayed significant differences in abundance (Table 1). Finally, α-diversity analyses reveal significantly lower diversity in E1 by several complementary measures (Fig. S1).
Table 1.
Average abundances of major genera (overall mean abundance >1%) between E1 and E2 enterotypes
| Major genera | E1_Bacteroides | E2_Robinsoniella | ANOVA P value (FDR corrected) |
| Bacteroides | 0.6026 | 0.1013 | 3.20E-15 |
| unclassified_Lachnospiraceae | 0.0297 | 0.1246 | 1.40E-07 |
| Robinsoniella | 0.0098 | 0.1356 | 5.52E-05 |
| Barnesiella | 0.0325 | 0.0944 | 9.55E-04 |
| Helicobacter | 0.0480 | 0.0364 | 6.13E-01 |
| Lactobacillus | 0.0513 | 0.0103 | 1.53E-01 |
| Oscillibacter | 0.0139 | 0.0423 | 5.52E-05 |
| Coprobacillus | 0.0273 | 0.0145 | 2.88E-01 |
| unclassified_Ruminococcaceae | 0.0157 | 0.0272 | 1.28E-02 |
| unclassified_Porphyromonadaceae | 0.0122 | 0.0300 | 1.39E-02 |
| Parabacteroides | 0.0257 | 0.0095 | 8.38E-02 |
| unclassified_Rikenellaceae | 0.0032 | 0.0317 | 9.55E-04 |
| Alistipes | 0.0053 | 0.0281 | 2.19E-03 |
| Mucispirillum | 0.0123 | 0.0204 | 2.77E-01 |
| unclassified_Prevotellaceae | 0.0082 | 0.0138 | 3.61E-01 |
Bold numbers denotes significant difference revealed by ANOVA [P < 0.05 after Benjamin–Hochberg correction (31)]. FDR, false-discovery rate.
Wild Mice Moved to a Laboratory Environment.
During the course of fieldwork conducted in 2009 (7), additional mice sampled in the Massif Central region of France were brought back to the breeding facility of the Max Planck Institute for Evolutionary Biology. In total, 10 mice captured from seven different farms were analyzed after 1 y of laboratory housing. To assess the possible influence of laboratory housing on the distribution of enterotype-like clusters among mice, we added these 10 cecal content samples to the 80 samples dissected on-site and repeated the evaluation of clustering performed above. This finding reveals nearly identical results as described above, with support for two clusters (Table S3). Intriguingly, however, all 10 laboratory-housed samples belong to E2. This finding is in stark contrast to the mice sampled from the same Massif Central region but dissected on-site, which display a proportion of 5:3 of E1:E2, respectively (Table 2), and is highly unlikely to be observed by chance (Fisher’s exact test, P = 0.001508). However, we lack knowledge of the status of these 10 mice at the time of capture. Thus, these results offer only circumstantial evidence that common environmental conditions experienced in the laboratory influence enterotype-like clustering.
Table 2.
Distribution of enterotypes with respect to sampling location and time
| Location | E1_Bacteroides | E2_Robinsoniella |
| AN* | 5 | 3 |
| CB | 1 | 4 |
| DB | 7 | 4 |
| ES | 4 | 10 |
| LO | 8 | 5 |
| MC | 5 | 3 |
| NA | 5 | 5 |
| SL | 8 | 4 |
| MC (after >1 y in laboratory) | 0 | 10 |
| CB (newly captured in 2012) | 8 | 6 |
Sampling locations and time are those given in Linnenbrink et al. (7), unless otherwise noted.
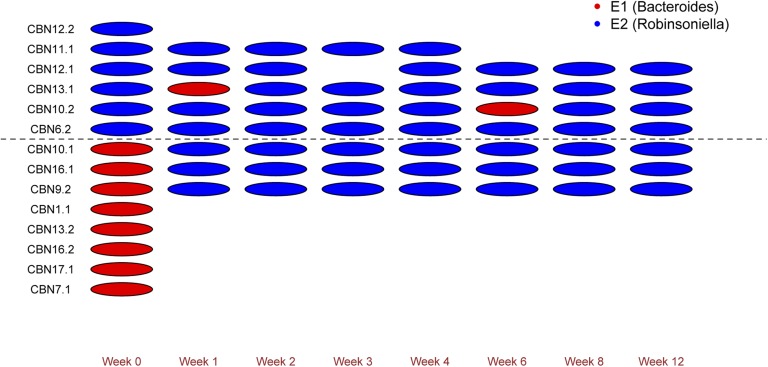
To directly document whether individuals shift their enterotype classification upon moving from the wild to a laboratory environment, additional sampling of wild mice was performed in the Cologne/Bonn region of Germany in 2012, which previously displayed an E1:E2 ratio of 1:4 in 2010 (Table 2) (7). In total, 14 mice were sampled (week 0), of which six died during the process of transportation and eight were transferred to the laboratory breeding facility to be monitored at regular intervals (1, 2, 3, 4, 6, 8, 12 wk) by sampling feces (Methods). One mouse was not sampled at week 3 because of treatment for a skin injury, whereas another was sampled only until week 4, after which it gave birth to a litter and was removed from the experiment. Thus, in total, 66 feces samples from eight different time points were analyzed.
