Abstract
Non-homologous end joining (NHEJ) is the major pathway for the repair of ionizing radiation induced DNA double strand breaks in human cells. Here, we discuss current insights into the mechanism of NHEJ and the interplay between NHEJ and other pathways for repair of IR-induced DNA damage.
Keywords: Non-homologous end joining, ionizing radiation, DNA double strand break repair
1. Introduction
Non-homologous end joining (NHEJ) is the major pathway for the repair of ionizing radiation (IR)-induced DNA double strand breaks (DSBs) in human cells and is essential for the repair of RAG-induced DSBs during V(D)J recombination [1-3]. Evidence is emerging that aberrant NHEJ is also a major source of genomic rearrangements and chromosomal translocations [4], leading to genomic instability, a hallmark of cancer [5]. Proteins shown to be required for NHEJ and V(D)J recombination in mammalian cells include the Ku70/80 heterodimer, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), Artemis, XRCC4, DNA ligase IV and XLF (XRCC4-like factor, also called Cernunnos). Loss of any of these core NHEJ factors is associated not only with radiation sensitivity but with defective V(D)J recombination and, in animals and patients, immune defects [1, 3, 6-11].
Much of what we know about the mechanism of NHEJ combines results from studies on the cellular responses to IR and other DNA damaging agents as well as studies on the mechanism of V(D)J recombination. V(D)J recombination occurs at discrete sites (recombination signal sequences) in immunoglobulin, T cell receptor and B cell receptor genes, and produces closed DNA hairpin ends (at the coding ends) that upon hairpin opening, processing and rejoining go on to form the coding sequence, and 5′-phosphorylated blunt signal ends that are sealed and lost from the system [3]. In contrast, IR induced lesions are produced stochastically and thought to be dispersed randomly throughout the genome. Also, due to the nature of IR-induced DNA damage, IR-induced DSBs are inherently complex and variable in nature. A characteristic of IR-induced DNA damage is damage or loss of bases, as well as production of DNA single strand breaks (SSBs) that frequently terminate in non-ligatable end groups such as 3′-phosphates, 3′-phosphoglycolates, or 5′-hydroxyl groups [12, 13]. The presence of two SSBs on opposite strands of the DNA approximately 10 bp apart yields a DSB, therefore IR-induced DSBs have short overhanging ends that terminate in non-ligatable end groups that must be processed or removed prior to ligation (reviewed in [13]). Moreover, IR-induced DSBs are frequently surrounded by other DNA lesions, such as base damage, that together make IR-induced DSBs difficult to repair and one of the most cytotoxic forms of DNA damage [13]. Consequently, NHEJ in response to IR-induced DNA damage requires flexibility to respond to complex and variable types of DSBs.
For simplicity, we can think of NHEJ of IR-induced DSBs as proceeding in three distinct stages. First is detection of the DSB by the Ku70/80 heterodimer and tethering of the DNA ends, by the Ku - DNA-PKcs complex. Second, is processing of the damaged DNA termini by a variety of enzymes including polynucleotide kinase phosphatase, PNKP, as well as gap-filling DNA polymerases mu and lambda. Lastly, is rejoining of the DSB ends by the XRCC4-DNA ligase IV complex [2, 14] (Fig. 1). However, apart from DSB detection by Ku, the order of recruitment of the different NHEJ proteins maybe quite flexible, and not all NHEJ components may be required for repair of all lesions [15, 16]. Thus, a model is emerging in which NHEJ may take place within a dynamic, multi-protein complex rather than in a stepwise, more sequential manner in which one component is released before another is recruited. In this review, we will discuss current thinking on the mechanism of NHEJ of IR-induced DSBs and speculate on how NHEJ and other repair pathways interact to repair IR-induced DNA damage throughout the cell cycle.
Figure 1. A model for NHEJ illustrating the central role of the Ku70/80 heterodimer.
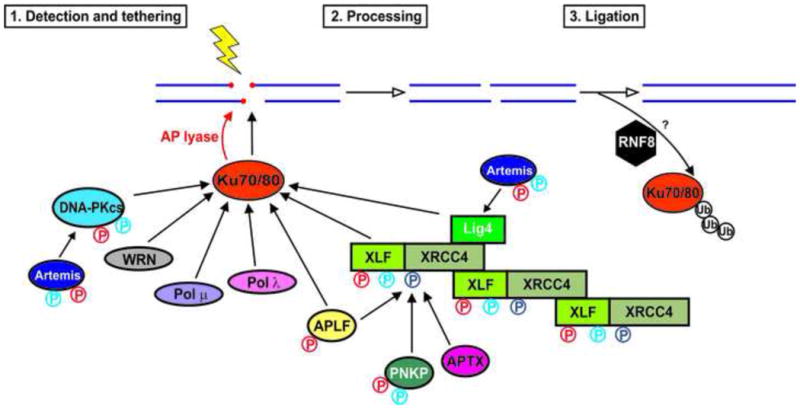
Adapted from [14] with permission. DSBs are detected by the Ku70/80 heterodimer, which interacts with multiple components of the NHEJ machinery, including DNA-PKcs, XLF, WRN and APLF, recruiting them to DSBs. Once the DSB is detected, various enzymes may be involved in processing of the DSB ends, for example, removal of non-ligatable end groups by PNKP, and gap filling by DNA polymerases mu and lambda. The break is resealed by DNA ligase IV, which exists in complex with XRCC4 and XLF. Recent studies suggest that the XRCC4-DNA ligase IV complex is required for DNA-PKcs autophosphorylation [79], suggesting that XRCC4 and DNA ligase IV may be present at the break as a multi-protein complex rather than recruited in the later stages of NHEJ. How Ku is removed from DSBs is unknown, but may involve proteolytic degradation [124-126] or Mre11 nuclease activity [99]. Direct protein-protein interactions are indicated by black lines. The AP lyase activity of Ku [30, 31] is shown by the red arrow. ATM and DNA-PK dependent phosphorylation events are shown in red and blue P symbols respectively. See text and [14] for details.
