Significance
Patients homozygous for the C-C chemokine receptor type 5 (CCR5) gene with 32-bp deletions (Δ32) are resistant to HIV infection. Using the piggyBac technology plus transcription activator-like effector nucleases or clustered regularly interspaced short palindromic repeats-Cas9, the authors report, to our knowledge, for the first time in induced pluripotent stem cells (iPSCs) the efficient and seamless derivation of a homozygous CCR5Δ32 mutation, exactly mimicking the natural mutation. Monocytes and macrophages differentiated from these mutated iPSCs in vitro are resistant to HIV infection. This approach can be applied in the future toward the functional cure of HIV infection. The findings are also of great interest to researchers in many fields who wish to correct or introduce mutations in specific genes.
Keywords: homologous recombination, TTAA site, off-site target, cellular therapy
Abstract
Individuals homozygous for the C-C chemokine receptor type 5 gene with 32-bp deletions (CCR5Δ32) are resistant to HIV-1 infection. In this study, we generated induced pluripotent stem cells (iPSCs) homozygous for the naturally occurring CCR5Δ32 mutation through genome editing of wild-type iPSCs using a combination of transcription activator-like effector nucleases (TALENs) or RNA-guided clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 together with the piggyBac technology. Remarkably, TALENs or CRISPR-Cas9–mediated double-strand DNA breaks resulted in up to 100% targeting of the colonies on one allele of which biallelic targeting occurred at an average of 14% with TALENs and 33% with CRISPR. Excision of the piggyBac using transposase seamlessly reproduced exactly the naturally occurring CCR5Δ32 mutation without detectable exogenous sequences. We differentiated these modified iPSCs into monocytes/macrophages and demonstrated their resistance to HIV-1 challenge. We propose that this strategy may provide an approach toward a functional cure of HIV-1 infection.
The C-C chemokine receptor type 5 (CCR5) is the major coreceptor used by HIV-1 to infect T cells, macrophages, and other cell types. Individuals who are heterozygous or homozygous for the CCR5∆32 mutation in the CCR5 gene have slower progression or resistance to HIV infections, respectively (1–3). One patient has apparently been cured of HIV infection following allogeneic hematopoietic stem cell transplants from a homozygous CCR5Δ32 donor (4). This finding suggests a promising avenue for developing stem cell therapy to treat HIV infection. However, although it is encouraging, allogeneic transplantation is not likely to be widely applicable because the low frequency of CCR5Δ32 homozygotes in the general population plus the logistics and feasibility of identifying suitable HLA-compatible donors with this mutation hinder practical applications. Furthermore, this approach requires full bone marrow ablation and immune suppression.
Autologous transplantations are less toxic, because they may not require complete bone marrow ablation or immune suppression for engraftment. In this setting, several studies have attempted to produce HIV resistance by disabling the CCR5 gene in CD34+ hematopoietic stem progenitor cells (HSPCs) or CD4+ T cells using shRNA or by gene disruption using zinc finger nucleases (ZFNs) (5–11). These reagents are delivered into CD34+ or T cells using lentiviral or adenoviral vectors. However, the effects of these approaches may not be long-lasting, because the modified cells may be unable to compete with the endogenous unmodified cells. Also, integration mutagenesis by lentiviral vectors could potentially cause other complications (12, 13), and the immunogenicity of adenoviral vectors could eliminate the transduced cells from the recipient (14).
Induced pluripotent stem cells (iPSCs) (15) could provide a rich source of patient-specific stem cells that can be genetically modified and differentiated into various cell lineages. Thus, iPSCs could be an ideal cell type for providing an autologous stem cell therapy as treatment for HIV-infected patients.
Recently, several engineered nucleases have been developed such as ZFNs (16), transcription activator-like effector nucleases (TALENs) (17), and the RNA-guided CRISPR-Cas9 nuclease for genome modifications (18, 19). These nucleases introduce double-strand breaks (DSBs) into the genome and greatly enhance genome targeting efficiency through the host’s cellular DNA repair machinery. In addition, piggyBac transposon technology can allow the insertion of selectable markers into the targeting construct to enrich cells that have undergone homologous recombination. Subsequently, the transposon can be removed by transient expression of transposase, leaving a seamless genome modification behind (20).
In the current study, we combined these technologies and devised a strategy that generated the naturally occurring CCR5Δ32 mutation without leaving any residual sequences. We also compared the efficiencies of the TALENs and CRISPR-Cas9 approaches in achieving this goal. Finally, we differentiated these CCR5-mutated iPSCs into monocytes and macrophages in vitro and demonstrated that the cells are resistant to HIV-1 infection.
Results
Construction of Targeting Vectors Using PiggyBac Transposon and TALENs Vectors.