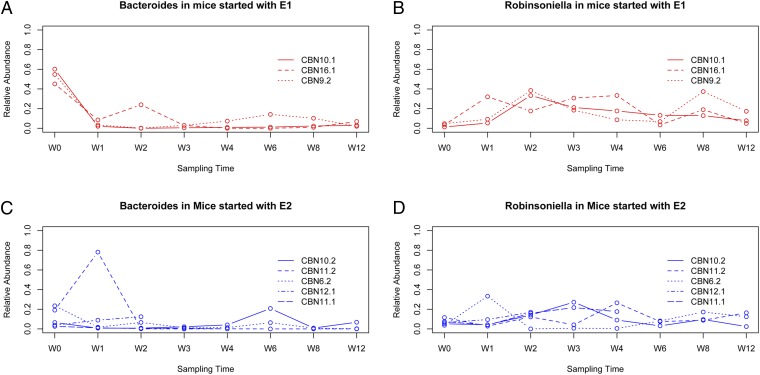
To remain consistent with our previous analysis we used the panel of 80 wild-caught samples as a reference for enterotype classification together with the 66 additional feces samples (Table S4). Again, this process yielded support for two clusters, which explain 32.11% of the variation in BC distance (adonis, P = 0.001). These effects are nearly threefold greater than the differences between fecal vs. cecal content sampling (adonis, 11.78% variation in BC distance explained, P = 0.001), which is unavoidable for the purpose of monitoring changes over time. Based on BC and pam clustering, eight E1 and six E2 classifications are present among the 14 mice at the initial point of sampling (Fig. 2). Subsequently, eight mice (three E1 and five E2) were sampled at regular intervals in the laboratory up to 12 wk postcapture. Among the five individuals initially classified as E2, all remained E2, with the exception of two individuals that displayed a short-term (i.e., a single sampling timepoint) transition to E1. In contrast, all three individuals classified as E1 at the onset of sampling quickly (i.e., after 1 wk) shifted to E2 and remained so throughout the remaining sampling period (Fig. 2). These changes are recapitulated by observing the differences in abundance of the signature taxa Bacteroides and Robinsoniella, where a drastic initial drop in abundance is observed for Bacteroides among the mice initially classified as E1 (Fig. 3A). Accordingly, temporary spikes in Bacteroides abundance are also associated with fluctuations between E1 and E2 in two mice initially classified as E2 (Fig. 3C).
Fig. 2.
Enterotype classification of mice transferred to the laboratory. Timepoints range from time of capture in the Cologne-Bonn (week 0; n = 14) through a 12-wk period of regular sampling in the laboratory (n = 8). Six mice died during transport to the laboratory and were thus sampled only at week 0. Each oval denotes one sample and colors correspond to enterotype classification (red for E1 and blue for E2).
Fig. 3.
Abundances of signature genera Bacteroides and Robinsoniella. A and B display Bacteroides and Robinsoniella abundance over time among mice classified as E1 at the time of capture (n = 3; red). C and D display Bacteroides and Robinsoniella abundance over time among mice classified as E2 at the time of capture (n = 5; blue).
Functional Metagenomic Analysis of Enterotype-Like Clusters.
