Abstract
We computationally designed a de novo protein-protein interaction between wild-type ubiquitin and a redesigned scaffold. Our strategy was to incorporate zinc at the designed interface to promote affinity and orientation specificity. A large set of monomeric scaffold surfaces were computationally engineered with three-residue zinc coordination sites, and the ubiquitin residue H68 was docked to the open coordination sphere to complete a tetrahedral zinc site. This single coordination bond was intended as a hotspot and polar interaction for ubiquitin binding, and surrounding residues on the scaffold were optimized primarily as hydrophobic residues using a rotamer-based sequence design protocol in Rosetta. From thousands of independent design simulations, four sequences were selected for experimental characterization. The best performing design, called Spelter, binds tightly to zinc (Kd < 10 nM) and binds ubiquitin with a Kd of 20 µM in the presence of zinc and 68 µM in the absence of zinc. Mutagenesis and NMR chemical shift perturbation experiments indicate that Spelter interacts with H68 and the target surface on ubiquitin, however, H68 does not form a hotspot as intended. Mutation of H68 to alanine tightens (five-fold) instead of weakens binding. While a 3/1 zinc coordination arrangement at an interface cannot be ruled out as a means to improve affinity, our study led us to conclude that 2/2 coordination arrangements or multiple-zinc designs are more likely to promote high-affinity protein interactions.
Keywords: computational interface design, de novo, heterodimer, metal coordination, zinc binding, protein-protein interaction
Introduction
Understanding the physical basis of protein-protein interaction is a continued pursuit in molecular biology. A ground-up approach for understanding protein binding will help clarify mechanisms of cellular functions and lead to new therapeutic and diagnostic uses of proteins in medicine. Studies of natural interactions have provided valuable insights into how proteins interact, from detailed dissection of individual binding partners such as barnase and barstar1–3, to broad studies of hundreds of complexes4–9. Although much research has been aimed at studying protein interactions observed in nature, a complementary approach is to rationally design and build new interactions10.
Redesigning existing interactions for improved affinity or altered specificity is a good test of current understanding of protein binding11–13; however, the most rigorous test of our understanding is to design new protein-protein interactions from scratch. De novo computational interface design is still a young endeavor but has already seen a number of successes. Many of these studies strategically use pre-existing knowledge of patterns of recognition by using sequence profiles14, augmenting a native complex15,16, using known binding grooves17–19, side-chain interaction motifs19–21, or backbone interaction motifs (strand-strand pairing, linear epitopes, GxxG helix-helix contact, helix stacking)21–24. Karanicolas et al. used ankyrin repeat protein as a known versatile binding protein for design, but they ambitiously avoided using pre-existing interaction motifs already observed in natural protein-protein interactions25.
Although efforts in computational interface design have been encouraging, there is a significant need for improvement for reliable computational engineering of new interactions. Broad conclusions cannot be reliably drawn from a small number of attempts in de novo interface design, so continued efforts that explore different approaches and different modes of interaction will be critical in accumulating deeper knowledge about the physical basis of protein-protein interactions26. One repeated lesson from protein-protein interaction studies is that a few hotspot residues dominate the binding event27,28 – hotspot-based approaches have been used to design new interfaces, and these hotspots can be grafted from natural interfaces19,20,29 and developed from scratch19,30. Here we designed a three-residue zinc site from scratch where the open coordination sphere was the intended hotspot for target protein binding.
Computational methods have been used to design new tetrahedral zinc binding sites31,32, and zinc sites have previously been shown to promote affinity and orientation specificity in designed homo-oligomeric interactions. For cytochrome cb562 self-assembly, metal-mediated binding modes have been determined empirically but not predicted rationally33–35. In our previous homodimer design, two metal sites were used to promote binding in the desired orientation36. In this work, our one-zinc approach is for a heterodimeric interaction with a wild-type target.
We chose ubiquitin as a target because it has one surface histidine that may participate in zinc binding and because it is a small, stable protein that has been structurally characterized by crystallography and NMR. We observed a moderate binding affinity between wild-type ubiquitin and our redesigned scaffold named Spelter, where the presence of zinc resulted in 3-fold increase in affinity (Kd = 20 µM in the presence of zinc, Kd = 68 µM in the absence of zinc). Despite successful zinc binding in the redesigned scaffold (Kd < 10 nM), we conclude that this engineered zinc site did not provide a robust hotspot for target binding. Moderate affinity in a one-sided de novo interface design is a significant achievement for computation-only protein interface design. However, micromolar affinity has thus far been an affinity barrier for successful small hydrophobic designs, and designing polar interface contacts from scratch remains a significant challenge in protein interface design.
Materials and Methods
Scaffolds and target for protein interface design
A set of 635 scaffold proteins from the Protein Data Bank37 were used in the computational design of a ubiquitin (target) binding protein. The PDB query required these scaffolds to be listed as monomeric, expressible in E. coli, <2.5 Å resolution, without disulfides, without ligands, and between 80–250 residues. The list of scaffolds can be obtained from the Supporting Material of our previous work36.
RosettaMatch
These 635 scaffolds were used in RosettaMatch to search for possible three-residue histidine/cysteine zinc binding sites on the scaffold surfaces. RosettaMatch is a protocol typically used in enzyme design38,39 within the Rosetta modeling suite40. It uses a transition state model (TS) to search for designable residue sets on a scaffold protein that might stabilize the TS and hence catalyze the reaction. In our case, the pseudo-TS consisted of a HisT (histidine from ‘target’) positioned at an optimal coordination distance and orientation with a zinc atom consistent with the geometry of zinc coordination.
Distance, angle, and torsion constraints were used to correctly build the HisT downstream of all possible histidine or cysteine rotamers at each allowed residue position. Each HisT placement was followed by clash detection between the HisT and the scaffold backbone, and the clash-free HisT locations were recorded in a 6-dimensional bin (3 dimensions for Cartesian position, 3 dimensions for rotational orientation) (Fig. S1). If HisT locations from three different residues were hashed into the same 6-D bin, this was a match.
After matching, a geometry-based evaluation of all output matches was performed to select the matches with coordination bond lengths, angles, and dihedrals that were close to ideal (Fig. S2). This was done using the ZincMatchFilter application in Rosetta. Among 635 scaffolds, ~2,000 high-quality tetrahedral three-residue zinc matches were identified. Half of these matches, however, featured cysteine residues at i, i+1 positions. We excluded these matches from the final list of 1,015 because we did not identify the Cys/Cys at i, i+1 as a naturally occurring zinc coordination motif. The Rosetta 2.3 version of RosettaMatch was used for this study, however, the current version of Rosetta (3.4) contains an updated implementation of the RosettaMatch algorithm. Input files and command lines for the equivalent Rosetta3 implementation of RosettaMatch are also given in the Supporting Material.