2. The Ku heterodimer - a multi-functional NHEJ protein
IR-induced DSBs are detected by the abundant, nuclear protein Ku70/80, composed of approximately 70 and 80 kDa subunits, that binds double stranded (ds) DNA ends with high affinity in a largely sequence independent manner [17]. Ku70 and Ku80 form a heterodimer with a preformed ring that encircles ends of dsDNA [18], making it ideally suited for binding to DSB ends. However, in recent years it has become apparent that the Ku70/80 heterodimer plays other critical roles in NHEJ, not only in the initial detection of the DSB but in the recruitment of essentially all of the core NHEJ factors to the DSB. Ku is required for recruitment of DNA-PKcs to DSBs [19] and stimulation of DNA-PKcs kinase activity [20]. Since DNA-PKcs interacts with Artemis [21], Ku may also be responsible, indirectly, for the recruitment of Artemis to DSBs. Ku80 also interacts with XLF, and Ku is required for recruitment of XLF to DSBs in vivo [22, 23]. Moreover, Ku70 interacts with XRCC4 and laser micro-irradiation studies suggest that Ku enhances retention of XRCC4 at DSBs [24]. In addition, Ku80 interacts with APLF (aprataxin and PNKP-like factor, also called PALF for PNKP and aprataxin-like factor), which enhances NHEJ, stimulating DSB end ligation by DNA ligase IV [25, 26]. Ku also interacts with the Werner's syndrome helicase [27] as well as with DNA polymerase mu and lambda [28, 29] suggesting roles in the recruitment or retention of additional end-processing factors at the break (Fig. 1).
3. Processing complex DNA damage at DSB ends
As discussed above, IR induced DSBs are considered complex, frequently containing base damage close to the DSB as well as short DNA overhangs and non-ligatable DNA termini [12, 13]. Accordingly, several enzymes have been implicated in DSB end-processing in NHEJ, but which enzymes are involved likely depends on the nature of the DNA damage incurred. In addition to its critical roles in DSB detection and recruitment of other NHEJ factors, Ku has recently been shown to possess 5′-RP/AP lyase activity, suggesting that it also plays a catalytic role in NHEJ, modifying damaged bases proximal to the DNA ends [30, 31]. PNKP has 3′-DNA phosphatase and 5′- DNA kinase activities, making it an ideal candidate for removing IR-induced non-ligatable DNA end groups at DSB termini [32]. PNKP interacts with scaffolding protein XRCC4 in a phosphorylation-dependent manner involving the N-terminal fork-head associated (FHA) domain of PNKP and CK2-Thr233 phosphorylated XRCC4 suggesting a mechanism by which it is recruited to DSBs [33]. The FHA domain of PNKP also interacts with CK2-phosphorylated XRCC1, conferring roles in repair of IR-induced SSBs as well as DSBs [34]. Accordingly, PNKP null cells are radiation sensitive, consistent with a role in repair of IR-induced strand breaks [35].
Recently, two other proteins have been identified that bear a PNKP-like FHA domain: APLF, and aprataxin (APTX). Like PNKP, APLF interacts with CK2-phosphorylated XRCC4 and XRCC1 via its N-terminal FHA domain [36, 37]. APLF also interacts directly with Ku80, through its central domain [25, 26] and enhances NHEJ and DNA ligase IV mediated ligation in NHEJ [25, 38]. Moreover, APLF has been reported to have nuclease activity, suggesting possible roles in DSB end processing [36, 39]. APLF also interacts with poly-ADP ribose and histones, providing links between NHEJ and chromatin [40, 41].
In contrast, APTX removes AMP from the 5′-termini of DNA breaks that result from abortive ligation events [42]. Like PNKP and APLF, APTX interacts with phosphorylated XRCC4 and XRCC1 [43]. Absence of APTX confers defects in DNA SSB repair and is associated with the neurological condition, ataxia with oculomotor apraxia 1 (AOA1) [44], however, to date, no defects in DSB repair have been reported (reviewed in [45]). Factors that regulate the ability of phosphorylated XRCC4 (and XRCC1) to interact with APLF, APTX and PNKP remain to be elucidated.
Additional end processing in NHEJ is carried out by DNA polymerases mu and lambda (and TdT in V(D)J recombination) which interact with Ku and the XRCC4-DNA ligase IV complex and perform gap filling roles [29]. Artemis, a structure specific endonuclease interacts with DNA-PKcs and plays a critical role in opening DNA hairpin coding ends in V(D)J recombination [21, 46]. Artemis was originally suggested to have exonuclease activity [21], but this has recently been questioned [47]. Artemis null cells are mildly radiation sensitive suggesting that it is required for repair of a subset of IR-induced DSBs [48], however, its DNA substrates in the DNA damage response are not known.
V(D)J recombination assays reveal that DNA-PKcs phosphorylation plays an important role in regulating the access of nucleases to DNA ends prior to ligation. DNA-PKcs is recruited to DSBs by Ku, and the protein kinase activity of DNA-PKcs is required for NHEJ [49, 50]. DNA-PKcs phosphorylates multiple NHEJ proteins in vitro [2, 51-53] and autophosphorylates on multiple sites in vitro [54]. DNA-PKcs is also highly phosphorylated in vivo (reviewed in [55]). The most well characterized phosphorylation sites in DNA-PKcs, located between threonines 2609 and 2647 are termed the ABCDE cluster [56, 57]. These sites are phosphorylated in a DNA-PK-dependent manner in vivo (consistent with autophosphorylation) [58, 59], but can also be phosphorylated by other members of the phosphatidylinositol-3 kinase-like protein kinase (PIKK) family, ataxia telangiectasia mutated (ATM) and ATM, Rad3-related (ATR) (reviewed in [2, 53, 55]). Ablation of three of the ABCDE sites in mice (corresponding to threonines 2609, 2638 and 2647 in human DNA-PKcs) results in severe bone marrow failure and lethality [60]. DNA-PKcs also autophosphorylates on serine 2056, located in the PQR cluster, in response to DNA damage [58, 59]. Ablation of ABCDE and PQR phosphorylation sites within DNA-PKcs affects the length of DSB end termini in a reciprocal manner [61] suggesting that phosphorylation of different sites within DNA-PKcs regulates accessibility of the DNA termini to nucleases (reviewed in [2, 53, 62]). DNA-PKcs phosphorylation has also been shown to play an important role in regulating homologous recombination repair (HRR) [63, 64] (discussed in more detail below). DNA-PKcs dependent regulation of both end processing and pathway choice has been proposed to occur through phosphorylation-dependent conformational changes that result in dissociation of DNA-PKcs from the DNA-Ku complex [55, 65]. Interestingly, a recent study suggests that DNA-PKcs is only required for the repair of complex DSBs i.e. DSBs which are surrounded by additional DNA lesions [66], consistent with the theme of flexibility within the NHEJ pathway. Also, DNA-PKcs is not found in yeast or in most lower multicellular eukaryotes (with the exception of Dictyostelium and possibly Ciona [67]), consistent with it being required for repair of a subset of breaks, and /or those in the genomes of a subset of eukaryotes.