To determine what functional genomic changes may be associated with different enterotype-like clusters, we sequenced the 14 mice captured in the Cologne/Bonn region as well as the week 1 and 12 time points of the eight mice brought back to the laboratory using an Illumina Hiseq platform (Methods and Table S5). Sequence data were analyzed using the MG-RAST pipeline (14). This pipeline provides four levels of functional categories, from most general to most specific, based on the results of mapping to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway maps (15). First, we evaluated community-level differences in functional genomic content after standardizing the abundances of each category to the total number of proteins predicted for each metagenomic sample (i.e., in a manner analogous to taxon abundance relative to the number of 16S rRNA gene sequences in a sample). Based on the application of adonis to each of the three applicable β-diversity measures [BC, JS, Jaccard (JA)], significant differences are apparent for both abundance-based measures (BC and JS) at all four levels of functional categories, for which enterotype status explains between 15% and 30% of the variation. Comparatively less variation was explained (∼10%) by a presence/absence measure (JA), although significant differences are present at three category levels (Table 3).
Table 3.
Differences in hierarchical functional categories (KEGG pathway levels) between E1 and E2 enterotypes
| KEGG pathway level | Dissimilarity Index | Adonis P value between E1 and E2 | Variances explained by enterotypes |
| Level 1 | Bray–Curtis | 0.011 | 0.13065 |
| Jensen–Shannon | 0.003 | 0.34091 | |
| Jaccard | NA* | NA | |
| Level 2 | Bray–Curtis | 0.007 | 0.14713 |
| Jensen–Shannon | 0.003 | 0.34037 | |
| Jaccard | 0.001 | 0.1011 | |
| Level 3 | Bray–Curtis | 0.003 | 0.1488 |
| Jensen–Shannon | 0.002 | 0.32386 | |
| Jaccard | 0.002 | 0.10017 | |
| Level 4 | Bray–Curtis | 0.001 | 0.14434 |
| Jensen–Shannon | 0.002 | 0.2893 | |
| Jaccard | 0.004 | 0.08484 |
Jaccard index not applicable at this level due to no differences in presence or absence.
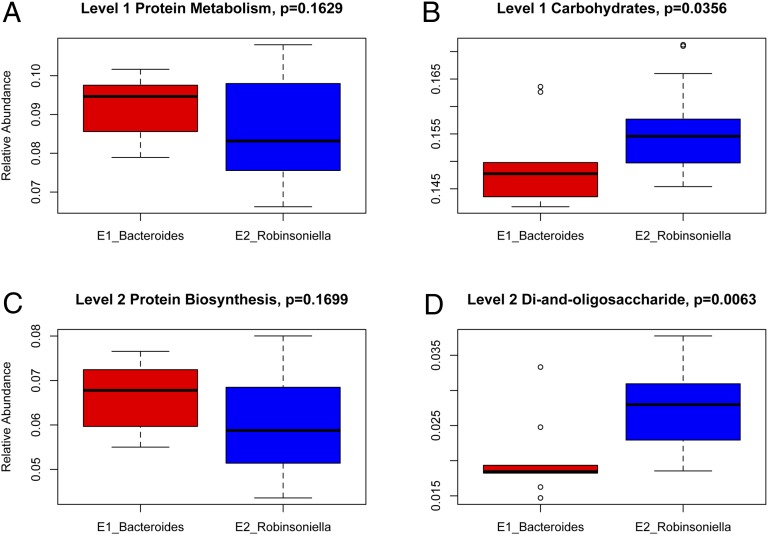
Next, we identified the major functional differences in the microbiome between the two enterotype classes using the SIMPER method in a manner analogous to that used to identify signature genera among 16S rRNA sequence data, but instead using the standardized functional categories from each level produced by MG-RAST. At both the first and second highest levels of functional categories, genes involved in protein metabolism/biosynthesis and carbohydrates are among the largest contributors to differences between E1 and E2, whereby E1 displays a higher proportion of protein-related genes and E2 a higher proportion of carbohydrate-related genes (Fig. 4 and Table 4). Furthermore, genes involved in bacterial motility appear to more frequent in E2 (Table 4). Similar patterns are observed at the remaining two levels of functional categories, although the contribution of each function becomes less prominent (Table S6), likely because of the increase in number of pathways.
Fig. 4.
Differences in hierarchical KEGG pathways between enterotypes. (A) Relative abundance of genes involved in level 1 protein metabolism. (B) Relative abundance of genes involved in level 1 carbohydrates. (C) Relative abundance of genes involved in level 2 protein biosynthesis. (D) Relative abundances of genes involved in level 2 di-and-oligo saccharides metabolism.
Table 4.