ZincHeterodimerDesign
ZincHeterodimerDesign is the name of the Rosetta protocol that was written for the protein interface design stage. Reorganization of the Rosetta code into discrete protocols simplified the addition of new design protocols40. The required inputs for ZincHeterodimerDesign are the scaffold, the three-residue zinc match, and ubiquitin. Other command-line options are given in the Supporting Material. In Step 1 of the protocol, the zinc match atomic coordinates were grafted onto the scaffold. In Step 2, the HisT transition state residue was replaced by the H68 residue of ubiquitin, which docked the ubiquitin to the scaffold with a zinc coordination site bridging the interface. This pseudo-docking step ignored protein complementarity, so in Step 3, a Monte Carlo rigid-body search was performed to relieve steric clashes between the scaffold and ubiquitin while preserving the zinc binding site. The zinc binding site was conserved by limiting the degrees of freedom to rotation about the H68-zinc coordination bond and the H68 chi 1 and chi 2 torsion angles. H68 torsion angles were limited to rotamers from the 2002 Dunbrack library used in Rosetta41,42, and “inverse rotamer” sampling moved the ubiquitin and kept the imidazole ring fixed in space. In this rigid-body search, both scaffold and ubiquitin side chains were scored as centroids to evaluate overall shape complementarity and ignore clashes between side chains that could be redesigned. These centroid protein representations were kinematically coupled to the full-atom representations required to explore the torsional rotations of the H68 side chain. The lowest-energy centroid docking arrangement was chosen from the Monte Carlo search, and Step 4 of the protocol was full-atom design of the scaffold interface residues and repacking of the ubiquitin interface residues. Interface residues were identified as those within 10 Å of the other partner based on Cβ-Cβ distances, and this design step used the standard fixed-backbone rotamer packing functionality in Rosetta. In summary, this protocol used zinc binding and emphasized docking and side-chain degrees of freedom but did not include backbone sampling. The best models were chosen based on computed binding energy per Å2 of interface surface area, followed by visual inspection.
Cloning, Expression, Purification, Mutagenesis
We synthesized genes of the four designs, the wild-type scaffold (PDB code 2D4X, the bacterial flageller hook-filament junction protein), and wild-type ubiquitin by oligo-assembly43. The genes were cloned into the pQE-80L vector (Qiagen) supplemented with an N-terminal MBP (maltose binding protein) fusion to aid expression and solubility. Ubiquitin with a G76C mutation for fluorescence polarization experiments was previously cloned into the pET21b+ vector (Novagen). Mutageneses were performed by overlapping PCR using internal primers encoding the desired mutations. These proteins were expressed in BL21(DE3)pLysS (Invitrogen) cells with induction using 333 µM IPTG for six hours at 25 °C. The 6xHis-MBP-design fusions were purified by Ni2+-NTA high-affinity chromatography (HisTrap columns, GE Healthcare Biosciences). The eluent was supplemented with 1 mM DTT to prevent disulfide formation and 0.5 mM EDTA to scavenge metal ions. The 6xHis-MBP tag was cleaved by overnight incubation by TEV protease. For ubiquitin G76C, the His-tag was cleaved by overnight thrombin proteolysis. The proteolyzed samples were dialyzed back into HisTrap column buffer, and a second HisTrap purification removed the 6xHis-MBP tag or uncleaved protein. The flow-through containing the desired protein was again supplemented with 1 mM DTT and 0.5 mM EDTA before concentration to <4 ml for gel filtration using column Superdex 75 Hiload 16/60 (Amersham Biosciences). The final protein buffer was 20 mM MOPS, pH 6.9, 50 mM NaCl, 0.5 mM TCEP (thiol-free reducing agent).
Isothermal Titration Calorimetry
Isothermal titration calorimetry experiments to measure zinc binding were performed using a MicroCal VP-ITC (GE Healthcare). 2.3 ml of 20 µM protein was loaded into the sample chamber, and 250 µM ZnSO4 injectant was diluted from a high concentration stock using the protein dialysis buffer. 29 titrations of 10 µl volume were performed with 150 seconds equilibration, and the resulting titration curves were fit using one-site binding in the Microcal Origin 5.0 software.
Circular Dichroism
Thermal denaturation experiments were performed using circular dichroism with a JASCO J-815 CD spectrometer. Temperature was ramped at 1 °C/min under the control of a JASCO Peltier device and water bath. Protein concentration for the 2D4X-variants was 15 µM in a 1-mm quartz cuvette, and for experiments containing zinc, ZnSO4 was added to 16.5 µM (protein to metal ratio of 1 to 1.1). Protein unfolding was monitored at wavelength 222 nm with units converted to mean molar ellipticity to provide apparent melting temperatures as a measure of thermostability.
Fluorescence Polarization
Wild-type ubiquitin was altered with a G76C mutation for covalent attachment of thiol-reactive Bodipy (507/545, Molecular Probes). Bodipy conjugation was performed as previously described17, and labeling efficiency of 20% was observed. Fluorescence polarization binding assays were performed using a SPEX FluoroLog-3 instrument. The 180 µl starting sample in a 3×3-mm quartz cuvette contained 5 µM ubiquitin (1 µM fluorescently labeled ubiquitin). Slits of 2.5 nm were used with excitation/emission wavelengths of 508/545 nm. Each polarization reading had 0.1 s integration time, and readings were taken in triplicate. Plots of polarization versus concentration of titrant were modeled with a single-site binding equation with nonlinear fitting using SigmaPlot 2001 Version 7.1 software to obtain apparent Kd values17.
NMR: HNCACB backbone assignment
Uniformly labeled 15N- and 13C- ubiquitin was expressed using minimal media as described in our previous work36 and was purified as described above. Ubiquitin concentration was 1.1 mM, and the sample buffer contained 20 mM MOPS, pH 6.9, 50 mM NaCl, 0.5 mM TCEP, and 10% D2O. To assign sequence-specific resonances of 1HN, 15N, 13Cα, and 13Cβ, we performed the three-dimensional HNCACB experiment44. Data collection proceeded for 70 hours on a Varian INOVA 700 MHz spectrometer with a cold probe. Data were processed using the NMRPipe/NMRDraw software45, and assignments were made using the Sparky software46. A subset of strips showing peak quartets spanning residues 63–73 and a list of chemical shift values are given in the Supporting Material (Table SII).