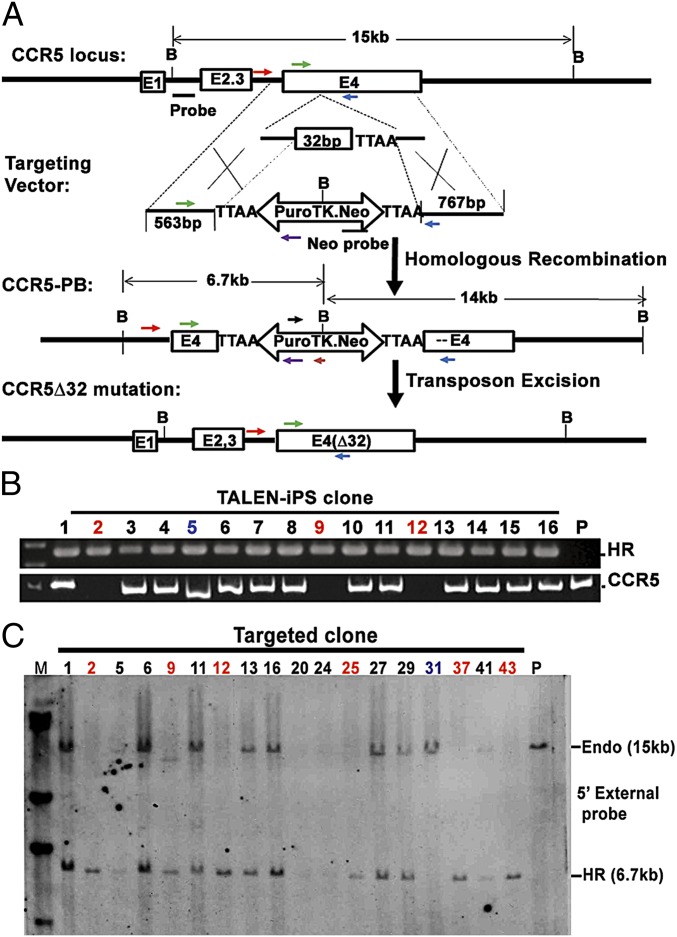
To reproduce the naturally occurring CCR5Δ32 mutation, we devised a strategy that could precisely generate the ∆32 mutation without leaving any residual sequences in the targeted genome. On examination of the sequences at exon 4 of the CCR5 gene in the region of the ∆32 mutation, we discovered a TTAA tetranucleotide sequence that piggyBac uses to insert into or be released from the host genome. The tetranucleotide is located immediately downstream of the naturally occurring 32-bp deletion. We made a targeting construct carrying the 32-bp mutation and a CAG-puroΔTK cassette flanked by piggyBac repeats into the TTAA site (Fig. 1A). The homologous sequence on the right arm starts from the TTAA. On the left, the homology begins 32 bp away from the genomic TTAA (Fig. 1A). By homologous recombination, the piggyBac vector can be inserted precisely at this TTAA site to allow selection of the cells with puromycin (Fig. 1A). The transposon containing the selection cassette inserted at the TTAA site can then be excised using the transient expression of transposase, leaving the ∆32 mutation behind (Fig. 1A). Further negative selection with FIAU [1-(2-deoxy-2-fluoro-b-d-arabinofuranosyl] can dispose of the cells that retain the piggyBacs carrying the TK gene (Fig. 1A).
Fig. 1.
Gene targeting in the CCR5 locus by TALENs in iPSCs. (A) Strategy for seamless genome modification using the piggyBac technology. (Top) Structure of the CCR5 gene; B, BamHI; E, exon. Arrows: PCR primers (P1, black; P2, purple; P3, green; P4, blue; P5, red, P6, brown). Box with 32 bp: the 32-bp deletion found in the CCR5∆32 gene. Open double-headed arrows: piggyBac transposon carrying a CAG promoter driven puroΔTK and Neo cassettes. (B) Genotyping of targeted clones by PCR analyses. (Upper) Detection of homologous recombination by P2 and P5 primers. (Lower) Biallelic knockout of the untargeted CCR5 gene by P3 and P4 primers. Clones 2, 9, and 12, biallelic targeted CCR5 gene indicated by the absence of amplification of both alleles owing to the insertion of the large targeting construct; clone 5, one allele with NHEJ, the other with homologous recombination effectively knocking out the function of both alleles; HR, homologous recombination; P, parental iPSCs without targeting. (C). Genotyping of targeted clones by Southern blot analysis. After BamHI digestion of genomic DNA, the endogenous allele without targeting produces a 15-kb band. A 6.7-kb (HR) band indicates targeting by the HR donor using a 5′ external probe. Absence of a 15-kb endogenous band indicates biallelic targeting (clone numbered in red); clone 31 (in blue) has no targeting.
We first used the TALENs-mediated DNA DSB strategy for enhancing gene targeting efficiency. The computer program ZIFIT targeter was used to search for potential targeting sites around the Δ32 mutation region of the CCR5 gene. The program identified the same two pairs of targeting sequences of TALENs (L538 and R557; L540 and R560) that had previously been shown by the Surveyor nuclease assay to give up to 27% efficiency in targeted site modification (17). To increase the specificity of TALENs binding to the targeting site, the L540 was extended 1 bp at the 5′ and 2 bp on the 3′ end to bind a total of 17 bp (Fig. S1). Each designed TALENs was driven by a CAG promoter and had the C-terminal segment shortened to 63 residues that retain in vitro cleavage activity (17).