4. Sealing the break
Once DNA ends have been processed, they are covalently joined by DNA ligase IV, which exists in complex with XRCC4, which, in turn, interacts with XLF [68]. The XRCC4 homodimer interacts with the BRCT domains of DNA ligase IV with high affinity via its coiled coil stalk domain [69, 70] and, with lower affinity, with the XLF homodimer [71]. XRCC4 homodimers can also interact with themselves, to form higher order complexes, and with dsDNA (reviewed in [72]). XRCC4 is required for stabilization of DNA ligase IV and stimulates its activity [73]. In vitro studies suggest that XLF is only required for repair at a subset of breaks, involving mismatched or non-cohesive ends [74]. XLF also stimulates the re-adenylation activity of DNA ligase IV, which is essential for re-charging DNA ligase IV, allowing a new catalytic cycle [71]. In vitro studies reveal that XRCC4 and XLF interact via their head domains to form long helical protein filaments that may be involved in bridging DNA ends or holding DNA ends in an orientation that facilitates ligation [75-78]. How DNA ligase IV and other XRCC4 interacting proteins such as PNKP and APLF have direct access to DSB ends within the context of XRCC4-XLF filaments remains to be determined (reviewed in [72]).
5. A Ku-dependent NHEJ complex?
Given the requirement of DNA ligase IV for ligation of the DNA, it has long been assumed that the XRCC4-DNA ligase IV complex functions in the final stage of NHEJ, however recent studies have revealed that DNA ligase IV deficient cells are unable to support phosphorylation of DNA-PKcs, suggesting that DNA-ligase IV is required for DNA-PKcs' activation and/or the function of DNA-PKcs at DSB ends [79]. These findings suggest that the XRCC4-DNA ligase IV complex assembles with Ku and DNA-PKcs at DSBs, which in turn suggests that Artemis (recruited by DNA-PKcs), XLF and APLF (recruited by Ku and XRCC4) and PNKP (thought to be recruited by XRCC4) may also be recruited to the DSB to form a multi-protein complex. DNA-ligase IV (which interacts with XRCC4 and is recruited to Ku at DSBs) also interacts with Artemis (which in turn interacts with DNA-PKcs) and this interaction is required for V(D)J recombination [80, 81]. Thus, a picture is emerging of multiple proteins, recruited as needed, in a Ku-dependent manner to the DSB and possibly stabilized by multiple additional protein-protein and protein-DNA interactions.
This model then raises further questions regarding how individual proteins within this putative complex would gain access to the actual DSB ends. One possibility is that DNA-PKcs-mediated autophosphorylation, phosphorylation of other proteins and resulting conformational changes remodel the NHEJ complex, regulating appropriate and timely access of NHEJ proteins to the DNA ends [53, 62]. It is also worth noting that DNA damage-induced phosphorylation of several NHEJ proteins, including Artemis [82-84], XLF [85], PNKP [86, 87], APLF [88-90] as well as some phosphorylation sites on DNA-PKcs [91, 92] requires ATM rather than DNA-PKcs. Since ATM is required for NHEJ at complex lesions and/or at DSBs in heterochromatic regions [93, 94], this suggests that ATM-dependent phosphorylation of NHEJ proteins may regulate NHEJ at a subset of DSBs.
6. Competition between NHEJ and other DNA repair and DNA damage response pathways
NHEJ is active throughout the cell cycle, whereas repair of DSBs via HRR repair occurs only in late S and G2 when a sister chromatid is available for accurate, template-mediated repair [95]. These observations raise important questions regarding pathway choice: specifically, how Ku and the MRN complex (composed of Mre11, Rad50 and Nbs1), compete for detection of DSBs in different stages of the cell cycle. For example, DSB detection by Ku would channel repair into NHEJ in all stages of the cell cycle, whereas, DSB detection by MRN channels repair into HRR in G2 but additionally, results in activation of ATM throughout the cell cycle (Fig. 2). Initiation of HRR requires the generation of long regions of ssDNA that are detected initially by RPA and subsequently bound by Rad51 to allow strand invasion [96]. Therefore, factors that control cell cycle associated DNA end resection are critical to the choice between NHEJ and HRR in G2 (discussed in detail in [97] and [98]). Interestingly, the nuclease activity of Mre11 is critical for end resection and initiation of HRR but not for activation of ATM [99, 100] revealing distinct structural and catalytic roles for the MRN complex in regulating pathway choice.
Figure 2. A simplified model for the cellular response to IR-induced DNA damage.
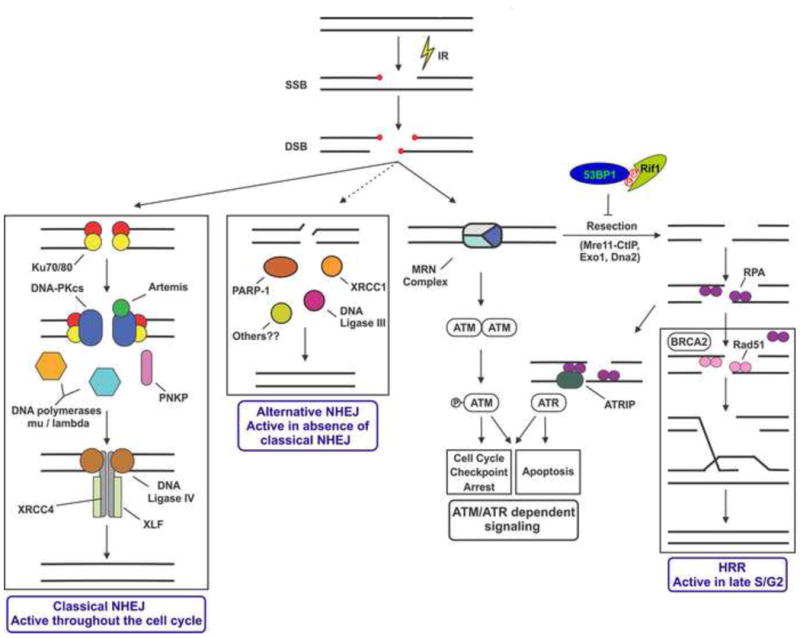
Adapted from [14] with permission. IR induces base damage and SSBs. Two SSBs on opposite strands of the DNA induce a DSB. Damaged, non-ligatable termini are represented by the small red circles. The DSB can be detected by the Ku heterodimer, initiating NHEJ, the major pathway for the repair of IR-induced DSBs, which is active in all stages of the cell cycle. In the absence of classical NHEJ, an alternative or back-up NHEJ pathway, that involves PARP1, XRCC1 and DNA ligases I or III may be initiated [127]. DSBs are detected by the Mre11, Rad50, Nbs1 (MRN) complex which leads to activation of the ATM protein kinase and downstream signaling events that induce cell cycle checkpoint arrest and other cellular responses to DSBs [128]. The MRN complex is required for 5′-3′ resection, likely involving Mre11, CtIP, Exo1, Dna2 and other proteins, which produces long 3′ ends that are recognized by ssDNA-binding protein, RPA. The 53BP1/RIF complex promotes NHEJ by blocking DSB ends from resection [102-106], but the mechanism by which it does so is poorly understood. RPA is displaced by BRCA2, allowing binding of Rad51 and initiation of homologous recombination repair (HRR), which is active only in late S and G2 phases of the cell cycle [96, 129]. Recruitment of RPA also initiates activation of ATR, which like ATM, contributes to activation of cell cycle checkpoints and other cellular responses [130]. Precisely how Ku and MRN compete or cooperate to initiate NHEJ, HRR and activation of ATM is an area of active investigation. See text for details.