NMR: 15N- and 13C-HSQC
2D [15N, 1H]- and [13C, 1H]-HSQC spectra were recorded at 25 °C on a Varian INOVA 700 MHz spectrometer with a cold probe. Titration of Spelter with equimolar zinc was performed to monitor chemical shift perturbations upon binding. Data were processed using the NMRPipe/NMRDraw software45, and peak-picking was performed using the NMRViewJ software47. Peaks in the 15N-HSQC spectra were assigned based on chemical shift values from our HNCACB 3-D backbone assignment. Peaks in the 13C-HSQC were assigned using CA and CB chemical shift values from our HNCACB experiment combined with published values for proton, CG, CD, and CE values48. Compound changes in chemical shift (Δδcomp) were measured as distances according to Eq. 149,50. In cases of two peaks from two protons on the same heavy atom – for example, two peaks for a CB side-chain atom in histidine – these measured distances were averaged. Due to peak broadening upon formation of a complex larger than 30 kDa, we compared chemical shifts from the ubiquitin-only spectrum with a limited titration in which Spelter was present at a 30% molar ratio with ubiquitin. Our limited titration along with intermediate exchange resulted in small changes in chemical shift (Eq. S1), so we also analyzed changes in peak intensities as a second method to determine the ubiquitin residues at the Spelter binding interface. Peak intensities were calculated using the NMRViewJ software47.
| Eq. 1 |
Results
Computational design
Choosing a target and scaffold for interface design
We aimed to bind a wild-type target protein using a computationally redesigned scaffold protein where one metal coordination group is supplied by the target protein and the other three coordinating groups are supplied by the redesigned scaffold (Fig. 1A). This 3/1 metal coordination arrangement seemed attractive for designing a heterodimeric interaction with a wild-type target because many target surfaces will have one or more metal-coordinating side chains (histidine, aspartate, glutamate, cysteine). The criterion for a good target protein was a surface histidine for metal binding near surface hydrophobic residues. Ubiquitin is a well-studied protein that has a surface histidine (H68) near residues V8, I44, and L70. I44 has been shown to be a hotspot residue for protein interactions with ubiquitin51.
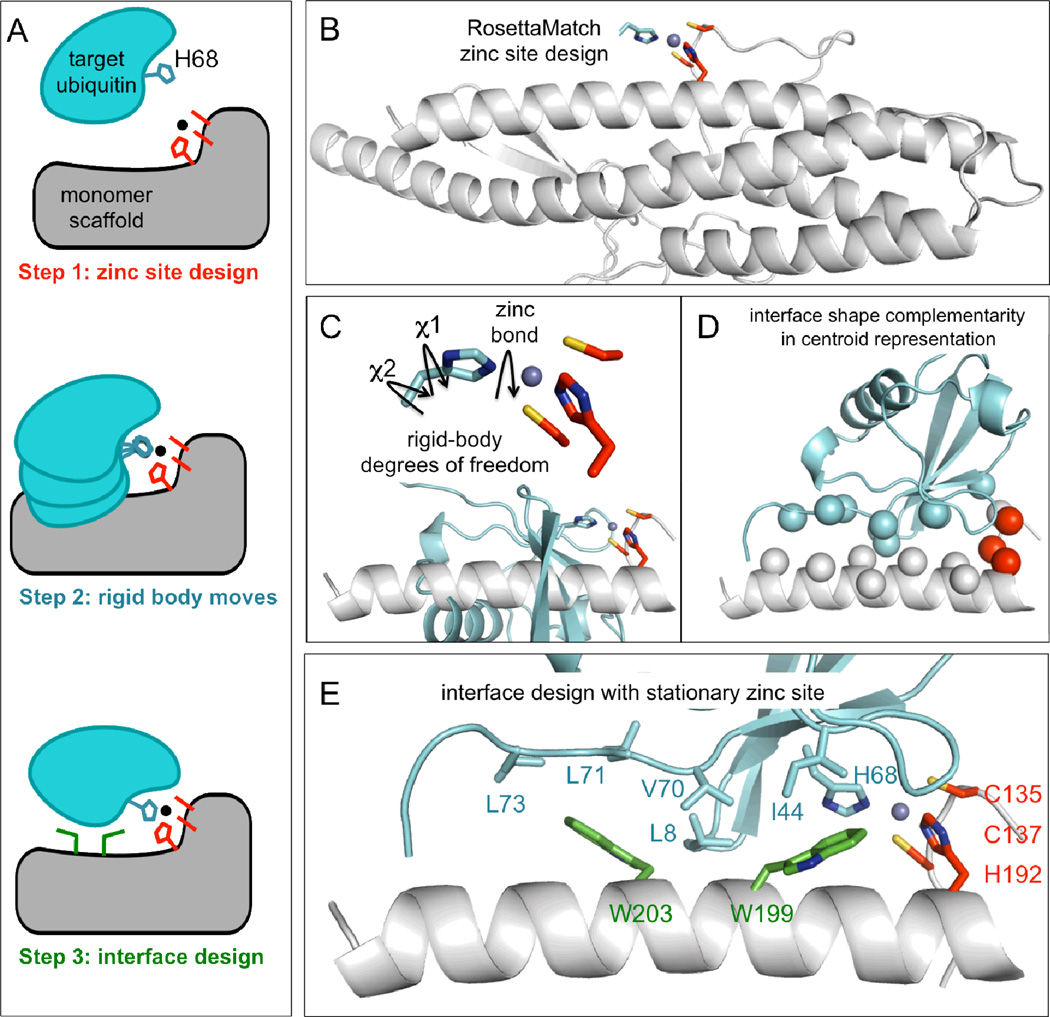
Fig. 1. Zinc heterodimer computational design protocol.