Gene Targeting of the CCR5 Locus in CCR5 Wild-Type iPSCs by TALENs.
To target the CCR5 locus in iPSCs, three combinations of the TALEN pairs (L538–R557, L540long–R560, and L540long–R557) expressing vectors were cotransfected with a targeting vector containing the piggyBac cassette (Fig. 1A, Middle) into three wild-type iPSC cell lines. These had been previously derived from peripheral blood mononuclear cells from two different donors (21). After a 2-wk selection with puromycin, resistant clones were picked and screened by PCR (Fig. 1B). Remarkably, up to 100% of the puromycin-resistant colonies were targeted in one CCR5 allele, of which an average of 14% was simultaneously targeted in both alleles (Table 1). No significant difference in the targeting efficiency was found for the three combinations of TALEN pairs. The PCR analysis also revealed that, in total, 15% of the clones targeted on one allele with homologous recombination (HR) also had nonhomologous end joining (NHEJ) on the other allele. Sequence analysis of four randomly chosen NHEJ clones revealed different degrees of deletions that could abolish all normal CCR5 gene functions (Fig. S2 A and B). Homologous recombination was further confirmed by Southern blot analyses (Fig. 1C). In 85.7% of biallelic knockout events, no extra integration in the genome was detected by Southern blot analysis using a probe inside the cassette (Neo probe) (Table S1).
Table 1.
Summary of targeting efficiency by PCR analyses
| Cell line | TALEN pair | Puror analyzed | HR (efficiency) | Biallelic targeting (efficiency) | NHEJ (frequency) |
| WT1-6 | L540long-R560 | 34 | 34 (100%) | 4 (11.7%) | 2 (5.9%) |
| WT1-6 | L538-R560 | 41 | 41 (100%) | 3 (7.3%) | 4 (9.3%) |
| WT1-11 | L540long-R560 | 27 | 26 (96%) | 6 (22.2%) | 6 (22.2%) |
| WT2-3 | L538-R557 | 20 | 20 (100%) | 4 (20%) | 6 (30%) |
| Total | 122 | 121 (99%) | 17 (14%) | 18 (15%) |
Three iPSC clones derived from two different donors were used for this study. HR, number of homologous recombination clones studied; NHEJ, number of nonhomologous end-joining clones evaluated; Puror analyzed, number of puromycin resistance clones being analyzed; WT1, from donor 1; WT2, from donor 2.
Excision of Transposon from Targeted Clones.
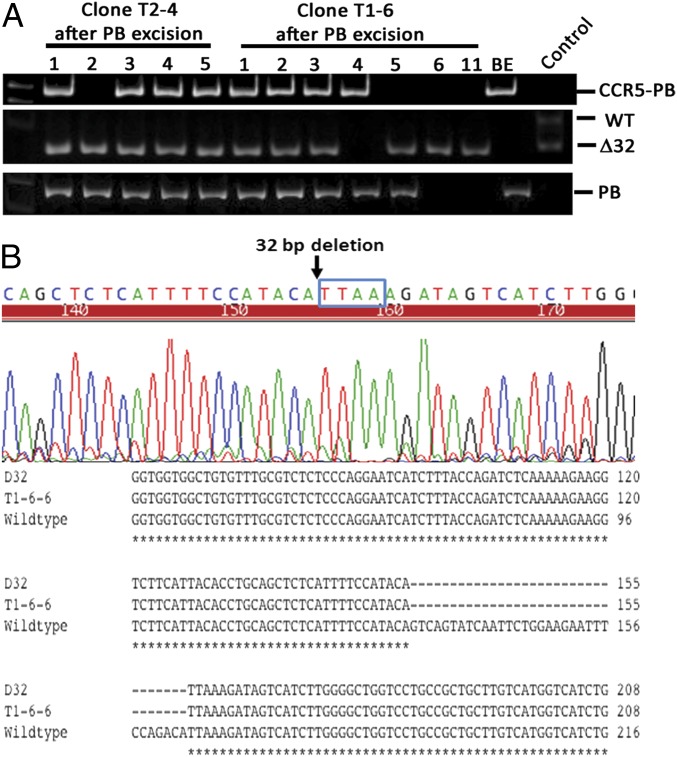
To remove exogenous DNA, we transiently transfected two homozygously targeted clones (T1-6 and T2-4) with hyperactive piggyBac transposase (pCMV-HyPBase), followed by negative selection with FIAU. We then picked 48 clones from each transfection, but only 11 and 6 clones could be expanded from T1-6 and T2-4, respectively. These 17 clones were genotype-analyzed by PCR. Two clones from T1-4 showed biallelic excision of transposon cassettes, but the cassettes reintegrated into other locations in the genome. Six clones from T1-6 showed biallelic excision of transposons from the CCR5 loci, and two of them (clones 6 and 11) did not show reintegration of the transposons into another place in the genome (Fig. 2A and Table 2). This finding was further confirmed by Southern blot analysis using the internal Neo probe (Fig. S3). Sequence analyses revealed that the Δ32 mutation was generated on both alleles in these two clones that carried no piggyBac after the transposon excision (Fig. 2B).
Fig. 2.