Level 1 and 2 KEGG pathways differing between E1 and E2 enterotypes
| Level | Pathways | Median in E1 (%) | Median in E2 (%) | Contribution to variance (%)* | Wilcoxon P value† |
| Level 1 | Protein metabolism | 9.11 | 8.31 | 10.78 | 0.1629 |
| Motility and chemotaxis | 0.99 | 1.58 | 9.22 | 0.0248 | |
| Carbohydrates | 14.77 | 15.35 | 8.48 | 0.0356 | |
| Phages prophages transposable elements/plasmids | 4.24 | 3.92 | 6.78 | 0.1288 | |
| Level 2 | Protein biosynthesis | 6.53 | 5.87 | 4.60 | 0.1699 |
| Di and oligosaccharides | 1.90 | 2.79 | 3.64 | 0.0063 | |
| Flagellar motility in Prokaryota | 0.81 | 1.27 | 3.41 | 0.0166 | |
| Phages prophages | 2.56 | 2.87 | 2.94 | 0.3238 | |
| Transposable elements | 0.96 | 0.52 | 2.86 | 0.0598 | |
| RNA processing and modification | 5.07 | 4.71 | 2.22 | 0.0858 | |
| Fermentation | 0.97 | 1.31 | 1.93 | 0.0063 | |
| Capsular and extracellular polysacchrides | 1.78 | 2.09 | 1.86 | 0.0063 | |
| Ton and Tol transport systems | 0.68 | 0.33 | 1.74 | 0.0166 | |
| Central carbohydrate metabolism | 3.62 | 3.28 | 1.74 | 0.0063 | |
| Plant/prokaryote | 6.37 | 6.27 | 1.73 | 0.1509 | |
| Monosaccharides | 2.28 | 2.27 | 1.46 | 0.4470 |
The contribution of each pathway to the total variance is calculated via SIMPER.
Significant differences [P < 0.05 after Benjamin–Hochberg correction (31)] are shown in bold.
Stable Osotope Analysis of Wild Mice.
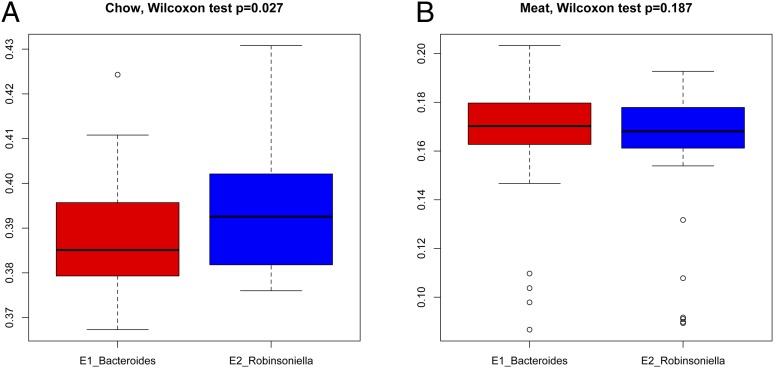
The results of the functional metagenomic analysis suggests that differences in diet (e.g., at the level of carbohydrate vs. protein intake), might contribute to the occurrence of enterotype-like clusters in wild mice. To test this hypothesis, we further analyzed stable isotopes (δ15N and δ13C) in muscle and liver tissues of original wild-caught mouse samples from Linnenbrink et al. (7), then quantified likely diets using the Bayesian mixing model FRUITS (16), which incorporates information from representative potential food sources (Methods and Table S7). We found that on average, food sources similar to standard laboratory chow (chow) are a major component of the long-term diet (39.95 ± 1.31%) of wild mice, followed by grain (36.42 ± 4.06%), meat (16.37 ± 2.67%), and insects (8.14 ± 2.90%). We also found significant differences in the major components of plant-derived food sources (chow) between the two enterotypes (median for E1 38.51% and E2 39.25%, Wilcoxon test P = 0.027), and the opposite pattern for animal-derived food sources (meat, median for E1 17.02% and E2 16.80%) although the difference is not significant (Wilcoxon test P = 0.187) (Fig. 5). No significant differences were found for grain or insects between enterotypes. Additionally, the application of linear models reveals Bacteroides abundance to be positively correlated with the percentage of meat and negatively correlated with the percentage of chow in the diet, whereas the opposite pattern is present for Robinsoniella abundance, although neither is significant (all P > 0.05). We do, however, observe a significant positive correlation between unclassified_Lachnospiraceae and the percentage of chow (r2 = 0.1089, P = 0.002), as well as a negative correlation between unclassified_Porphyromonadaceae and the percentage of meat (r2 = 0.0964, P = 0.005), both consistent with their differences in abundance between enterotypes (Table 1).