A) Sketch to summarize the full computational interface design protocol. B) RosettaMatch design of a zinc binding site with an open coordination site within an existing protein backbone. Among 635 scaffolds, 1015 high-quality zinc binding sites were identified. Shown in cartoon is the bacterial flagellar hook-filament junction protein (PDB code 2D4X), a 3-residue zinc binding designed by RosettaMatch (C135, C137, H192), and the target histidine (HisT) supplied by the target binding protein. This designed zinc binding protein is called 2D4X-CCH. C) The H68 residue of ubiquitin (blue sticks) replaced the ideally positioned histidine from the RosettaMatch simulations. This starting orientation satisfied ideal zinc coordination and ignored protein orientation. Protein orientation was optimized while the zinc coordination geometry remained fixed. Degrees of freedom were rotation about the H68-zinc coordination bond and H68 chi 1 and chi 2 side-chain torsion angles (black arrows). D) After rigid-body optimization, the lowest-energy centroid docked orientation was chosen for interface design. E) During interface design, the proteins were modeled in full-atom representation. The scaffold interface side chains could change identity to favorably interact with the ubiquitin native side chains, which could only change conformation. In the design called Spelter, the prominent interactions were formed between W199 and W203 (green sticks) of Spelter with the hydrophobic patch of ubiquitin (blue sticks).
RosettaMatch for zinc site design
We used RosettaMatch to find potential three-residue zinc binding sites on the surfaces of 635 small, monomeric scaffold proteins from the Protein Data Bank (Fig. 1B). Using histidine and cysteine side chains as zinc-coordinating groups, 1,015 high-quality zinc-binding matches consisting of His/Cys triplets were identified among these 635 scaffolds. The RosettaMatch algorithm has been previously described38,39 and is summarized in the Methods section of this work.
Interface design
The second step after RosttaMatch was to rebuild the target ubiquitin (PDB code 1UBQ)52 starting from its native surface histidine H68, replacing the idealized histidine (HisT) from the RosettaMatch transition state. This pseudo-docking step completed the four-coordination of zinc and resulted in one zinc coordination bond bridging the protein interface. The coordinating bond, chi 1, and chi 2 angles of the ubiquitin H68 residue (Fig. 1C) were degrees of freedom for rigid-body optimization. The orientation with the best overall complementarity (Fig. 1D) was the starting point for full-atom design. During design, scaffold protein interface residues could change to any amino acid except histidine and cysteine, and the wild-type ubiquitin residues could change conformation but not identity (Fig. 1E). Thus, this protocol used zinc binding and emphasized docking and side-chain degrees of freedom but did not include backbone sampling. For all 1,015 zinc matches on different scaffolds, 10 ZincHeterodimerDesign design trajectories were performed, resulting in ~10,000 total designs.
Selection criteria for designed interfaces
The 10,000 designed interfaces were evaluated based on binding energy calculations using the Rosetta energy function. The zinc binding site was ignored in these calculations – favorable zinc coordination geometry was established by RosettaMatch and was left intact during the interface design protocol, and a histidine-zinc coordination bond was assumed to offer a similar contribution to affinity across different designs. The binding energy density (binding energy / buried surface area), number of unsatisfied hydrogen bonding groups, and number of scaffold mutations were considered for determining the quality of the designed interface. After visual inspection, four designs using three different starting scaffolds (PDB codes 2D4X, 2FZ4, 2ONU) were selected for experimental characterization (Table SI, Fig. S3).
Experimental validation
We experimentally tested four designs: one was insoluble and could not be purified, and the other three designs showed moderate to low affinity for ubiquitin. The low-affinity interaction was not further investigated, and the two moderate-affinity interactions used the same starting scaffold (PDB code 2D4X53) and the same zinc-binding motif but featured a different ubiquitin orientation (Fig. S3). We chose to only investigate the higher affinity interaction in further detail, and we refer to this design as Spelter. In addition to the zinc binding site, the major interface contacts in this design model feature two designed tryptophan residues that make hydrophobic contacts with ubiquitin, especially I44, a known hotspot residue for ubiquitin binding51. This design model does not resemble naturally-occurring ubiquitin heterocomplexes54 (Fig. S4).
Validation of the designed zinc binding site
To validate the designed zinc binding motif on the scaffold, we generated a mutant called 2D4X-CCH (2D4X is the starting scaffold PDB code) that contained all wild-type residues except for the C135, C137, and H192 mutations (CCH) for zinc binding (we renumbered such that the first residue in the 2D4X crystal structure was residue 1). Isothermal titration calorimetry (ITC) experiments titrating zinc (ZnSO4) into 2D4X-CCH indicated a binding affinity <10 nM with a molar ratio of 1.0 (Fig. 2A). Point mutants reverting each zinc coordinating residue back to the wild-type residue (denoted with an x) showed substantial decrease in binding affinity for zinc: the 2D4X-CCx mutant bound to zinc approximately 10-fold weaker, and the other two mutants, 2D4X-xCH and 2D4X-CxH did not show any evidence of zinc binding (Table I). As expected, 2D4X-CCH and Spelter showed similar affinities for zinc in ITC (Fig. S5).
Fig. 2. Zinc binding by a 2-Cys/1-His mutant, 2D4X-CCH.
A) Titration of ZnSO4 (250 µM) into 2D4X-CCH protein (20 µM) measured by isothermal titration calorimetry indicates a binding affinity (Kd) of <10 nM and molar ratio of 1.1. B) Thermal denaturation monitored by circular dichroism at 222 nm indicates a 4°C increase in melting temperature (Tm) upon addition of equimolar zinc (16.5 µM). See Table II for additional zinc binding data upon mutagenesis of each zinc binding residue.
Table I.
The designed 3-residue zinc binding site
| Protein | CD, ΔTm with zinc | ITC, Kd for zinc |
|---|---|---|
| 2D4X-CCH* | 4°C | <10 nM |
| 2D4X-xCH** | 0°C | not detectable |
| 2D4X-CxH | 0°C | not detectable |
| 2D4X-CCx | 0°C | 68 nM |
2D4X-CCH is the wild-type scaffold (2D4X) with three zinc-binding residues (CCH)
x indicates mutation back to the wild-type residue
Zinc binding stabilized 2D4X-CCH and Spelter as shown by an increase in melting temperature (Tm). We performed thermal denaturation of the protein using far-UV circular dichroism (CD) using a 222 nm wavelength. In response to zinc, 2D4X-CCH showed a 4°C increase in melting temperature (Fig. 2B). Individual point mutations reverting each Cys or His to the wild-type residue abolished the stabilizing effect of zinc as measured by thermal denaturation, indicating that the designed 2-Cys/1-His motif binds zinc (Table I). As expected, 2D4X-CCH and Spelter showed similar stabilization upon addition of zinc (Fig. S5). The zinc binding site was also specific for zinc: CaCl2, MgCl2, MnSO4, CoCl2, NiSO4 did not change the Tm by more than 1°C in CD thermal denaturation experiments (Fig. S6).