Generation of the seamless CCR5∆32 mutation by piggyBac transposase excision. (A) PCR analyses after transposon excision. (Top) Absence of a band indicates biallelic excision of the transposon at the CCR5 locus using P2 and P5 primers. (Middle) Presence of the Δ32 band indicates successful generation of the 32-bp deletion using P3 and P4 primers. (Bottom) Absence of a band indicates complete removal of transposon from the genome, using P1 and P6. (B) Sequence analyses for seamless excision of transposon. (Upper) Sequences of excision clone reveals a 32-bp deletion before the TTAA site. (Lower) Alignment of excision clone (T1-6-6) sequences with wild type and Δ32 mutation of the CCR5 gene.
Table 2.
Summary of transposon excision
| Cell line | Clones analyzed | Single excision | Biallelic excision without reintegration, no. of clones (frequency) | Biallelic excision with reintegration, no. of clones (frequency) |
| T2-4 | 6 | 4 | 0 (0%) | 2 (33.3%) |
| T1-6 | 11 | 6 | 2 (18%) | 2 (18%) |
| Mean frequency | 9% | 25.7% |
T1, targeting clone used L540long-R560 TALEN pair; T2, targeting clone used L538-R557 TALEN pair.
Pluripotency of Genetically Modified Homozygous CCR5Δ32 iPSCs and Normal Karyotype.
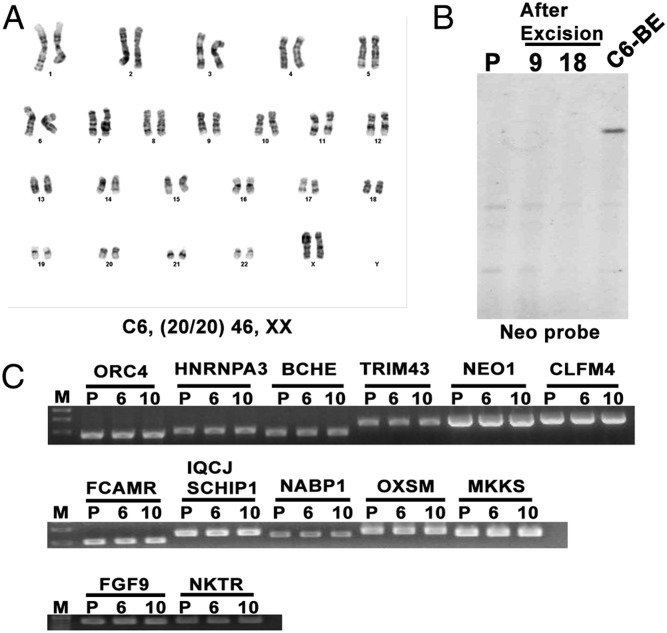
We next examined whether the genetically modified iPSC clones maintained pluripotency. They stained positively with the pluripotent markers NANOG, SSEA3, TRA-1-81, and TRA-1-60 before and after transposon excision (Fig. S4A). After transposon excision, they also randomly differentiated into cells that stained positively with the markers representing the three germ layers in vitro (Fig. S4B) and formed teratomas that contained the three germ layers in immunodeficient mice (Fig. S4C). The modified iPSCs had, as well, the expected normal female karyotype (Fig. S4D). These results demonstrate that, after two rounds of transfection to produce seamlessly the homozygous CCR5∆32 mutation, the iPSC lines maintained their pluripotency in vitro and in vivo.
Specific Genome Modification Without Off-Target Effects by TALENs.
To test for potential nonspecific mutations induced by the introduction of the TALENs, we studied 22 potential off-target loci of TALEN pairs (Table S2), predicted using a bioinformatics-based approach, the ranking algorithm PROGNOS (22). All of the potential off-target sites were located in intron and intergenic regions; 21 out of the 22 consisted of inverted repeats of the right TALEN target sites, making these sites vulnerable to cleavage by the wild-type Fok1 homodimer (23). Notably, 16 out of these 21 loci that could be amplified by PCR from the genomic DNA of both the wild-type parental iPSCs and the modified iPSCs showed no mutations by Sanger sequence analysis, as summarized in Table S2. However, amplification of the remaining five loci was not successful in both the parental and modified iPSCs, most likely owing to repeat sequences or genome annotation problems.
RNA-Guided Cas9 Nuclease-Mediated Gene Targeting of the CCR5 Gene.
Recently, an engineered type II CRISPR-Cas9 system has been developed for genome editing (18, 19). It uses a guide RNA (gRNA) having 20 nt complementary to genome target sequences followed by a trinucleotide (5′-NGG-3′) called protospacer adjacent motif (PAM) that can be recognized by Cas9 nuclease. The simplicity and ease of construction make this system an attractive alternative to TALENs for genome editing. Hence, we compared the efficiencies of RNA-guided Cas9 nuclease to TALENs in introducing the ∆32 mutation into the CCR5 gene.
