Large assemblages of bacteria are turning up in the strangest of places, but perhaps none is more interesting than the communities that inhabit the human body. The compositions of these microbiota are far from random: only some bacteria are able to thrive in the host environment, and ecological interactions preclude many of all possible combinations of co-occurring species. Because the makeup of a microbial community dictates how it functions, hosts preferentially accommodate those bacterial cohorts that supply properties beneficial to their own health and physiology. Despite these factors, which serve to restrict the composition of bacterial communities, the gut microbiota remains one of the most complex assemblages surveyed to date. The microbiome is dominated by just a few bacterial phyla—mostly Firmicutes and Bacteriodetes, which together typically constitute 90% of the classifiable gut flora (1)—but surveys of human populations have revealed substantial variation among gut microbial communities at finer taxonomic levels. Arumugam et al. (2) proposed that human gut microbiomes can be grouped into multiple classes, termed “enterotypes,” based on the relative abundances of the resident bacterial genera. In PNAS, Wang et al. (3) adopt an experimental approach and provide strong evidence that alternative enterotype structures in the gut are ancient features of mammalian microbiomes driven by dietary variation.
Soon after their discovery, the causes and consequences—and even the existence—of human gut enterotypes came under scrutiny. Originally, the different enterotypes were not found to be associated with age, sex, body mass index, or provenance (2), and some studies failed to recognize the same number of enterotype groupings (often two instead of three) (4). Moreover, it was argued that community compositions might be better described as a continuum rather than clusters (5).
Based on functional profiling, there was an early hint that enterotypes were linked to host diet (2). The enterotype characterized by high frequencies of Prevotella contained genes for thiamine and folate biosynthesis, whereas the enterotype with high frequencies of Bacteroides was enriched in genes that synthesize biotin, riboflavin, pantothenate, and ascorbate. That the enterotypes were specialized for the production of different sets of vitamins suggested connections between enterotypes and host nutrition. This jibed with a previous comparison of children whose diets differed in the amounts of fiber and animal protein and whose gut microbiomes displayed corresponding differences in the frequencies of Prevotella and Bacteroides (6).
To further explore the relationship between diet and enterotype, Wu et al. (4) characterized the gut microbiomes of nearly 100 individuals of known dietary habits and showed that the Bacteroides-enriched enterotype was more common in individuals who regularly consumed protein- and fat-based diets, whereas the Prevotella/Lachnospiraceae-enriched enterotype predominated in individuals who had carbohydrate-based diets. These authors then attempted to elicit changes in host enterotype through controlled feeding experiments; however, none occurred after 10-d dietary treatments. Their failure to induce a single switch suggested that enterotypes were the product of long-term dietary habits.
Enter Wang et al. (3), who provide both retrospective and experimental evidence that the maintenance of enterotypes in host populations is driven by dietary variation. First, they corroborate previous reports of enterotype clusters in laboratory-reared mouse lines (7) and show that wild mice harbor enterotypes that mirror those detected in humans. One of the mouse enterotypes is enriched in Lachnospiraceae and in metabolic pathways that process carbohydrates, and the other enterotype is overrepresented by Bacteroides and by genes involved in the digestion of proteins. Through stable isotope probing of the sampled mice to determine levels of protein and carbohydrate intake, they then demonstrate that mouse enterotypes, also, reflect the dietary histories of their hosts. Mice consuming plant-based diets tended to possess the enterotype enriched in Lachnospiraceae, whereas mice consuming animal-derived foodstuffs more commonly possess the enterotype overrepresented by Bacteroides.
However, the contributions of Wang et al. (3) go beyond correlations between enterotype and diet. They make use of their model system to show experimentally that dietary change can induce enterotype switches within hosts. On capture, eight of the surveyed wild mice were moved to the laboratory and fed identical chow-based diets. Initially, three of the eight contained the Bacteroides-enriched enterotype, whereas the other five contained the Lachnospiraceae-enriched enterotype. Within 1 wk, the microbiomes of each of these newly domesticated mice converged on the same enterotype: that dominated by Lachnospiraceae and specialized for carbohydrate digestion. Over the next 12 wk, enterotypes remained unchanged as hosts continued to consume diets composed primarily of plant material. The rapid acclimation of the gut microbiome to dietary intervention suggests that hosts have the remarkable ability to switch enterotypes in response to fluctuating food abundances.
In addition to elucidating the causes of enterotype clustering in wild mice, Wang et al. (3) take on the task of dispelling some of the controversy regarding the existence of enterotypes. Previous studies have argued that statistical support for enterotypes wanes when certain clustering algorithms and distance metrics are applied (5). Wang et al. show that detection of enterotypes depends on the level at which sequence reads are assigned to taxonomic groups. When reads are assigned to broad, genus-level groupings, as when enterotypes were originally recognized (2), enterotypes are consistently supported regardless of the distance metric or clustering method used. However, when reads are assigned to finer-scale units [e.g., 95% or 97% operational taxonomic units (OTUs) (5)], support for enterotypes becomes contingent on the clustering method and the distance metric used. Thus, enterotypes are most readily identifiable when reads are assigned to known bacterial taxa, not to ad hoc OTU cutoffs.
Finally, the detection of enterotype clusters in wild mice raises new questions about the evolutionary origins of these alternative community configurations. It was recently reported that wild populations of chimpanzees harbor enterotypes that are compositionally analogous to those identified in humans (8), suggesting that enterotypes originated before the divergence of humans and chimpanzees. The discovery by Wang et al. (3) that similarly structured enterotypes are maintained in wild populations of house mice (Fig. 1) suggests that the existence of enterotypic variation predates the diversification of the major orders of mammals, about 100 Mya. These results put forward the tantalizing possibility that enterotypes are features common across mammalian microbiomes; if this turns out to be the case, the enterotype concept may prove an invaluable framework for understanding the evolution of community structure in the gut microbiome.
Fig. 1.
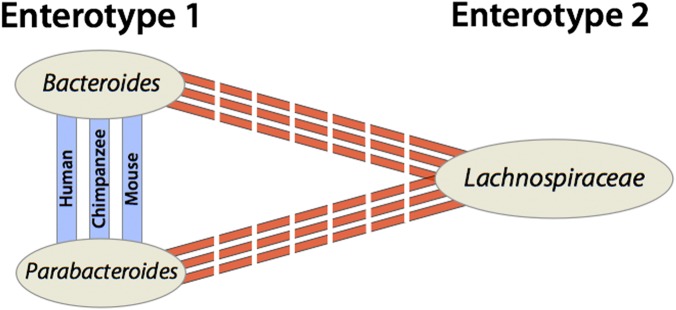
Mice, humans, and chimpanzees contain enterotypes defined by the same bacterial taxa. Colored lines connect bacterial taxa that co-occur within hosts (blue) or that exclude one another (red).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
See companion article on page E2703.
References
- 1.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arumugam M, et al. MetaHIT Consortium Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, et al. Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. Proc Natl Acad Sci USA. 2014;111:E2703–E2710. doi: 10.1073/pnas.1402342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koren O, et al. A guide to enterotypes across the human body: Meta-analysis of microbial community structures in human microbiome datasets. PLOS Comput Biol. 2013;9(1):e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107(33):14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hildebrand F, et al. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013;14(1):R4. doi: 10.1186/gb-2013-14-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moeller AH, et al. Chimpanzees and humans harbour compositionally similar gut enterotypes. Nat Commun. 2012;3:1179. doi: 10.1038/ncomms2159. [DOI] [PMC free article] [PubMed] [Google Scholar]