Fig. 5.
Abundance of major dietary components reconstructed for the two enterotypes. (A) Relative abundances of “chow” and (B) “meat” in the diet (see Methods for details on food standards).
Discussion
Our study provides several points of biological insight into the origin and functionality of enterotype-like community clusters. In particular, we describe the state of enterotypes in natural populations of house mice, and therefore identify the taxa and structuring of bacterial communities with which the mouse lineage most likely coevolved. Furthermore, we provide detailed information on the circumstances in which communities change their enterotype classification as well as the functional genomic consequences of such changes. Finally, the results of stable isotope analysis are consistent with underlying functional genomic changes (i.e., carbohydrate vs. protein metabolism), providing additional evidence that differences in recent dietary history/behavior and their metabolic consequences may contribute to the existence of different enterotype states.
Overall, our study adds to a growing picture of similarity in signature taxa across host species. Following the nomenclature in humans (3) and chimps (9), the major contributing genus to E1 in our study, Bacteroides, is also a significant contributor to “enterotype 1” in humans and chimps, as is Parabacteroides (Table 1). Similarly, unclassified Lachnospiraceae are significantly more abundant in E2 in our study and follow the same pattern in the human/chimp “enterotype 2.” Finally, the human/chimp “enterotype 3” is characterized in part by Ruminococcus. Although we identify no equivalent of a third enterotype in mice, Robinsoniella, a signature taxon of E2, is a close relative of Ruminococcus. One exception is that a Prevotella-dominated enterotype is identified only in humans. In laboratory and wild mice this may in part be the result of an overall 20- to 40-fold reduction in Prevotella abundance compared with humans (Table S8), but remains an interesting unexplained difference between the composition of human and chimpanzee enterotypes, as its overall abundance in chimpanzees is higher than in humans (9).
The study of Hildebrand et al. (11) identifies somewhat different signature taxa, which can possibly be attributed to investigating inbred laboratory mice. Nevertheless, laboratory and wild mice are still more similar to each other in overall composition than either is to humans (Fig. S2A and Table S8), and we identify the same number of enterotypes and similarities in the signature taxa between the two mouse studies. In particular, although a different nomenclature is used, their “ET1” displays increased Lachnospiraceae as does our E2. Interestingly, however, the other major contributor to our E2, Robinsoniella, appears absent in the Hildebrand et al. (11) study. We previously identified this genus as the most consistent and abundant member of the Firmicutes in wild mice (7), suggesting an important biological role that may be either absent or replaced by functional equivalents in the communities of laboratory mice. In addition, we identify one enterotype with reduced α-diversity, E1, for which a similar phenomenon is present in the enterotype “ET2” from Hildebrand et al. (11). A possible reason for this correspondence may be the increase in Bacteroides and Bacteroidaceae in the current study (E1) and Hildebrand et al. (11) (ET2), respectively, at the expense of other community members. Indeed, when the overall similarity in genus-level composition is compared taking enterotype status into consideration, the enterotype classes display more similarity with each other than either does to the other enterotype class within its respective laboratory- or wild mouse hosts (Fig. S2B).
Soon after the first report of enterotypes in humans (3), Wu et al. (8) reported that long-term diets were strongly associated with enterotype status, where individuals with more animal fat and protein intake were more likely to present the Bacteroides-dominated enterotype compared with those with more carbohydrate intake being associated with a Prevotella-dominated enterotype. The most compelling evidence for dietary influences on enterotypes given by our study is that the Bacteroides-dominated enterotype in wild mice, E1, quickly shifts to the alternative state of E2 after being transferred to the laboratory. Presumably, the standard chow fed to the mice in our breeding facility is in contrast to the greater variety of food sources available in the wild, which includes animal material, such as insects, worms, and so forth. Thus, a laboratory chow-restricted diet would be expected to deliver a more consistent source of plant-derived nutrients. Indeed, when comparing the major functional genomic categories that differentiate mouse enterotypes by shotgun metagenomic sequencing, the Robinsoniella-dominated E2 displays a significantly greater proportion of categories attributed to carbohydrate metabolism. Finally, by reconstructing dietary patterns via stable isotope analysis of liver and muscle tissue together with surrogates of food sources similar to the range available in the wild, we revealed a pattern of diet composition that is consistent with a more chow-like food intake in E2. The fact that the wild mice included in our study change their enterotype status within 1 wk after being transferred to the laboratory is in contrast to the 10-d dietary intervention conducted in humans by Wu et al. (8), where no change was observed. This finding may in part be explained by the higher metabolic rate of small mammals such as the mouse. Furthermore, although the mice in our study display significant patterns with respect to differences in carbohydrate intake- and functional metagenomic content between enterotypes, the role of other dietary components is less clear. Both protein metabolic function and the proportion of meat-derived intake are elevated in E1 in a direction consistent with the human Bacteroides-dominated enterotype (8), although the differences are not significant. This result suggests that a wider survey of nutritional sources and their combination is warranted to explore the circumstances under which mice transition from E2 to E1.