Recent studies suggest that DNA-PKcs and Ku play important roles in regulating the balance between NHEJ and HRR in cells. Loss of either Ku or DNA-PKcs results in enhanced DNA end resection in G2 phase cells, which consequently promotes repair by HR [63]. Moreover, inability of DNA-PKcs to undergo autophosphorylation at the ABCDE cluster causes delay in Rad51 foci formation [63]. One explanation for these observations is that abrogation of DNA-PKcs' ability to undergo autophosphorylation at the ABCDE sites prevents or delays its release from DSB ends, thus delaying initiation of HRR [63]. From these studies, Jeggo and colleagues suggest that NHEJ attempts to repair IR-induced DSBs in all stages of the cell cycle. If successful, the DSB is repaired rapidly. In G2, if NHEJ is unsuccessful, resection ensues, committing the cell to HRR [63, 97].
In vitro studies from the Paull laboratory demonstrate that the presence of DNA-PKcs blocks Exonuclease 1 (Exo1) activity and that DNA-PKcs' autophosphorylation or phosphorylation of DNA-PKcs by ATM, is required to relieve inhibition of Exo1 by DNA-PKcs [101], again consistent with autophosphorylation of DNA-PKcs being required for its release from DNA ends and subsequent pathway progression [55]. Interestingly, the authors also showed that the MRN complex promotes DNA-PKcs autophosphorylation in vitro [101], raising the interesting possibility that MRN may facilitate removal of DNA-PKcs from DNA ends, to facilitate HRR.
Another significant recent development has been the generation by the Tainer laboratory of small molecule inhibitors specific for Mre11 endo- or exonuclease activities [99]. Mre11 nuclease activity is critical for initiation of HRR but not for activation of ATM [100]. Through the use of Mre11 endo- and exo-nuclease specific inhibitors, Tainer and Jeggo propose a two-step process in which the endonuclease activity of Mre11 first introduces a nick in the 5′ strand, upstream of the DSB, which is followed by bi-directional resection utilizing Mre11 3′-5′ exonuclease activity in concert with the 5′-3′ exonuclease activity of the Exo1/BLM complex [99].
Another important regulator of pathway choice is 53BP1, which, with its interacting partner Rif1, acts to promote NHEJ in G1 by protecting DSB ends from DNA end-resection [102-106] (Fig. 2). Elucidation of how 53BP1 carries out this important function however, the mechanism by which is achieves this is unclear. One of the obstacles to our ability to address pathway choice has been the inability to detect Ku at DSBs in cells. The recent development of methods to visualize individual Ku heterodimers at DSB termini may afford new ways to directly address the relative positioning and timing of Ku, MRN, 53BP1 and other DNA damage sensing proteins at DSBs in cells [107].
Since IR induces damage to DNA bases as well as DNA single strand breaks, much also remains to be learned about how NHEJ interfaces with base excision repair and DNA single strand break repair pathways as well as alternative NHEJ pathways [108, 109]. Since DSBs are regarded to be more toxic than single strand breaks or base lesions, it seems probable that DSBs would be repaired first with high priority, whereas repair of base lesions and single strand breaks in proximity to the DSB may be repaired more slowly, once the DSB ends have been ligated. Interestingly, however, base damage upstream of a DSB has been shown to impair NHEJ [110], thus IR-induced base damage may directly impinge on the ability NHEJ to repair IR-induced DSBs. Unraveling the complexities of pathway choice will clearly continue to be a major focus for future investigations.
7. Concluding remarks
It has been over twenty years since DNA-PKcs was identified [111, 112] and Ku and DNA-PKcs were shown to form the DNA-PK complex [20, 113, 114]. Evidence for their involvement in DSB repair quickly followed [115-118]. The discovery of additional components of the NHEJ pathway, such as XRCC4 [119], the XRCC4-DNA ligase IV complex [120], Artemis [48] and XLF [68, 121] followed and our understanding of the mechanisms of DSB repair in eukaryotes as well as V(D)J recombination has increased dramatically in past decades (reviewed in [3, 122]). Recent findings, described here, have begun to reveal how these proteins interact with each other and with their target, damaged DNA. Significant progress is also being made in elucidating the structural biology of individual NHEJ components, and pictures of NHEJ interactions in multi-protein-DNA complexes is beginning to emerge (reviewed in [123]). As we embark upon the third decade of research into NHEJ, there is still much to learn about how the NHEJ complex is regulated, how it interfaces with chromatin and other DNA damage response pathways and how these pathways can be targeted for therapeutic advantage in cancer therapy.
Acknowledgments
Work in the author's laboratory is supported by the Canadian Institutes of Health Research (MOP13639) and the National Institutes of Health Program Project Grant (P01-CA92584). SPLM is a Scientist of Alberta Innovates Health Solutions, and holds a Killam Annual Professorship and the Engineered Air Chair in Cancer Research.