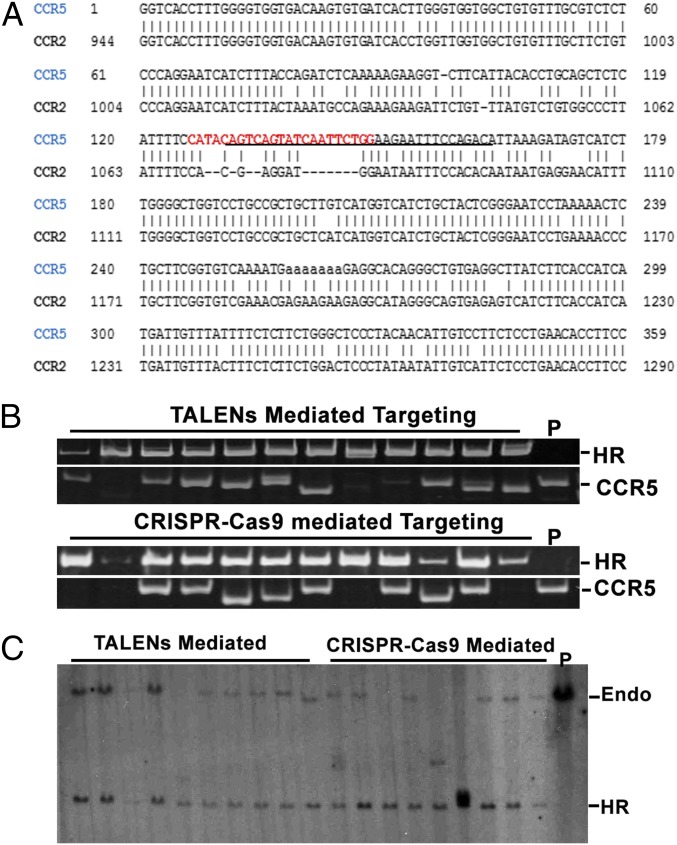
The 10–12 bp proximal to the PAM sequence are the most important determinant of targeting specificity (18, 19). Owing to the high homology between the CCR5 and CCR2 genes around the mutation region, only a few target sites met these requirements. We chose a targeting sequence with the least homology (eight mismatches within 12 bp of the PAM proximal region) to CCR2 (Fig. 3A). The gRNA was cotransfected with Cas9 and the targeting construct used in the TALEN study. In parallel, the TALENs combinations with the targeting construct were also transfected into the same iPSC line for comparison. Four times more colonies were obtained after puromycin selection from the gRNA transfections than from TALENs. From each transfection, 20 clones were picked and analyzed by PCR to detect the homologous recombination and biallelic targeting. The gRNA- and TALENs-mediated targeting both yielded 100% homologous recombination, of which biallelic targeting was 10% with TALENs and 33% with gRNA by PCR analysis (Fig. 3B and Table 3). NHEJ was detected in 45% of the clones with TALENs-mediated targeting and 25% in gRNA-mediated targeting (Table 3). To confirm homologous recombination, Southern blot analysis was performed using an external probe. All of the targeted clones by the TALENs and gRNA approaches showed the same patterns and correct sizes in CCR5 locus modification consistent with homologous recombination (Fig. 3C). Thus, the frequency of homologous recombination with CRISPR-Cas9 is four times greater than with TALENs. Moreover, although they could both target a single allele of the CCR5 locus with 100% efficiency, the biallelic targeting efficiency by the CRISPR-Cas9 system was three times higher. Like the homologous recombination mediated by TALENs, the karyotype remained normal and the cassette was removed seamlessly, as confirmed by Southern blot analysis (Fig. 4 A and B).
Fig. 3.
Targeting the CCR5 locus using RNA-guided CRISPR-Cas9 nuclease compared with TALENs. (A) Schematic representation of the location of target DNA sequences in the CCR5 gene aligned with the CCR2 gene. The underlined sequences are the Δ32 mutation; sequences in red are gRNA targeting sequences. (B) Genotyping of targeted clones by PCR analysis. The same sets of primers were used as in the TALENs study. Homologous recombination (HR) was determined by the amplification within the P2 and P5 primers. Biallelic targeting was determined by no amplification with the P3 and P4 primers. NHEJ was detected by the amplification with the P3 and P4 primers with the size either larger or smaller than that of wild-type parental control (P). (C) Genotyping of targeted clones mediated by TALENs and CRISPR-Cas9 system using Southern blot analysis. Genomic DNAs from TALENs-targeted and CRISPR-Cas9–targeted iPSC clones were digested by BamHI and separated side by side on 0.8% agarose gel and then hybridized with the external probe indicated in Fig. 1A. The endogenous allele without targeting produces a 15-kb band. The allele with HR produces a 6.7-kb band. P, parental control without targeting.
Table 3.
Comparison of TALENs and CRISPR-Cas9 system on targeting of the CCR5 locus
| Method | HR efficiency | Biallelic targeting efficiency | NHEJ efficiency |
| TALEN (L538-R557) | 100% | 10% | 45% |
| CRISPR-Cas9 | 100% | 33.3% | 25% |
The comparison was conducted using the same parental clone, WT2-3. Twenty clones were analyzed by PCR for each experiment.
Fig. 4.
