Abstract
The Drosophila syncytial embryo is a powerful developmental model system for studying dynamic coordinated cytoskeletal rearrangements. Confocal microscopy has begun to reveal more about the cytoskeletal changes that occur during embryogenesis. Total Internal Reflection Fluorescence (TIRF) microscopy provides a promising new approach for the visualization of cortical events with heightened axial resolution. We have applied TIRF microscopy to the Drosophila embryo to visualize cortical microtubule and actin dynamics in the syncytial blastoderm. Here we describe the details of this technique, and report qualitative assessments of cortical microtubules and actin in the Drosophila syncytial embryo. In addition, we identified a peak of cortical microtubules during anaphase of each nuclear cycle in the syncytial blastoderm, and using images generated by TIRF microscopy we quantitatively analyzed microtubule dynamics during this time.
Keywords: Total Internal Reflection Fluorescence (TIRF) microscopy, Drosophila syncytial development, time-lapse imaging, cytoskeleton, actin, microtubules
Introduction
Temporal and spatial control of cytoskeletal organization is critical for many cellular processes. Cytoskeletal dynamics have been studied primarily during cell migration and cytokinesis using cell culture and yeast (reviewed in (Wehrle-Haller and Imhof, 2003; Pearson and Bloom, 2004; Moseley and Goode, 2006; Mattila and Lappalainen, 2008). However, in vivo cytoskeletal dynamics are not well understood during the development of a complex organism. The Drosophila syncytial blastoderm is an excellent model system for studying dynamic, coordinated cytoskeletal rearrangements in a complex, in vivo system due to the synchronous, cell cycle dependent changes of both the microtubule and actin networks.
In early Drosophila development, the first 14 nuclear divisions occur without cytokinesis (reviewed in (Sullivan and Theurkauf, 1995, Schejter, 1993). The initial nuclear divisions occur in the central cytoplasm, and during nuclear cycles 7, 8 and 9 the nuclei migrate to the cortex. The cortical migration of the nuclei is completed by interphase of nuclear cycle 10, resulting in the syncytial blastoderm. During the four subsequent rounds of synchronous, cortical nuclear divisions, the actin and microtubule cytoskeletons undergo dramatic, cyclic rearrangements. In interphase, actin is organized into “caps” cortical to each nucleus that appear to help maintain even nuclear spacing in the blastoderm (Fig 1B) (reviewed in (Schejter and Wieschaus, 1993)). The centrosome associated with each nucleus is located between the nucleus and the actin cap, and the associated microtubules extend into the embryo, enveloping each nucleus. As the nuclei progress into prophase, microtubules reorganize into spindles. The actin caps expand in a SCAR-and Arp2/3-dependent manner (Stevenson et al., 2002; Zallen et al., 2002). Ultimately, the actin extends into the embryo during late prophase and metaphase to form pseudocleavage furrows. The extension of the furrows requires the Adenomatous Polyposis Coli (APC) family protein APC2 and the formin Diaphanous (Webb et al., 2009). As viewed from the surface of the embryo, the tops of the pseudocleavage furrows appear as a tight actin rings surrounding each spindle. The furrows function as physical barriers between nuclei to prevent spindle collisions (Fig 1B). As the spindle poles begin to separate during anaphase, the actin furrows regress. During telophase, the actin caps begin to reform, while the microtubule spindles complete nuclear division and the microtubules begin to reorient into the interior of the embryo.
Figure 1. Schematics illustrating the setup for TIRF microscopy and cytoskeletal organization in the syncytial embryo.