Footnotes
Conflict of interest: The authors declare there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helmink BA, Sleckman BP. The response to and repair of RAG-mediated DNA double-strand breaks. Annu Rev Immunol. 2012;30:175–202. doi: 10.1146/annurev-immunol-030409-101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunting SF, Nussenzweig A. End-joining, translocations and cancer. Nat Rev Cancer. 2013;13:443–454. doi: 10.1038/nrc3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Zha S, Guo C, Boboila C, Oksenych V, Cheng HL, Zhang Y, Wesemann DR, Yuen G, Patel H, Goff PH, Dubois RL, Alt FW. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature. 2011;469:250–254. doi: 10.1038/nature09604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Burg M, van Dongen JJ, van Gent DC. DNA-PKcs deficiency in human: long predicted, finally found. Curr Opin Allergy Clin Immunol. 2009;9:503–509. doi: 10.1097/ACI.0b013e3283327e41. [DOI] [PubMed] [Google Scholar]
- 8.Woodbine L, Neal JA, Sasi NK, Shimada M, Deem K, Coleman H, Dobyns WB, Ogi T, Meek K, Davies EG, Jeggo PA. PRKDC mutations in a SCID patient with profound neurological abnormalities. J Clin Invest. 2013;123:2969–2980. doi: 10.1172/JCI67349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee PP, Woodbine L, Gilmour KC, Bibi S, Cale CM, Amrolia PJ, Veys PA, Davies EG, Jeggo PA, Jones A. The many faces of Artemis-deficient combined immunodeficiency - Two patients with DCLRE1C mutations and a systematic literature review of genotype-phenotype correlation. Clin Immunol. 2013;149:464–474. doi: 10.1016/j.clim.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Girard PM, Kysela B, Harer CJ, Doherty AJ, Jeggo PA. Analysis of DNA ligase IV mutations found in LIG4 syndrome patients: the impact of two linked polymorphisms. Hum Mol Genet. 2004;13:2369–2376. doi: 10.1093/hmg/ddh274. [DOI] [PubMed] [Google Scholar]
- 11.Nijnik A, Dawson S, Crockford TL, Woodbine L, Visetnoi S, Bennett S, Jones M, Turner GD, Jeggo PA, Goodnow CC, Cornall RJ. Impaired lymphocyte development and antibody class switching and increased malignancy in a murine model of DNA ligase IV syndrome. J Clin Invest. 2009;119:1696–1705. doi: 10.1172/JCI32743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cadet J, Ravanat JL, TavernaPorro M, Menoni H, Angelov D. Oxidatively generated complex DNA damage: tandem and clustered lesions. Cancer Lett. 2012;327:5–15. doi: 10.1016/j.canlet.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Georgakilas AG, O'Neill P, Stewart RD. Induction and repair of clustered DNA lesions: what do we know so far? Radiat Res. 2013;180:100–109. doi: 10.1667/RR3041.1. [DOI] [PubMed] [Google Scholar]
- 14.Wang C, Lees-Miller SP. Detection and Repair of Ionizing Radiation-Induced DNA Double Strand Breaks: New Developments in Nonhomologous End Joining. Int J Radiat Oncol Biol Phys. 2013;86:440–449. doi: 10.1016/j.ijrobp.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yano K, Chen DJ. Live cell imaging of XLF and XRCC4 reveals a novel view of protein assembly in the non-homologous end-joining pathway. Cell Cycle. 2008;7:1321–1325. doi: 10.4161/cc.7.10.5898. [DOI] [PubMed] [Google Scholar]
- 16.Gu J, Lieber MR. Mechanistic flexibility as a conserved theme across 3 billion years of nonhomologous DNA end-joining. Genes Dev. 2008;22:411–415. doi: 10.1101/gad.1646608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat Rev Mol Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 18.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 19.Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 21.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 22.Yano KI, Morotomi-Yano K, Lee KJ, Chen DJ. Functional significance of the interaction with Ku in DNA double-strand break recognition of XLF. FEBS Lett. 2011;585:841–846. doi: 10.1016/j.febslet.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008;9:91–96. doi: 10.1038/sj.embor.7401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, Houtsmuller AB, van Gent DC. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci U S A. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grundy GJ, Rulten SL, Zeng Z, Arribas-Bosacoma R, Iles N, Manley K, Oliver A, Caldecott KW. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 2013;32:112–125. doi: 10.1038/emboj.2012.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirodkar P, Fenton AL, Meng L, Koch CA. Identification and functional characterization of a Ku-binding motif in aprataxin polynucleotide kinase/phosphatase-like factor (APLF) J Biol Chem. 2013;288:19604–19613. doi: 10.1074/jbc.M112.440388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orren DK, Machwe A, Karmakar P, Piotrowski J, Cooper MP, Bohr VA. A functional interaction of Ku with Werner exonuclease facilitates digestion of damaged DNA. Nucleic Acids Res. 2001;29:1926–1934. doi: 10.1093/nar/29.9.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol Cell Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramsden DA, Asagoshi K. DNA polymerases in nonhomologous end joining: are there any benefits to standing out from the crowd? Environ Mol Mutagen. 2012;53:741–751. doi: 10.1002/em.21725. [DOI] [PubMed] [Google Scholar]
- 30.Roberts SA, Strande N, Burkhalter MD, Strom C, Havener JM, Hasty P, Ramsden DA. Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010;464:1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strande N, Roberts SA, Oh S, Hendrickson EA, Ramsden DA. Specificity of the dRP/AP lyase of Ku promotes nonhomologous end joining (NHEJ) fidelity at damaged ends. J Biol Chem. 2012;287:13686–13693. doi: 10.1074/jbc.M111.329730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JN. Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem Sci. 2011;36:262–271. doi: 10.1016/j.tibs.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koch CA, Agyei R, Galicia S, Metalnikov P, O'Donnell P, Starostine A, Weinfeld M, Durocher D. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. Embo J. 2004;23:3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, Weinfeld M, Caldecott KW. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 35.Rasouli-Nia A, Karimi-Busheri F, Weinfeld M. Stable down-regulation of human polynucleotide kinase enhances spontaneous mutation frequency and sensitizes cells to genotoxic agents. Proc Natl Acad Sci U S A. 2004;101:6905–6910. doi: 10.1073/pnas.0400099101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanno S, Kuzuoka H, Sasao S, Hong Z, Lan L, Nakajima S, Yasui A. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. Embo J. 2007;26:2094–2103. doi: 10.1038/sj.emboj.7601663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iles N, Rulten S, El-Khamisy SF, Caldecott KW. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol Cell Biol. 2007;27:3793–3803. doi: 10.1128/MCB.02269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rulten SL, Fisher AE, Robert I, Zuma MC, Rouleau M, Ju L, Poirier G, Reina-San-Martin B, Caldecott KW. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol Cell. 2011;41:33–45. doi: 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 39.Li S, Kanno S, Watanabe R, Ogiwara H, Kohno T, Watanabe G, Yasui A, Lieber MR. Polynucleotide kinase and aprataxin-like forkhead-associated protein (PALF) acts as both a single-stranded DNA endonuclease and a single-stranded DNA 3′ exonuclease and can participate in DNA end joining in a biochemical system. J Biol Chem. 2011;286:36368–36377. doi: 10.1074/jbc.M111.287797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 41.Mehrotra PV, Ahel D, Ryan DP, Weston R, Wiechens N, Kraehenbuehl R, Owen-Hughes T, Ahel I. DNA repair factor APLF is a histone chaperone. Mol Cell. 2011;41:46–55. doi: 10.1016/j.molcel.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 43.Clements PM, Breslin C, Deeks ED, Byrd PJ, Ju L, Bieganowski P, Brenner C, Moreira MC, Taylor AM, Caldecott KW. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair (Amst) 2004;3:1493–1502. doi: 10.1016/j.dnarep.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 44.Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, Mendonca P, Costa M, Barros J, Yanagisawa T, Watanabe M, Ikeda Y, Aoki M, Nagata T, Coutinho P, Sequeiros J, Koenig M. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 45.Rulten SL, Caldecott KW. DNA strand break repair and neurodegeneration. DNA Repair (Amst) 2013;12:558–567. doi: 10.1016/j.dnarep.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 46.Ma Y, Schwarz K, Lieber MR. The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair (Amst) 2005;4:845–851. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 47.Pawelczak KS, Turchi JJ. Purification and characterization of exonuclease-free Artemis: Implications for DNA-PK-dependent processing of DNA termini in NHEJ-catalyzed DSB repair. DNA Repair (Amst) 2010;9:670–677. doi: 10.1016/j.dnarep.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, Fischer A, de Villartay JP. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 49.Kurimasa A, Kumano S, Boubnov NV, Story MD, Tung CS, Peterson SR, Chen DJ. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol Cell Biol. 1999;19:3877–3884. doi: 10.1128/mcb.19.5.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kienker LJ, Shin EK, Meek K. Both V(D)J recombination and radioresistance require DNA-PK kinase activity, though minimal levels suffice for V(D)J recombination. Nucleic Acids Res. 2000;28:2752–2761. doi: 10.1093/nar/28.14.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lees-Miller SP, Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 52.Meek K, Gupta S, Ramsden DA, Lees-Miller SP. The DNA-dependent protein kinase: the director at the end. Immunol Rev. 2004;200:132–141. doi: 10.1111/j.0105-2896.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- 53.Meek K, Dang V, Lees-Miller SP. DNA-PK: the means to justify the ends? Adv Immunol. 2008;99:33–58. doi: 10.1016/S0065-2776(08)00602-0. [DOI] [PubMed] [Google Scholar]
- 54.Douglas P, Sapkota GP, Morrice N, Yu Y, Goodarzi AA, Merkle D, Meek K, Alessi DR, Lees-Miller SP. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem J. 2002;368:243–251. doi: 10.1042/BJ20020973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobbs TA, Tainer JA, Lees-Miller SP. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst) 2010;9:1307–1314. doi: 10.1016/j.dnarep.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, Lees-Miller SP, Meek K. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol Cell Biol. 2003;23:5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, Chen DJ. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, Botvinick E, Qin J, Chen DJ. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem. 2005;280:14709–14715. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- 59.Douglas P, Cui X, Block WD, Yu Y, Gupta S, Ding Q, Ye R, Morrice N, Lees-Miller SP, Meek K. The DNA-dependent protein kinase catalytic subunit (DNA-PKcs) is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Mol Cell Biol. 2007;27:1581–1591. doi: 10.1128/MCB.01962-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang S, Yajima H, Huynh H, Zheng J, Callen E, Chen HT, Wong N, Bunting S, Lin YF, Li M, Lee KJ, Story M, Gapud E, Sleckman BP, Nussenzweig A, Zhang CC, Chen DJ, Chen BP. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J Cell Biol. 2011;193:295–305. doi: 10.1083/jcb.201009074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cui X, Yu Y, Gupta S, Cho YM, Lees-Miller SP, Meek K. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol Cell Biol. 2005;25:10842–10852. doi: 10.1128/MCB.25.24.10842-10852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neal JA, Meek K. Choosing the right path: does DNA-PK help make the decision? Mutat Res. 2011;711:73–86. doi: 10.1016/j.mrfmmm.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shibata A, Conrad S, Birraux J, Geuting V, Barton O, Ismail A, Kakarougkas A, Meek K, Taucher-Scholz G, Lobrich M, Jeggo PA. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 2011;30:1079–1092. doi: 10.1038/emboj.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neal JA, Dang V, Douglas P, Wold MS, Lees-Miller SP, Meek K. Inhibition of homologous recombination by DNA-dependent protein kinase requires kinase activity, is titratable, and is modulated by autophosphorylation. Mol Cell Biol. 2011;31:1719–1733. doi: 10.1128/MCB.01298-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, So S, Abolfath RM, Chen DJ, Lees-Miller SP, Tainer JA. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem. 2010;285:1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reynolds P, Anderson JA, Harper JV, Hill MA, Botchway SW, Parker AW, O'Neill P. The dynamics of Ku70/80 and DNA-PKcs at DSBs induced by ionizing radiation is dependent on the complexity of damage. Nucleic Acids Res. 2012;40:10821–10831. doi: 10.1093/nar/gks879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Block WD, Lees-Miller SP. Putative homologues of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and other components of the non-homologous end joining machinery in Dictyostelium discoideum. DNA Repair (Amst) 2005;4:1061–1065. doi: 10.1016/j.dnarep.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 68.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 69.Sibanda BL, Critchlow SE, Begun J, Pei XY, Jackson SP, Blundell TL, Pellegrini L. Crystal structure of an Xrcc4-DNA ligase IV complex. Nat Struct Biol. 2001;8:1015–1019. doi: 10.1038/nsb725. [DOI] [PubMed] [Google Scholar]