Karyotype of CRISPR-Cas9–mediated, genome-modified iPSC seamless modification and off-target analysis for CRISPR-Cas9–mediated targeting. (A) Karyotype of the CRISPR-Cas9–mediated targeted clone after transposon excision; 20/20 indicates that 20 metaphases examined for each line had the same normal female karyotype. (B) Southern blot analysis of BamHI-digested genomic DNA with Neo internal probe after transposon excision. C6-BE, clone 6 before transposon excision; P, parental line without targeting. (C) Off-target analysis for CRISPR-Cas9–mediated targeting. Thirteen potential off-target sites were PCR-amplified from two biallelic targeted clones (6, 10) and the parental line (P). The closest gene name is used to indicate the amplification presented on top of the lines. These amplifications were subjected to Sanger sequence analysis to reveal any mutation between the targeted clones and the parental line. The results are summarized in Table S3. M, size marker.
Specific Targeting of the CCR5 Gene Without Off-Target Modifications by CRISPR-Cas9.
Because the gRNAs typically target up to 20-bp DNA sequences, these short sequences may exist potentially in multiple off-target sites in the whole genome. Therefore, the 20-nt sequence of gRNA was BLAST-searched in the NCBI database of the human genome and many partially matching sequences were identified. Because the base pairing with the 10–12 bp 5′ of the PAM (PAM-proximal) is the primary determinant of Cas9 specificity, we chose the potential off-target sites based on this concept. We analyzed the seven perfect matches on the PAM-proximal region ranging from 10 to 13 bp as well as two with single mismatches and one with three mismatches within the 15 bp of the PAM-proximal region (Table S3). Previous studies have found that targeting sequences followed by a 5′-NAG PAM also demonstrate off-target modifications (24). Thus, we added three other 20-bp highly homologous sites that were followed by a PAM with 5′-NAG sequences for analysis (Table S3). These 13 loci were amplified by PCR in two targeted clones and the parental cell line (Fig. 4C). No mutation was found by Sanger sequence analysis (Table S3).
Resistance of Monocytes/Macrophages Derived from CCR5 Mutant iPSC to HIV-1 Infection.
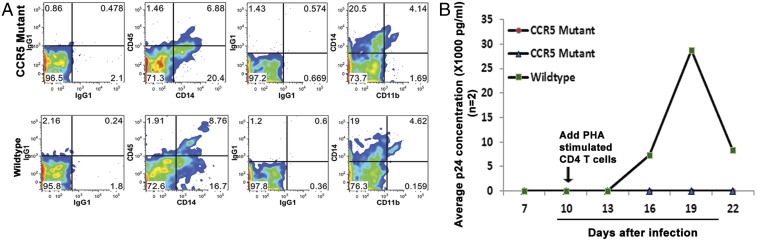
To investigate whether the CCR5∆32 mutated from the wild type could confer resistance to HIV-1 infection just like the naturally occurring one, mutant and wild-type parental iPSC lines were differentiated into monocytes/macrophages using a two-step differentiation protocol similar to that described (25, 26). After 24 d of culture, we were able to obtain a comparable number of monocytes/macrophages (CD45+CD14+ and CD14+CD11b+) derived from both the mutant and parental iPSCs (Fig. 5A). The differentiated cells from two mutants and the parental iPSCs were inoculated with a CCR5-tropic virus isolate, HIV-1SF170, for 1 d and then the virus was removed by washing. Ten days after infection, we amplified the virus by adding phytohemagglutinin (PHA)-stimulated primary CD4+ cells to the culture. Beginning from day 16, virus replication was detected in the monocytes/macrophages derived from the wild-type parental line but not in the cells derived from the mutant lines (Fig. 5B). These results demonstrate that wild-type iPSCs engineered to be homozygous for the CCR5∆32 gene mutation can impart resistance to HIV-1 infection in monocytes/macrophages differentiated from them.
Fig. 5.
Resistance of monocytes/macrophages derived from modified iPSCs to HIV-1 infection. (A) Flow cytometry analysis of iPSCs-derived monocytes and macrophages. Monocytes and macrophages derived from CCR5 mutant (Upper) and wild-type (Lower) iPSCs were stained with CD14, CD45, and CD11b. (B) HIV-1 challenge of CCR5 mutant and wild-type iPSC-derived monocytes/macrophages. A CCR5-tropic virus isolate, HIV-1SF170, was inoculated onto three monocyte/macrophage cultures (two mutants and one wild type) at day 0. One day later, the virus was removed by washing and medium was changed every 2–3 d. On day 10, PHA-stimulated CD4+ T cells were added to the culture to amplify the virus for detection. Cell culture supernatants were sampled from various days postinfection and analyzed for p24 antigen by ELISA. The experiment was performed in duplicate. A representative study is presented.
Discussion
Individuals homozygous for the CCR5∆32 genotype have no discernable deleterious clinical effects (1, 27). This observation makes CCR5 an attractive target for gene therapy for AIDS. Toward this objective, certain groups have reported CCR5 gene modification in HSPC, CD4+ T lymphocytes, and iPSCs (6–10, 28). Most studies focused on disrupting the CCR5 gene by delivering ZFNs or TALENs. Whether cells with complete disruption of the CCR5 gene could have some yet-undiscovered deleterious effect is not known. Because individuals with the homozygous CCR5Δ32 mutation are apparently healthy, we focused on reproducing this genotype.