Mutagenesis studies show zinc-mediated interaction of Spelter with ubiquitin
We measured the binding affinity of Spelter for ubiquitin using fluorescence polarization (Table II). The ubiquitin G76C mutant was covalently labeled with thiol-reactive Bodipy, and titration of the designed protein led to an increase in fluorescence polarization due to heterocomplex formation. Curves were fit to a single-site binding equation17. The 2D4X-CCH protein, which contained only the metal coordinating motif and no other designed residues, did not bind ubiquitin in fluorescence polarization (Fig. 3), demonstrating that the residues designed in Spelter for protein-protein contacts with ubiquitin were critical for binding. The affinity (Kd) of Spelter for ubiquitin was 20 µM with zinc and 68 µM without zinc. Thus, zinc provided a ~3-fold increase in binding affinity, corresponding to ~0.7 kcal/mol of binding energy (Fig. 3). A mutant of Spelter in which the two cysteines were reverted to their wild-type identity (Spelter-xxH) binds with a similar affinity as Spelter in the absence of zinc (Kd = 51 µM). The affinity of this mutant was unaffected by zinc, as expected. Individual tryptophan to alanine point mutations were tested for ubiquitin affinity. In the presence of zinc, W199A weakened affinity by 2-fold, but W203A did not change binding affinity. In the absence of zinc, binding affinities were 2-fold weaker for both tryptophan point mutants (Kd = 110 µM, Fig. S7a). Adding both tryptophan residues to the 2D4X wild-type scaffold (2D4X-WW) resulted in weak binding (Kd ≈ 300 µM), so while these tryptophan residues offer a starting point for binding, additional interface mutations also contribute. Overall, the mutagenesis studies indicate that the designed surface patch on Spelter participates in ubiquitin binding, though details of the binding orientation were not elucidated and no Spelter residues were identified as hotspots.
Table II.
The designed zinc binding site and other designed residues contribute to ubiquitin binding
| Titrated protein | Fluorescently- labeled protein |
Kd with zinc (µM) |
Kd without zinc (µM) |
|---|---|---|---|
| Spelter | Ubiquitin | 20 | 68 |
| Spelter-xxH | Ubiquitin | 51 | 51 |
| Spelter-W199A | Ubiquitin | 42 | 107 |
| Spelter-W203A | Ubiquitin | 18 | 110 |
| Spelter-E136A | Ubiquitin | 59 | 138 |
| Spelter-E136A | Ubiquitin-H68A | 62 | 79 |
| Spelter | Ubiquitin-H68A | biphasic* | 50 |
| 2D4X-CCH | Ubiquitin | > 500 | > 500 |
| 2D4X-WW | Ubiquitin | ~250 | ~400 |
| 2D4X-CCHEWW | Ubiquitin | ~250 | ~200 |
biphasic, apparent binding affinity <10 µM
Fig. 3. Binding affinities of Spelter and Spelter mutants for Bodipy-labeled ubiquitin measured by fluorescence polarization.
Bodipy-labeled ubiquitin was present at 1.1 µM starting concentration, and polarization increases upon heterocomplex formation with titrated protein. Assays were performed in buffer containing 10 mM MOPS pH 6.9, 25 mM NaCl, and 0.5 mM TCEP. Fluorescence polarization titration curves were fit to a single-site binding equation to determine apparent Kd values17.
Structural study of Spelter-ubiquitin interface
To investigate whether the correct surface patch of ubiquitin participates in binding, we performed NMR chemical shift perturbation analysis of ubiquitin upon titration of Spelter. Changes to the binding mode upon mutagenesis can lead to unclear interpretation of the role of each residue, but the HSQC titration study does not require any mutation to assess the binding contributions of individual residues in the designed interaction. Ubiquitin residues have already been assigned by NMR, but due to subtle differences between our spectra and published spectra from differing experimental conditions48, we performed a 3D-HNCACB experiment to ensure the correct assignment of all backbone peaks in our 15N-HSQC spectra (Fig. S8, Table SII). Additionally, C-alpha (CA) and C-beta (CB) chemical shifts from our 3D-HNCACB experiment helped identify peaks in our 13C-HSQC spectra when combined with previously published proton chemical shifts. A subset of C-gamma, C-delta, and C-epsilon (CE) peaks were assigned based on previously published chemical shifts48. We analyzed the 15N- and 13C-HSQC spectra before and after titration of Spelter with zinc to discern which residues experience a change in environment upon complex formation (Fig. 4A). Changes in peak intensity (Fig. 4B) and changes in chemical shift (Fig. 4C) were used to identify the binding surface of ubiquitin (Eq. 1, Methods). Most of the ubiquitin residues that experience changes in peak intensity and chemical shift are located at the computationally predicted protein interface in both the 15N-HSQC and 13C-HSQC spectra (Fig. 4D). The largest changes in chemical shift occur in the H68 CE and CB side-chain atoms, implicating this residue as a key component of the designed interface. However, H68 could either interact directly with zinc or with an unintended Spelter protein residue to cause this chemical shift change.
Fig. 4. NMR peak intensity and chemical shift changes of 15N- and 13C-labeled ubiquitin upon titration of zinc-bound Spelter. 15N, left; 13C, right.
Isotopically enriched ubiquitin was present at 1.1 mM concentration, and Spelter with zinc was titrated to 0.3 mM. A) Regions of the 2D-HSQC spectra showing peaks that change and do not change upon Spelter titration. B) Ratio of HSQC peak intensities before and after titration of Spelter. Peaks with the largest intensity changes are colored in red and orange. C) Compound chemical shift changes before and after titration of Spelter (Eq. 1, Methods). Peaks with the largest chemical shift changes are colored in red and orange. D) Computational model of the Spelter-ubiquitin interaction where ubiquitin is colored according to chemical shift change (red/orange indicate larger changes in compound chemical shift). Predicted interface residues including L8, I44, and H68 have higher changes in chemical shift, supporting the computational model.