- 70.Wu PY, Frit P, Meesala S, Dauvillier S, Modesti M, Andres SN, Huang Y, Sekiguchi J, Calsou P, Salles B, Junop MS. Structural and functional interaction between the human DNA repair proteins DNA ligase IV and XRCC4. Mol Cell Biol. 2009;29:3163–3172. doi: 10.1128/MCB.01895-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riballo E, Woodbine L, Stiff T, Walker SA, Goodarzi AA, Jeggo PA. XLF-Cernunnos promotes DNA ligase IV-XRCC4 re-adenylation following ligation. Nucleic Acids Res. 2009;37:482–492. doi: 10.1093/nar/gkn957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mahaney BL, Hammel M, Meek K, Tainer JA, Lees-Miller SP. XRCC4 and XLF form long helical protein filaments suitable for DNA end protection and alignment to facilitate DNA double strand break repair. Biochem Cell Biol. 2013;91:31–41. doi: 10.1139/bcb-2012-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 74.Tsai CJ, Kim SA, Chu G. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc Natl Acad Sci U S A. 2007;104:7851–7856. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hammel M, Rey M, Yu Y, Mani RS, Classen S, Liu M, Pique ME, Fang S, Mahaney B, Weinfeld M, Schriemer DC, Lees-Miller SP, Tainer JA. XRCC4 interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double-strand break repair. J Biol Chem. 2011;286:32638–32650. doi: 10.1074/jbc.M111.272641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Andres SN, Vergnes A, Ristic D, Wyman C, Modesti M, Junop M. A human XRCC4-XLF complex bridges DNA. Nucleic Acids Res. 2012;40:1868–1878. doi: 10.1093/nar/gks022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu Q, Ochi T, Matak-Vinkovic D, Robinson CV, Chirgadze DY, Blundell TL. Non-homologous end-joining partners in a helical dance: structural studies of XLF-XRCC4 interactions. Biochem Soc Trans. 2011;39:1387–1392. doi: 10.1042/BST0391387. suppl 1382 p following 1392. [DOI] [PubMed] [Google Scholar]
- 78.Ropars V, Drevet P, Legrand P, Baconnais S, Amram J, Faure G, Marquez JA, Pietrement O, Guerois R, Callebaut I, Le Cam E, Revy P, de Villartay JP, Charbonnier JB. Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining. Proc Natl Acad Sci U S A. 2011;108:12663–12668. doi: 10.1073/pnas.1100758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cottarel J, Frit P, Bombarde O, Salles B, Negrel A, Bernard S, Jeggo PA, Lieber MR, Modesti M, Calsou P. A noncatalytic function of the ligation complex during nonhomologous end joining. J Cell Biol. 2013;200:173–186. doi: 10.1083/jcb.201203128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Ioannes P, Malu S, Cortes P, Aggarwal AK. Structural basis of DNA ligase IV-Artemis interaction in nonhomologous end-joining. Cell Rep. 2012;2:1505–1512. doi: 10.1016/j.celrep.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Malu S, De Ioannes P, Kozlov M, Greene M, Francis D, Hanna M, Pena J, Escalante CR, Kurosawa A, Erdjument-Bromage H, Tempst P, Adachi N, Vezzoni P, Villa A, Aggarwal AK, Cortes P. Artemis C-terminal region facilitates V(D)J recombination through its interactions with DNA Ligase IV and DNA-PKcs. J Exp Med. 2012;209:955–963. doi: 10.1084/jem.20111437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Poinsignon C, de Chasseval R, Soubeyrand S, Moshous D, Fischer A, Hache RJ, de Villartay JP. Phosphorylation of Artemis following irradiation-induced DNA damage. Eur J Immunol. 2004;34:3146–3155. doi: 10.1002/eji.200425455. [DOI] [PubMed] [Google Scholar]
- 83.Zhang X, Succi J, Feng Z, Prithivirajsingh S, Story MD, Legerski RJ. Artemis is a phosphorylation target of ATM and ATR and is involved in the G2/M DNA damage checkpoint response. Mol Cell Biol. 2004;24:9207–9220. doi: 10.1128/MCB.24.20.9207-9220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen L, Morio T, Minegishi Y, Nakada S, Nagasawa M, Komatsu K, Chessa L, Villa A, Lecis D, Delia D, Mizutani S. Ataxia-telangiectasia-mutated dependent phosphorylation of Artemis in response to DNA damage. Cancer Sci. 2005;96:134–141. doi: 10.1111/j.1349-7006.2005.00019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu Y, Mahaney BL, Yano K, Ye R, Fang S, Douglas P, Chen DJ, Lees-Miller SP. DNA-PK and ATM phosphorylation sites in XLF/Cernunnos are not required for repair of DNA double strand breaks. DNA Repair (Amst) 2008;7:1680–1692. doi: 10.1016/j.dnarep.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zolner AE, Abdou I, Ye R, Mani RS, Fanta M, Yu Y, Douglas P, Tahbaz N, Fang S, Dobbs T, Wang C, Morrice N, Hendzel MJ, Weinfeld M, Lees-Miller SP. Phosphorylation of polynucleotide kinase/phosphatase by DNA-dependent protein kinase and ataxia-telangiectasia mutated regulates its association with sites of DNA damage. Nucleic Acids Res. 2011;39:9224–9237. doi: 10.1093/nar/gkr647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Segal-Raz H, Mass G, Baranes-Bachar K, Lerenthal Y, Wang SY, Chung YM, Ziv-Lehrman S, Strom CE, Helleday T, Hu MC, Chen DJ, Shiloh Y. ATM-mediated phosphorylation of polynucleotide kinase/phosphatase is required for effective DNA double-strand break repair. EMBO Rep. 2011;12:713–719. doi: 10.1038/embor.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fenton AL, Shirodkar P, Macrae CJ, Meng L, Koch CA. The PARP3- and ATM-dependent phosphorylation of APLF facilitates DNA double-strand break repair. Nucleic Acids Res. 2013;41:4080–4092. doi: 10.1093/nar/gkt134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Macrae CJ, McCulloch RD, Ylanko J, Durocher D, Koch CA. APLF (C2orf13) facilitates nonhomologous end-joining and undergoes ATM-dependent hyperphosphorylation following ionizing radiation. DNA Repair (Amst) 2008;7:292–302. doi: 10.1016/j.dnarep.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 90.Bekker-Jensen S, Fugger K, Danielsen JR, Gromova I, Sehested M, Celis J, Bartek J, Lukas J, Mailand N. Human Xip1 (C2orf13) is a novel regulator of cellular responses to DNA strand breaks. J Biol Chem. 2007;282:19638–19643. doi: 10.1074/jbc.C700060200. [DOI] [PubMed] [Google Scholar]