Using a combination of double-strand cleavage with TALENs or CRISPR-Cas9 and piggyBac technology, with positive and negative selections, we generated biallelic Δ32 mutations precisely matching the naturally occurring homozygous CCR5Δ32 genotype. Our findings also indicate the high efficiency of TALENs and RNA-guided CRISPR-Cas9–induced homologous recombination. We could obtain in iPSCs close to 100% efficiency of homologous recombination for a single allele and up to 22% and 33% for double alleles using the TALENs and RNA-guided CRISPR-Cas9 approaches, respectively (Tables 1 and 3). Our results are similar to the frequency Choi et al. obtained with TALENs-mediated homologous recombination for correcting alpha-1 antitrypsin deficiency (29). Moreover, the monocytes/ macrophages derived from biallelic CCR5∆32 mutation iPSCs became resistant to HIV-1 infection (Fig. 5).
Recently, Cho et al. (30) targeted a CCR5 locus about 500 bp upstream of ours using RNA-guided Cas9 nuclease in HEK293T cells. This approach resulted in an 18% cleavage efficiency as detected by the T7 endonuclease mutation detection assay. In another study on human iPSCs, the Cas9 gRNAs or TALENs were combined with single-strand oligodeoxynucleatides (ssODN) to generate a 2-bp substitution with scarless genome editing in the CCR5 locus (31). Whether the ssODN strategy could bring about the biallelic 32-bp deletion that we produced in the CCR5 locus needs further investigation.
Using iPSCs as target cell sources for potential HIV gene and cellular therapy has been explored (28). In that study, the CCR5 gene was modified by knockdown using a lentiviral vector that delivered short hairpin RNA targeting CCR5. Our present study demonstrates that the permanently mutated CCR5 gene in iPSCs and the monocytes/macrophages and likely other susceptible progenies derived from the CCR5∆32 mutated iPSCs are resistant to HIV-1 infection (Fig. 5).
A recent paper using ZFNs delivered by adenoviral vectors modified the CCR5 gene in CD4+ T cells from HIV-infected patients (32). The results after transplantation of these cells were encouraging, particularly in the one patient heterozygous for CCR5Δ32, who remained free of viremia after 3 mo without antiretroviral therapy. However, the number of engrafted T cells was limited, as was their half-life in the patients. Thus, repeated T-cell transplants would be needed and the adenoviral vectors, being immunogenic, could reduce the success of subsequent transplantation. Moreover, after ZFN cleavage, the CCR5 gene is repaired by a variety of different insertions or deletions. It is not clear whether some of these changes could be deleterious and raise safety issues for long-term treatment. Using iPSC-derived CCR5−/− stem cells for an autologous transplant has advantages over using primary CD4+ T cells. First, generation of iPSCs only requires a very small amount of blood (21). Second, unlimited iPSC production would overcome the limited amount of primary T cells available for transplantation. Third, unlike primary T cells, the individual cells can be checked for any genomic alteration after modification. Moreover, the individual iPSC clone can be sequenced to confirm the genome integrity and ensure a safe transplantation. Finally, using iPSC provides reconstruction of the entire hematopoietic cell systems including macrophages, not just CD4+ T cells.
The genetic manipulation of iPSCs is a crucial approach for the development of autologous cell-based therapy. Maintaining genome integrity with minimal genetic alteration is highly demanded for ensuring the value of iPSCs. Potential off-target cleavages by engineered nucleases raise concerns both for clinical application and biological studies. We have intensively examined the potential off-target sites for the TALENs pairs and the gRNA we used and found no sequence changes in the parental and modified iPSC lines (Tables S2 and S3 and Fig. 4C). These findings demonstrate that the appropriate gRNA choice could target the specific CCR5 locus and at a higher HR rate than TALENs. Moreover, the TALENs pairs also could be modified by introducing newly developed FokI heterodimer obligate versions to minimize cleavage at off-target sites consisting of homodimers of either target half-site (23). The Cas9 nickase combined with paired gRNAs strategy could enhance the specific targeting in the genome (33).
We have demonstrated seamless excision by piggyBac technology. However, reintegration of the transposon catalyzed by piggyBac transposase occurs in 90% of cells (Table 2). Thus, the excision might be improved by a newly developed excision-competent/integration-defective transposase (34).
In summary, we demonstrate that piggyBac technology in combination with engineered nucleases can mediate homologous recombination that generates the naturally occurring CCR5Δ32 mutation in iPSCs with high frequencies. Monocytes/macrophages derived from these iPSCs are resistant to HIV infection. These derived iPSC lines could form the source of cells used for autologous therapy in HIV infection.
Materials and Methods
Construction of the Targeting Vectors, TALEN Vectors, and gRNA Vector.
The piggyBac transposon vector pCAG-puroTK.Neo was modified from 5′-PTK-3′ plasmid, a gift from Allen Bradley (The Wellcome Trust Sanger Institute, Cambridge, UK). Detailed procedures are given in SI Materials and Methods.