Ambiguity in the role of ubiquitin H68 at the designed interface
Unexpectedly, Spelter bound the ubiquitin-H68A mutant tighter than wild-type ubiquitin (Kd value not determined, Fig. S7b). Moreover, binding affinity for this ubiquitin mutant was still modulated by addition of zinc. The increase in binding affinity upon mutation to alanine suggests that the interactions H68 is making at the interface are not large enough to overcome the desolvation cost for burying a histidine. This could be because the zinc-histidine contact is weak or not present at all. In either scenario, the open coordination sphere of the zinc site is not providing a robust hot spot for H68 as designed. One contributing factor to a weak or absent zinc-histidine contact could be that the design process was not able to identify a back side hydrogen bond for H68, which is partially buried at the interface. In naturally occurring zinc binding sites, zinc-coordinating side chains often feature a hydrogen bond with side chain or backbone groups from the protein55. Secondly, the sensitivity of ubiquitin-H68A to zinc could be from a direct interaction across the interface or could be due to pre-ordering in Spelter. Interestingly, a variant of Spelter with E136 mutated to alanine does not show zinc dependent binding to ubiquitin-H68A (Fig. S9a, Table II). E136 is adjacent to the metal binding site in a loop region, and in the design model E136 is predicted to form a salt bridge with K6 from ubiquitin (Fig. S9b) – K6 does experience a change in chemical shift upon binding to ubiquitin (Fig. 4C). Zinc binding could be pre-ordering this interaction. Though the role of ubiquitin H68 remains ambiguous, we conclude that the intended single coordination bond did not provide a robust hotspot.
Discussion
We cannot rule out 3/1 zinc coordination as a useful strategy for protein interface design. The hydrophobic interaction of Spelter with ubiquitin likely occurred with a different orientation than the predicted model, and the preferred orientation without zinc may not have been compatible with H68 zinc coordination. This hypothesis is indirectly supported by the weak affinity with and without zinc of the minimalist mutant 2D4X-CCHEWW (Fig. 3, Table II) – these six residues are predicted to be the most important in the computational model. Thus, although 3/1 coordination may be useful in other designs, here we discuss drawbacks to the 3/1 approach and suggest that 2/2 coordination arrangements or multiple-zinc designs will be more effective for de novo interface design.
The “chelate effect” states that polydentate ligands have a cooperative energetic advantage over an equal number of monodentate ligands56; thus, a monodentate interaction (H68 from ubiquitin) is at a disadvantage to a 2/2 bidentate interaction. An example of a zinc-mediated heterodimer from nature is the human growth hormone and prolactin receptor complex (PDB code 1BP3), and the (His/Glu)-(His/Asp) site contributes two coordination bonds per protein subunit to increase the affinity 8000-fold57,58. In previous interface design studies, metal-based strategies used multiple metal sites and did not feature three coordination bonds to one metal ion contributed by one protein chain. In, our previously designed homodimer36. Cytochrome cb562 helix bundles have been engineered to self-associate in response to metal ions, these metal sites do not feature three coordination bonds from one protein subunit. These design studies also feature multiple metal sites for additional cooperative benefit of metal binding35,36,59. Thus, while metals can be used as powerful agents for protein assembly, devoting three scaffold interface residues to bind zinc and gaining only one coordinating bond across an interface may not be a robust route to high affinity interactions.
In addition to lack of cooperativity from a 3/1 coordination approach, perhaps our open zinc coordination sphere was not an effective energy contributor for protein binding due desolvation penalties of the histidine residue and zinc ion. The backside delta-nitrogen of histidine is partially buried in the computational model and does not have a hydrogen bond partner (Fig. S10). Furthermore, three-residue zinc coordination sites often contain a zinc-bound water molecule60, and this water would also have to be removed in this 3/1 coordination approach. Minor errors in chemical complementarity such as desolvation of a single polar atom can cripple a binding interaction yet go overlooked in an overall computed score of a protein interface. Overcoming the desolvation penalty and satisfying the hydrogen bonding potential of both interacting partners is an outstanding challenge in protein interface design, and this is why successful designs have been mostly hydrophobic61. We attempted to complement a heterodimeric hydrophobic interface with a single metal coordination site, rather than attempt accurate design of multiple hydrogen bonds, but our 3/1 coordination approach did not produce the intended hotspot interaction. Although a 3/1 coordination site may be capable of providing an interface hotspot, we suggest that 2/2 coordination arrangements or multiple-zinc designs will be more effective.
Conclusion
In summary, why is computational interface design so challenging, what was our approach, and how well did it work? De novo interface design can be an overwhelming endeavor given the vast degrees of freedom to sample. Assuming a target protein has been chosen, there are still hundreds of possible scaffolds to consider, and for each scaffold an extensive search of docking orientation is required. These docking searches are exacerbated by the vast sequence space to explore during design, especially if backbone flexibility is allowed. Furthermore, unavoidable inaccuracies in energy evaluation will mislead the conformational sampling. To address these challenges, effective strategies for protein interface design must limit the conformational search and provide reliable energetically favorable hotspots. Furthermore, designs can be mostly hydrophobic, but small hydrophobic designed interactions seem to have a micromolar affinity barrier17,19,62, and polar residues often specify binding orientation22,36 and promote solubility of the unbound partners63. Our strategy used a zinc binding site to limit the conformational search, to provide a hotspot, and to specify the binding orientation. The results show a partial success. Spelter binds tightly to zinc and binds to the targeted ubiquitin surface with an affinity, 20 µM, which is tighter than many naturally occurring ubiquitin binders54 (Fig. S11). Thus, we designed a zinc site and added to the small number of de novo computational designs that bind a wild-type target17–19. However, our initial hypothesis that a zinc mediated interaction with H68 would significantly stabilize and orient the interface is not well supported, and we conclude that a more effective strategy is to include two points of contact with the metal. By continuing to pursue new strategies for computational interface design, we hope to find reliable strategies that lead to strong interactions for applications in cell biology and biotechnology.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health [GM073960 and T32GM008570]; the National Science Foundation graduate research fellowship [2009070950 to B.D. and 2008072760 to P.T.]; and the University of North Carolina Royster Society Pogue fellowship to S.L. and B.D. We also thank Dr. Ashutosh Tripathy at the UNC Macromolecular Interactions Facility, Dr. Greg Young at the UNC Biomolecular NMR Laboratory Core Facility, and Dr. Mike Miley and Dr. Mischa Machius at the UNC Molecular X-ray Crystallography Core Facility for their valuable contributions.