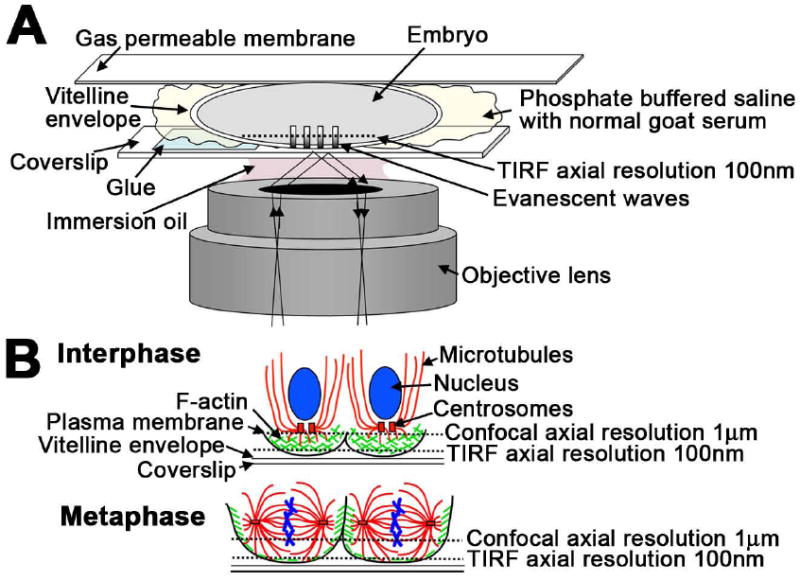
(A) Diagram illustrating the setup for TIRF imaging. Approximately one third of the length of the embryo was glued to the coverslip and mounted on a gas permeable membrane in phosphate buffered saline with normal goat serum. (B) The cytoskeletal organization of the syncytial embryo during interphase and metaphase. At interphase, actin is organized into caps cortical to each nucleus, and microtubules extend around each nucleus and into the embryo. At metaphase, actin has reorganized into rings that extend to form the actin pseudocleavage furrows and microtubules have reorganized into the spindle (modified from Webb et al., 2009).
The most commonly used approach for understanding cytoskeletal organization during Drosophila embryonic development has been fixing wild type and mutant embryos and visualizing actin and microtubules using probes and antibodies. This approach was instrumental in determining the cyclic changes in actin and microtubule organization during syncytial development, and in identifying many of the proteins involved in these changes (Sullivan et al., 1993; Rothwell et al., 1998; Afshar et al., 2000; Zallen et al., 2002; Webb et al., 2009). Live imaging of cytoskeletal changes using confocal microscopy has begun to reveal more about dynamic events during embryogenesis, such as actin cap expansion and microtubule spindle elongation in syncytial embryos (Foe et al., 2000; Sharp et al., 2000a; Stevenson et al., 2001), and filopodial dynamics in dorsal closure (Jankovics and Brunner, 2006; Gates et al., 2007). However, our understanding of cytoskeletal dynamics in a living embryo is limited.
To better understand the coordinated cortical cytoskeletal rearrangements in the Drosophila syncytial embryo, we applied Total Internal Reflection Fluorescence (TIRF) microscopy to the Drosophila embryo to visualize cortical microtubule and actin dynamics. While spinning disc confocal microscopy allows for an axial resolution of approximately one micron (Egner et al., 2002), TIRF microscopy is a powerful imaging technique that significantly improves the signal-to-noise ratio by capturing information within 100 nm of the cortex and consequently improves spatial resolution (reviewed in (St. Croix, 2005)). Furthermore, as a result of improved spatial resolution, this technique allows for a decrease in the exposure time during image acquisition, thus improving temporal resolution and potentially reducing photon-induced damage to the embryo. This report details the technique for using TIRF microscopy to examine microtubules and actin, and for quantitatively analyzing microtubule dynamics in Drosophila syncytial embryos. In addition to the analysis of cytoskeletal dynamics, TIRF microscopy can be used to examine other dynamic processes at the cortex of the embryo, including the organization and polarization of the plasma membrane (Mavrakis et al., 2009) and vesicle trafficking (Riggs et al., 2003; Albertson et al., 2008; Sokac and Wieschaus, 2008), with high spatial and temporal resolution. Thus, the application of this technique to the Drosophila embryo extends the utility of this valuable genetic and developmental model system.