The clear presence of a single enterotype among the wild mice transferred to our laboratory facility raises the question as to why two enterotypes are observed among inbred laboratory mice that are also fed a standard diet. In their study, Hildebrand et al. (11) identify “low-grade” inflammation as a possible contributing factor. Other authors found that laboratory mice may have abnormal metabolic functioning because of their diet or environment (17), as well as decreased immune functioning compared with their wild counterparts (18), which may play a role in the observations made by Hildebrand et al. (11). An evaluation of inflammatory status was not performed on the mice in our study, although our data strongly suggest that inflammation alone is not responsible for the observed clustering. Furthermore, determining whether elevated inflammation contributes to, or is rather a consequence of enterotype status, will require thorough experiments, particularly in wild mice because they display greater variability in immune function compared with laboratory mice (18). In addition to low-grade inflammation, an alternative possibility is that related enterotypes may emerge over time among the altered species assemblages present in laboratory mice. Future studies incorporating additional laboratory facilities, time series, and gnotobiotic experiments may help shed light on these questions.
Conclusions
In summary, we provide evidence for the existence of two functionally different enterotype-like clusters present in wild house mice. Remarkably, these clusters display several characteristics in common with those of the distantly related human and chimpanzee hosts, suggesting the existence of ancient shared traits among the bacterial communities that assemble within mammalian hosts. Finally, we provide additional evidence that dietary habits may be the most important contributing factor to changes in enterotype status, which warrants more intensive future research to understand the impact of diet–microbiome interactions on human health and disease.
Methods
Animal Material and Sampling.
The cecal contents of 80 wild-caught mice described by Linnenbrink et al. (7) and an additional 10 mice transferred from the Massif Central region of France were separated from cecal tissue and preserved in 4 mL RNALater according to manufacturer’s instructions until further processing, as previously described (7). Of the 10 mice from the Massif Central that were transferred to the breeding facility of the Max Planck Institute for Evolutionary Biology, six were housed in individual cages and four were maintained as two breeding pairs to generate offspring to be included in an outbred colony derived from this region (19). The four mice involved in breeding were maintained as pairs (i.e., cohoused) for 1 to 2 mo, but were housed separately for >6 mo before being killed.
To directly monitor changes in bacterial communities associated with transfer to the laboratory, 14 mice were newly sampled in 2012 from the same Cologne/Bonn region of Germany sampled in 2010 (7). Feces were collected from all mice within several hours of retrieving the traps. Six mice failed to survive transportation to the laboratory and were thus only included as “week 0” samples. The remaining eight surviving mice were brought back to the laboratory breeding facility and monitored at regular intervals from the point of capture (1, 2, 3, 4, 6, 8, 12 wk) by sampling feces. One mouse was not sampled at week 3 because of treatment for a skin injury, and another was sampled only until week 4, after which it gave birth to a litter and was removed from the experiment. Thus, in total, 66 feces samples from seven different time points were analyzed. The transportation, handling, and maintenance of mice was conducted according to the German animal welfare law (Tierschutzgesetz) and Federation of European Laboratory Animal Science Associations guidelines. The approval for mouse husbandry was obtained from the local veterinary office “Veterinäramt Kreis Plön” (Permit 1401-144/PLÖ-004697).
DNA Extraction.
Bacterial DNA from cecal contents and feces was extracted using the QIAmp DNA stool mini kit (QIAGEN). Approximately 200 mg of material was transferred to 2-mL screw-cap tubes containing 50 mg each of 0.1-mm, 0.5-mm, and 1-mm glass beads (BioSpec Products). The tubes/beads were treated with UV exposure for 2 h before performing the extraction. After adding 1.4 mL ASL lysis buffer, bead beating was performed using the Precellys (Peqlab) for 3 × 15 s at 4,723 × g. Samples were then heated to 95 °C for 10 min, after which the manufacturer’s protocol was followed.