Abbreviations
- CD
circular dichroism
- HSQC
heteronuclear single quantum coherence
- ITC
isothermal titration calorimetry
- TS
transition state
- HisT
target histidine
References
- 1.Hartley RW. Barnase and barstar: two small proteins to fold and fit together. Trends Biochem Sci. 1989;14(11):450–454. doi: 10.1016/0968-0004(89)90104-7. [DOI] [PubMed] [Google Scholar]
- 2.Frisch C, Schreiber G, Johnson CM, Fersht AR. Thermodynamics of the interaction of barnase and barstar: changes in free energy versus changes in enthalpy on mutation. J Mol Biol. 1997;267(3):696–706. doi: 10.1006/jmbi.1997.0892. [DOI] [PubMed] [Google Scholar]
- 3.Hoefling M, Gottschalk KE. Barnase-Barstar: from first encounter to final complex. J Struct Biol. 2010;171(1):52–63. doi: 10.1016/j.jsb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Xu D, Tsai CJ, Nussinov R. Hydrogen bonds and salt bridges across protein-protein interfaces. Protein Engineering. 1997;10(9):999–1012. doi: 10.1093/protein/10.9.999. [DOI] [PubMed] [Google Scholar]
- 5.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. Journal of Molecular Biology. 1999;285(5):2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 6.Brinda KV, Kannan N, Vishveshwara S. Analysis of homodimeric protein interfaces by graph-spectral methods. Protein Engineering. 2002;15(4):265–277. doi: 10.1093/protein/15.4.265. [DOI] [PubMed] [Google Scholar]
- 7.Bahadur RP, Chakrabarti P, Rodier F, Janin J. Dissecting subunit interfaces in homodimeric proteins. Proteins-Structure Function and Genetics. 2003;53(3):708–719. doi: 10.1002/prot.10461. [DOI] [PubMed] [Google Scholar]
- 8.Nooren IMA, Thornton JM. Structural characterisation and functional significance of transient protein-protein interactions. Journal of Molecular Biology. 2003;325(5):991–1018. doi: 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- 9.Chakrabarti P, Janin J. Dissecting protein-protein recognition sites. Proteins-Structure Function and Genetics. 2002;47(3):334–343. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- 10.Kortemme T, Baker D. Computational design of protein-protein interactions. Current Opinion in Chemical Biology. 2004;8(1):91–97. doi: 10.1016/j.cbpa.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Karanicolas J, Kuhlman B. Computational design of affinity and specificity at protein-protein interfaces. Current Opinion in Structural Biology. 2009;19(4):458–463. doi: 10.1016/j.sbi.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts KE, Cushing PR, Boisguerin P, Madden DR, Donald BR. Computational Design of a PDZ Domain Peptide Inhibitor that Rescues CFTR Activity. Plos Computational Biology. 2012;8(4) doi: 10.1371/journal.pcbi.1002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapp GT, Liu S, Stein A, Wong DT, Remenyi A, Yeh BJ, Fraser JS, Taunton J, Lim WA, Kortemme T. Control of protein signaling using a computationally designed GTPase/GEF orthogonal pair. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(14):5277–5282. doi: 10.1073/pnas.1114487109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen TS, Reinke AW, Keating AE. Design of Peptide Inhibitors That Bind the bZIP Domain of Epstein-Barr Virus Protein BZLF1. Journal of Molecular Biology. 2011;408(2):304–320. doi: 10.1016/j.jmb.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King NP, Sheffler W, Sawaya MR, Vollmar BS, Sumida JP, Andre I, Gonen T, Yeates TO, Baker D. Computational Design of Self-Assembling Protein Nanomaterials with Atomic Level Accuracy. Science. 2012;336(6085):1171–1174. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grigoryan G, Kim YH, Acharya R, Axelrod K, Jain RM, Willis L, Drndic M, Kikkawa JM, DeGrado WF. Computational Design of Virus-Like Protein Assemblies on Carbon Nanotube Surfaces. Science. 2011;332(6033):1071–1076. doi: 10.1126/science.1198841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jha RK, Leaver-Fay A, Yin SY, Wu YB, Butterfoss GL, Szyperski T, Dokholyan NV, Kuhlman B. Computational Design of a PAK1 Binding Protein. Journal of Molecular Biology. 2010;400(2):257–270. doi: 10.1016/j.jmb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sammond DW, Bosch DE, Butterfoss GL, Purbeck C, Machius M, Siderovski DP, Kuhlman B. Computational Design of the Sequence and Structure of a Protein-Binding Peptide. Journal of the American Chemical Society. 2011;133(12):4190–4192. doi: 10.1021/ja110296z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, Strauch EM, Wilson IA, Baker D. Computational Design of Proteins Targeting the Conserved Stem Region of Influenza Hemagglutinin. Science. 2011;332(6031):816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S, Liu SY, Zhu XL, Liang HH, Cao AN, Chang ZJ, Lai LH. Nonnatural protein-protein interaction-pair design by key residues grafting. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(13):5330–5335. doi: 10.1073/pnas.0606198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azoitei ML, Correia BE, Ban YE, Carrico C, Kalyuzhniy O, Chen L, Schroeter A, Huang PS, McLellan JS, Kwong PD, Baker D, Strong RK, Schief WR. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science. 2011;334(6054):373–376. doi: 10.1126/science.1209368. [DOI] [PubMed] [Google Scholar]
- 22.Stranges PB, Machius M, Miley MJ, Tripathy A, Kuhlman B. Computational design of a symmetric homodimer using beta-strand assembly. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(51):20562–20567. doi: 10.1073/pnas.1115124108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sievers SA, Karanicolas J, Chang HW, Zhao A, Jiang L, Zirafi O, Stevens JT, Munch J, Baker D, Eisenberg D. Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature. 2011;475(7354):96–U117. doi: 10.1038/nature10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanci CJ, MacDermaid CM, Kang SG, Acharya R, North B, Yang X, Qiu XJ, DeGrado WF, Saven JG. Computational design of a protein crystal. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(19):7304–7309. doi: 10.1073/pnas.1112595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karanicolas J, Corn JE, Chen I, Joachimiak LA, Dym O, Peck SH, Albeck S, Unger T, Hu WX, Liu GH, Delbecq S, Montelione GT, Spiegel CP, Liu DR, Baker D. A De Novo Protein Binding Pair By Computational Design and Directed Evolution. Molecular Cell. 2011;42(2):250–260. doi: 10.1016/j.molcel.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Der BS, Kuhlman B. From Computational Design to a Protein That Binds. Science. 2011;332(6031):801–802. doi: 10.1126/science.1207082. [DOI] [PubMed] [Google Scholar]