- 91.Lee BS, Gapud EJ, Zhang S, Dorsett Y, Bredemeyer A, George R, Callen E, Daniel JA, Osipovich O, Oltz EM, Bassing CH, Nussenzweig A, Lees-Miller S, Hammel M, Chen BP, Sleckman BP. Functional intersection of ATM and DNA-dependent protein kinase catalytic subunit in coding end joining during V(D)J recombination. Mol Cell Biol. 2013;33:3568–3579. doi: 10.1128/MCB.00308-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Krempler A, Yajima H, Lobrich M, Shiloh Y, Chen DJ. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J Biol Chem. 2007;282:6582–6587. doi: 10.1074/jbc.M611605200. [DOI] [PubMed] [Google Scholar]
- 93.Goodarzi AA, Jeggo PA. The heterochromatic barrier to DNA double strand break repair: how to get the entry visa. Int J Mol Sci. 2012;13:11844–11860. doi: 10.3390/ijms130911844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 95.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 97.Kakarougkas A, Jeggo P. DNA DSB repair pathway choice: an orchestrated handover mechanism. Br J Radiol. 2013 doi: 10.1259/bjr.20130685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ferretti LP, Lafranchi L, Sartori AA. Controlling DNA-end resection: a new task for CDKs. Front Genet. 2013;4:99. doi: 10.3389/fgene.2013.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, Maity R, van Rossum-Fikkert S, Kertokalio A, Romoli F, Ismail A, Ismalaj E, Petricci E, Neale MJ, Bristow RG, Masson JY, Wyman C, Jeggo PA, Tainer JA. DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou Y, Paull TT. DNA-dependent Protein Kinase regulates DNA end resection in concert with the Mre11-Rad50-Nbs1 (MRN) complex and Ataxia-Telangiectasia-Mutated (ATM) J Biol Chem. 2013 doi: 10.1074/jbc.M113.514398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zimmermann M, de Lange T. 53BP1: pro choice in DNA repair. Trends Cell Biol. 2013 doi: 10.1016/j.tcb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Feng L, Fong KW, Wang J, Wang W, Chen J. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J Biol Chem. 2013;288:11135–11143. doi: 10.1074/jbc.M113.457440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J, Cook MA, Rosebrock AP, Munro M, Canny MD, Xu D, Durocher D. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 105.Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell. 2013;49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2013 doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 107.Britton S, Coates J, Jackson SP. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J Cell Biol. 2013;202:579–595. doi: 10.1083/jcb.201303073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schipler A, Iliakis G. DNA double-strand-break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic Acids Res. 2013;41:7589–7605. doi: 10.1093/nar/gkt556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat Res. 2011;711:61–72. doi: 10.1016/j.mrfmmm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 110.Datta K, Purkayastha S, Neumann RD, Pastwa E, Winters TA. Base damage immediately upstream from double-strand break ends is a more severe impediment to nonhomologous end joining than blocked 3′-termini. Radiat Res. 2011;175:97–112. doi: 10.1667/RR2332.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lees-Miller SP, Chen YR, Anderson CW. Human cells contain a DNA-activated protein kinase that phosphorylates simian virus 40 T antigen, mouse p53, and the human Ku autoantigen. Mol Cell Biol. 1990;10:6472–6481. doi: 10.1128/mcb.10.12.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Carter T, Vancurova I, Sun I, Lou W, DeLeon S. A DNA-activated protein kinase from HeLa cell nuclei. Mol Cell Biol. 1990;10:6460–6471. doi: 10.1128/mcb.10.12.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dvir A, Peterson SR, Knuth MW, Lu H, Dynan WS. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc Natl Acad Sci U S A. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dvir A, Stein LY, Calore BL, Dynan WS. Purification and characterization of a template-associated protein kinase that phosphorylates RNA polymerase II. J Biol Chem. 1993;268:10440–10447. [PubMed] [Google Scholar]
- 115.Taccioli GE, Gottlieb TM, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann AR, Alt FW, Jackson SP, Jeggo PA. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 116.Blunt T, Finnie NJ, Taccioli GE, Smith GC, Demengeot J, Gottlieb TM, Mizuta R, Varghese AJ, Alt FW, Jeggo PA, et al. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 117.Lees-Miller SP, Godbout R, Chan DW, Weinfeld M, Day RS, 3rd, Barron GM, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 118.Kirchgessner CU, Patil CK, Evans JW, Cuomo CA, Fried LM, Carter T, Oettinger MA, Brown JM. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science. 1995;267:1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 119.Li Z, Otevrel T, Gao Y, Cheng HL, Seed B, Stamato TD, Taccioli GE, Alt FW. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 120.Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 121.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, Fischer A, Durandy A, de Villartay JP, Revy P. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 122.Jeggo P, Lavin MF. Cellular radiosensitivity: how much better do we understand it? Int J Radiat Biol. 2009;85:1061–1081. doi: 10.3109/09553000903261263. [DOI] [PubMed] [Google Scholar]
- 123.Williams GJ, Hammel M, Radhakrishnan SK, Ramsden D, Lees Miller SP, Tainer JA. Structural Insights into non-homologous end joining: building up a picture of the DNA double strand break repair complex one component at a time. DNA Repair (Amst) doi: 10.1016/j.dnarep.2014.02.009. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol. 2012;19:201–206. doi: 10.1038/nsmb.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Postow L. Destroying the ring: Freeing DNA from Ku with ubiquitin. FEBS Lett. 2011;585:2876–2882. doi: 10.1016/j.febslet.2011.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Postow L, Ghenoiu C, Woo EM, Krutchinsky AN, Chait BT, Funabiki H. Ku80 removal from DNA through double strand break-induced ubiquitylation. J Cell Biol. 2008;182:467–479. doi: 10.1083/jcb.200802146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chiruvella KK, Liang Z, Wilson TE. Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol. 2013;5:a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 129.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]