Bacterial 16S rRNA Gene Sequencing and Processing.
The 27F-338R primer pair spanning the hypervariable regions V1 and V2 was used for PCR and barcoded pyrosequencing on the 454 GS-FLX platform with Titanium sequencing chemistry, as previously described (2). Sequences were filtered using MOTHUR v1.22.2 (20) with the inclusion criteria of mean quality score >20 and length ≥250 bp. Sequences were assigned to samples by exact matches of 10-bp barcodes. For each sample, random subsets of 1,000 sequences were extracted to normalize coverage and taxonomical classification was performed using RDP Classifier (21). USEARCH/UCHIME v5.2.32 (22, 23) was used to identify chimeric sequences and perform sequencing clustering into operational taxonomic units using default parameters (OTU-based analyses are presented in Table S2).
Bacterial Community Analysis.
Bacterial community analyses including β-diversity metrics (BC, JA) and analysis of dissimilarity (adonis, which performs a multidimensional ANOVA on distance matrices and was applied to β-diversity metrics) were carried out using the VEGAN R package (24). The JS distance was calculated using the “flexmix” R package (25). Weighted and unweighted UniFrac distance matrices as well as phylogenetic diversity (PD) (26) were calculated from a maximum-likelihood tree constructed by FastTree (27) as implemented in MOTHUR. α-Diversity indices (Chao1 richness, Shannon) were calculated using the “VEGAN” R package (24), based on a normalized (subsampled) dataset (1,000 reads per sample).
To evaluate the presence of enterotype-like clustering, we used the original criteria of Arumugam et al. (3), which uses the Calinski–Harabasz index as an indicator for best clustering, in addition to the silhouette score and prediction strength suggested by Hildebrand et al. (11) and Koren et al. (10), respectively. The following clustering algorithms implemented in R packages were used: pam (R package “cluster”), kmeans (R package “flexclust”), and hierarchical clustering using “single,” “complete,” “average,” and “ward” linkage (R function “hclust”). To identify taxa contributing to groups based on BC distance, we applied the SIMPER method (13), which identifies taxa contributing to similarity within- and dissimilarity between groups and ranks their contribution.
Shotgun Metagenomic Sequencing and Analysis.
Bacterial DNA from fecal samples was subject to full metagenomic sequencing on the Illumina HiSEq. 2000 platform with paired-end 100-bp reads. Each sample was sequenced on a one-quarter lane, resulting in an average 8.54E9 bp of sequence per sample. After stringent quality filtering using the Fastq Tool Kit (each sequence required to have >99% of its nucleotides with a quality score ≥ 30), sequences were assembled using Meta-Velvet (28) under default parameters, resulting in an average of 1.36E8 bp of assembled reads per sample and then submitted to MG-RAST (http://metagenomics.anl.gov) (14) for further analysis. Assembly before submission to was necessary because of the Fastq data exceeding the limits of MG-RAST. The automated pipeline provided by MG-RAST was used to obtain taxonomic classification and functional annotation of genes predicted by FragGeneScan using the BLAT program referencing the M5NR database. For each sample, taxonomical and functional information was extracted and subjected to the same statistical analyses applied to 16S rRNA gene data using the VEGAN package in R (29).
Stable Isotope Analysis and Diet Reconstruction.
Approximately 200 mg each of liver and muscle tissue were recovered from enthanol-preserved carcasses subjected to extraction with a methanol/dichloromethane mixture to remove lipids (30) and dried at 70 °C for 48 h. The stable isotopes and concentrations of nitrogen and carbon were analyzed simultaneously with a THERMO/Finnigan MAT V isotope ratio mass spectrometer and THERMO Flash EA 1112 elemental analyzer at Braford University, United Kingdom. The ratios of stable isotopes are given using the conventional δ notion (δ15N and δ13C), with respect to atmospheric nitrogen and Vienna PeeDee Belemnite standards. Additionally we included four main foods groups that represent the range of possible dietary sources of wild mice. These groups included wheat grains from a rat food mixture and standard mouse laboratory chow (chow) as plant-derived food sources, as well as dried meal worms (insect) and meat-based dog food (meat). To reconstruct the most probable diet structure with respect to the relative proportion of each food standard listed above, we combined the isotope signatures from both muscle and liver and obtained dietary estimates using the Bayesian mixing model FRUITS (16) (http://sourceforge.net/projects/fruits).