- 27.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267(5196):383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 28.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280(1):1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 29.Azoitei ML, Ban YE, Julien JP, Bryson S, Schroeter A, Kalyuzhniy O, Porter JR, Adachi Y, Baker D, Pai EF, Schief WR. Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope. J Mol Biol. 2012;415(1):175–192. doi: 10.1016/j.jmb.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleishman SJ, Corn JE, Strauch EM, Whitehead TA, Karanicolas J, Baker D. Hotspot-centric de novo design of protein binders. J Mol Biol. 2011;413(5):1047–1062. doi: 10.1016/j.jmb.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regan L, Clarke ND. A Tetrahedral Zinc(Ii)-Binding Site Introduced into a Designed Protein. Biochemistry. 1990;29(49):10878–10883. doi: 10.1021/bi00501a003. [DOI] [PubMed] [Google Scholar]
- 32.Clarke ND, Yuan SM. Metal Search - a Computer-Program That Helps Design Tetrahedral Metal-Binding Sites. Proteins-Structure Function and Genetics. 1995;23(2):256–263. doi: 10.1002/prot.340230214. [DOI] [PubMed] [Google Scholar]
- 33.Salgado EN, Radford RJ, Tezcan FA. Metal-directed protein self-assembly. Acc Chem Res. 2010;43(5):661–672. doi: 10.1021/ar900273t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brodin JD, Ambroggio XI, Tang C, Parent KN, Baker TS, Tezcan FA. Metal-directed, chemically tunable assembly of one-, two- and three-dimensional crystalline protein arrays. Nat Chem. 2012;4(5):375–382. doi: 10.1038/nchem.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salgado EN, Faraone-Mennella J, Tezcan FA. Controlling protein-protein interactions through metal coordination: Assembly of a 16-helix bundle protein. Journal of the American Chemical Society. 2007;129(44) doi: 10.1021/ja075261o. 13374-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Der BS, Machius M, Miley MJ, Mills JL, Szyperski T, Kuhlman B. Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer. Journal of the American Chemical Society. 2012;134(1):375–385. doi: 10.1021/ja208015j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richter F, Leaver-Fay A, Khare SD, Bjelic S, Baker D. De Novo Enzyme Design Using Rosetta3. Plos One. 2011;6(5) doi: 10.1371/journal.pone.0019230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zanghellini A, Jiang L, Wollacott AM, Cheng G, Meiler J, Althoff EA, Rothlisberger D, Baker D. New algorithms and an in silico benchmark for computational enzyme design. Protein Sci. 2006;15(12):2785–2794. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popovic Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bower MJ, Cohen FE, Dunbrack RL. Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: A new homology modeling tool. Journal of Molecular Biology. 1997;267(5):1268–1282. doi: 10.1006/jmbi.1997.0926. [DOI] [PubMed] [Google Scholar]
- 42.Shapovalov MV, Dunbrack RL. A Smoothed Backbone-Dependent Rotamer Library for Proteins Derived from Adaptive Kernel Density Estimates and Regressions. Structure. 2011;19(6):844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164(1):49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 44.Wittekind M, Mueller L. Hncacb, a High-Sensitivity 3d Nmr Experiment to Correlate Amide-Proton and Nitrogen Resonances with the Alpha-Carbon and Beta-Carbon Resonances in Proteins. Journal of Magnetic Resonance Series B. 1993;101(2):201–205. [Google Scholar]
- 45.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. Journal of Biomolecular Nmr. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 46.Goddard TD, Kneller DG. SPARKY 3. not published;UCSF. [Google Scholar]
- 47.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods in Molecular Biology. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 48.Cornilescu G, Marquardt JL, Ottiger M, Bax A. Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. Journal of the American Chemical Society. 1998;120(27):6836–6837. [Google Scholar]
- 49.Mulder FAA, Schipper D, Bott R, Boelens R. Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. Journal of Molecular Biology. 1999;292(1):111–123. doi: 10.1006/jmbi.1999.3034. [DOI] [PubMed] [Google Scholar]
- 50.Otten R, Chu B, Krewulak KD, Vogel HJ, Mulder FAA. Comprehensive and Cost-Effective NMR Spectroscopy of Methyl Groups in Large Proteins. Journal of the American Chemical Society. 2010;132(9):2952–2960. doi: 10.1021/ja907706a. [DOI] [PubMed] [Google Scholar]
- 51.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10(10):659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vijaykumar S, Bugg CE, Cook WJ. Structure of Ubiquitin Refined at 1.8 a Resolution. Journal of Molecular Biology. 1987;194(3):531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 53.Imada K, Matsunami H, Yamane M, Samatey AF, Nagashima S, Namba K. PDB ID: 2D4X. Structure of the bacterial flagellar hook-filament junction. 2005 [Google Scholar]
- 54.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399(3):361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Namuswe F, Berg JM. Secondary interactions involving zinc-bound ligands: roles in structural stabilization and macromolecular interactions. J Inorg Biochem. 2012;111:146–149. doi: 10.1016/j.jinorgbio.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Page MI, Jencks WP. Entropic Contributions to Rate Accelerations in Enzymic and Intramolecular Reactions and Chelate Effect. Proceedings of the National Academy of Sciences of the United States of America. 1971;68(8) doi: 10.1073/pnas.68.8.1678. 1678-&. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cunningham BC, Bass S, Fuh G, Wells JA. Zinc mediation of the binding of human growth hormone to the human prolactin receptor. Science. 1990;250(4988):1709–1712. doi: 10.1126/science.2270485. [DOI] [PubMed] [Google Scholar]
- 58.Somers W, Ultsch M, De Vos AM, Kossiakoff AA. The X-ray structure of a growth hormone-prolactin receptor complex. Nature. 1994;372(6505):478–481. doi: 10.1038/372478a0. [DOI] [PubMed] [Google Scholar]
- 59.Radford RJ, Brodin JD, Salgado EN, Tezcan FA. Expanding the utility of proteins as platforms for coordination chemistry. Coordination Chemistry Reviews. 2011;255(7–8):790–803. [Google Scholar]
- 60.Lee YM, Lim C. Physical basis of structural and catalytic Zn-binding sites in proteins. J Mol Biol. 2008;379(3):545–553. doi: 10.1016/j.jmb.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 61.Stranges PB, Kuhlman B. A comparison of successful and failed protein interface designs highlights the challenges of designing buried hydrogen bonds. Protein Sci. 2012 doi: 10.1002/pro.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang PS, Love JJ, Mayo SL. A de novo designed protein protein interface. Protein Sci. 2007;16(12):2770–2774. doi: 10.1110/ps.073125207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jacak R, Leaver-Fay A, Kuhlman B. Computational protein design with explicit consideration of surface hydrophobic patches. Proteins. 2012;80(3):825–838. doi: 10.1002/prot.23241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.