TALENs constructs were derived from the pC-Goldy TALEN vector (38143; Addgene) backbone and custom-made by the University of California, San Francisco ES core facility. The U6 target gRNA empty vector (41824; Addgene) was used for gRNA constructing.
iPSC culture, gene targeting, transposon excision, TALEN, and Cas9 off-target site analysis, macrophage/monocyte differentiation, and HIV-1 challenge are described in SI Materials and Methods. Primer sequences are shown in Tables S4 and S5.
Supplementary Material
Acknowledgments
We thank Lianxing Liu for help with the HIV-1 challenge experiment and Chong Park from the University of California, San Francisco (UCSF) ES core facility for excellent work in the synthesis of the plasmids for TALENs. This work was supported by National Institutes of Health Grants AI 102825 and P01DK088760 and the UCSF Helmut Horten fund.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407473111/-/DCSupplemental.
References
- 1.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: Roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 2.Marmor M, et al. HIV Network for Prevention Trials Vaccine Preparedness Protocol Team Homozygous and heterozygous CCR5-Delta32 genotypes are associated with resistance to HIV infection. J Acquir Immune Defic Syndr. 2001;27(5):472–481. doi: 10.1097/00126334-200108150-00009. [DOI] [PubMed] [Google Scholar]
- 3.Samson M, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382(6593):722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 4.Hütter G, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360(7):692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 5.Anderson J, Akkina R. Complete knockdown of CCR5 by lentiviral vector-expressed siRNAs and protection of transgenic macrophages against HIV-1 infection. Gene Ther. 2007;14(17):1287–1297. doi: 10.1038/sj.gt.3302958. [DOI] [PubMed] [Google Scholar]
- 6.Anderson JS, Walker J, Nolta JA, Bauer G. Specific transduction of HIV-susceptible cells for CCR5 knockdown and resistance to HIV infection: A novel method for targeted gene therapy and intracellular immunization. J Acquir Immune Defic Syndr. 2009;52(2):152–161. doi: 10.1097/QAI.0b013e3181b010a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiGiusto DL, et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med. 2010;2(36):36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holt N, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28(8):839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, et al. Genomic Editing of the HIV-1 Coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther. 2013;21(6):1259–1269. doi: 10.1038/mt.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang M, et al. Inhibition of HIV-1 infection by a unique short hairpin RNA to chemokine receptor 5 delivered into macrophages through hematopoietic progenitor cell transduction. J Gene Med. 2010;12(3):255–265. doi: 10.1002/jgm.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilen CB, et al. Engineering HIV-resistant human CD4+ T cells with CXCR4-specific zinc-finger nucleases. PLoS Pathog. 2011;7(4):e1002020. doi: 10.1371/journal.ppat.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavazzana-Calvo M, et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467(7313):318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woods NB, Bottero V, Schmidt M, von Kalle C, Verma IM. Gene therapy: Therapeutic gene causing lymphoma. Nature. 2006;440(7088):1123. doi: 10.1038/4401123a. [DOI] [PubMed] [Google Scholar]
- 14.Tripathy SK, Black HB, Goldwasser E, Leiden JM. Immune responses to transgene-encoded proteins limit the stability of gene expression after injection of replication-defective adenovirus vectors. Nat Med. 1996;2(5):545–550. doi: 10.1038/nm0596-545. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25(7):778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 17.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 18.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yusa K, et al. Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478(7369):391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye L, et al. Blood cell-derived induced pluripotent stem cells free of reprogramming factors generated by Sendai viral vectors. Stem Cells Transl Med. 2013;2(8):558–566. doi: 10.5966/sctm.2013-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fine EJ, Cradick TJ, Zhao CL, Lin Y, Bao G. An online bioinformatics tool predicts zinc finger and TALE nuclease off-target cleavage. Nucleic Acids Res. 2014;42(6):e42. doi: 10.1093/nar/gkt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyon Y, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8(1):74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 24.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muench MO, Ohkubo T, Smith CA, Suskind DL, Bárcena A. Maintenance of proliferative capacity and retroviral transduction efficiency of human fetal CD38(-/CD34(++) stem cells. Stem Cells Dev. 2006;15(1):97–108. doi: 10.1089/scd.2006.15.97. [DOI] [PubMed] [Google Scholar]
- 26.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451(7175):141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 27.Martinson JJ, Chapman NH, Rees DC, Liu YT, Clegg JB. Global distribution of the CCR5 gene 32-basepair deletion. Nat Genet. 1997;16(1):100–103. doi: 10.1038/ng0597-100. [DOI] [PubMed] [Google Scholar]
- 28.Kambal A, et al. Generation of HIV-1 resistant and functional macrophages from hematopoietic stem cell-derived induced pluripotent stem cells. Mol Ther. 2011;19(3):584–593. doi: 10.1038/mt.2010.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi SM, et al. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology. 2013;57(6):2458–2468. doi: 10.1002/hep.26237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 31.Yang L, et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41(19):9049–9061. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tebas P, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370(10):901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, et al. piggyBac transposase tools for genome engineering. Proc Natl Acad Sci USA. 2013;110(25):E2279–E2287. doi: 10.1073/pnas.1305987110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.