Supplementary Material
Acknowledgments
We thank Silke Carstensen, Andrew Gledhill, Carl Heron, and Markus Schilhabel for excellent technical assistance and Almut Nebel and Guntram Grassl for helpful discussion. This work was supported by the Deutsche Forschungsgemeinschaft Grant BA 2863/2-2 (to J.F.B.), Excellence Cluster “Inflammation at Interfaces” (Nucleotide Laboratory) (P.R. and J.F.B.), and the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Bacterial 16S rRNA gene sequences are deposited at the European Nucleotide Archive, www.ebi.ac.uk (accession nos. ERP001970 and ERP004395). Metagenomic data are available under MG-RAST project (http://metagenomics.anl.gov/?page=MetagenomeProject&project=5130).
See Commentary on page 9372.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402342111/-/DCSupplemental.
References
- 1.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rausch P, et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci USA. 2011;108(47):19030–19035. doi: 10.1073/pnas.1106408108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arumugam M, et al. MetaHIT Consortium Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson J, et al. NIH HMP Working Group The NIH Human Microbiome Project. Genome Res. 2009;19(12):2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochman H, et al. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8(11):e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips CD, et al. Microbiome analysis among bats describes influences of host phylogeny, life history, physiology and geography. Mol Ecol. 2012;21(11):2617–2627. doi: 10.1111/j.1365-294X.2012.05568.x. [DOI] [PubMed] [Google Scholar]
- 7.Linnenbrink M, et al. The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Mol Ecol. 2013;22(7):1904–1916. doi: 10.1111/mec.12206. [DOI] [PubMed] [Google Scholar]
- 8.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moeller AH, Degnan PH, Pusey AE, Wilson ML, Hahn BH, et al. Chimpanzees and humans harbour compositionally similar gut enterotypes. Nat Commun. 2012;3:1179. doi: 10.1038/ncomms2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koren O, et al. A guide to enterotypes across the human body: Meta-analysis of microbial community structures in human microbiome datasets. PLOS Comput Biol. 2013;9(1):e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hildebrand F, et al. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013;14(1):R4. doi: 10.1186/gb-2013-14-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arthur JC, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338(6103):120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke KR. Nonparametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18(1):117–143. [Google Scholar]
- 14.Meyer F, et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogata H, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandes R, Millard AR, Brabec M, Nadeau MJ, Grootes P. Food reconstruction using isotopic transferred signals (FRUITS): A Bayesian model for diet reconstruction. PLoS ONE. 2014;9(2):e87436. doi: 10.1371/journal.pone.0087436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin B, Ji S, Maudsley S, Mattson MP. “Control” laboratory rodents are metabolically morbid: Why it matters. Proc Natl Acad Sci USA. 2010;107(14):6127–6133. doi: 10.1073/pnas.0912955107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abolins SR, Pocock MJO, Hafalla JCR, Riley EM, Viney ME. Measures of immune function of wild mice, Mus musculus. Mol Ecol. 2011;20(5):881–892. doi: 10.1111/j.1365-294X.2010.04910.x. [DOI] [PubMed] [Google Scholar]
- 19.Ihle S, Ravaoarimanana I, Thomas M, Tautz D. An analysis of signatures of selective sweeps in natural populations of the house mouse. Mol Biol Evol. 2006;23(4):790–797. doi: 10.1093/molbev/msj096. [DOI] [PubMed] [Google Scholar]
- 20.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 24.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14(6):927–930. [Google Scholar]
- 25.Grun B, Leisch F. FlexMix version 2: Finite mixtures with concomitant variables and varying and constant parameters. J Stat Softw. 2008;28(4):1–35. [Google Scholar]
- 26.Faith D. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61(1):1–10. [Google Scholar]
- 27.Price MN, Dehal PS, Arkin AP. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26(7):1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Namiki T, Hachiya T, Tanaka H, Sakakibara Y. MetaVelvet: An extension of Velvet assembler to de novo metagenome assembly from short sequence reads. Nucleic Acids Res. 2012;40(20):e155. doi: 10.1093/nar/gks678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 30.Cequier-Sánchez E, Rodríguez C, Ravelo AG, Zárate R. Dichloromethane as a solvent for lipid extraction and assessment of lipid classes and fatty acids from samples of different natures. J Agric Food Chem. 2008;56(12):4297–4303. doi: 10.1021/jf073471e. [DOI] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc, B. 1995;57(1):289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.