Results and Discussion
TIRF imaging setup
Live imaging of Drosophila embryos using confocal microscopy typically entails adhering the entire length of the dechorionated embryo to a coverslip and covering it with halocarbon oil to prevent desiccation and allow for gas exchange (Hazelrigg, 2000). With TIRF microscopy, the evanescent excitation wave is propagated at the interface between two materials with different refractive indices (RI) and results in the excitation of fluorophores within 100 nm of the interface (reviewed in (Thompson and Lagerholm, 1997; Axelrod, 2001)). For TIRF imaging of cultured cells, the interface is typically the coverslip (RI = 1.52) and the cells (RI ≌ 1.36) or the aqueous media surrounding the cells (RI = 1.33). The evanescent wave field only penetrates the cortex of cells that are very close to the coverslip. Thus, to visualize cortical dynamics in the embryo with TIRF microscopy, the vitelline membrane of the embryo must be in contact with the coverslip without an intermediate layer of adhesive between the coverslip and embryo, and the embryos must be mounted in an aqueous media. To achieve this, only one third of each dechorionated embryo was affixed to a coverslip (Fig 1A and Fig S1). This mounting scheme decreases the movement of the embryo during imaging, while allowing the central portion of the embryo to be accessible to the evanescent wave. Further, the embryos were mounted in phosphate buffered saline with 1% normal goat serum to prevent desiccation (Fig 1A), as the halocarbon oil typically used has a high refractive index (RI = 1.52) (reviewed in (Axelrod, 2001)). The coverslip was then mounted on a dish with a gas permeable membrane, which allowed for gas exchange and slightly flattened the embryo, increasing the imaging surface area (Fig 1A and Fig S1). The embryos were imaged on an inverted microscope where surface tension held the coverslip in place. These modifications in the preparation of embryos for live imaging were instrumental in our ability to successfully image embryos using TIRF microscopy.
Cortical actin dynamics visualized with confocal and TIRF microscopy
We examined actin dynamics with confocal and TIRF microscopy using a GFP-tagged actin binding protein, MoesinGFP (Edwards et al., 1997), a commonly used reagent for visualizing actin dynamics during Drosophila development (Kiehart et al., 2000; Riggs et al., 2007; Cao et al., 2008). MoesinGFP specifically labels F-actin (Edwards et al., 1997; Cao et al., 2008). The size and intensity of the actin objects observed with both types of microscopy compared to published accounts of individual fluorescent filaments in vitro (Amann and Pollard, 2001) suggests that the objects could be individual actin filaments and not bundles of filaments. However, neither imaging approach can resolve individual 6 nm thick actin filaments. For the sake of convenience in discussing the data, the elongated discrete actin objects we observed are called actin filaments throughout this report.
Following nuclear migration, the actin organizes into an actin cap cortical each nucleus in the syncytial blastoderm (Fig 1B). As the cell cycle progresses, actin caps expand, and actin rings and pseudocleavage furrows are formed around each nucleus. Both the normal development and the expansion of the actin cap require the branched actin nucleator Arp2/3, and its activator SCAR (Stevenson et al., 2002; Zallen et al., 2002), while the extension of the pseudocleavage furrows requires the unbranched actin nucleator, the formin Diaphanous (Afshar et al., 2000; Webb et al., 2009). Time-lapse imaging has shown the general dynamics of actin cap expansion (Stevenson et al., 2001; Foe et al., 2000). We have begun to evaluate actin organization and dynamics using both confocal and TIRF microscopy.
Using confocal and TIRF microscopy, cortical actin was visualized throughout the nuclear division cycle. While some filaments can be seen with both types of microscopy, there is improved spatial and temporal resolution with TIRF imaging. In comparison to confocal microscopy, with TIRF microscopy actin caps in interphase cycle 10 appear less dense (Fig 2A, B, respectively), allowing for the observation of individual filaments in some places in the interior of the cap, as well as at the edge of the cap (Fig 2B, B’ red and green arrowheads, respectively). In the confocal images, only individual filaments at the edge of the cap, where the actin is less dense, can be clearly resolved (Fig 2A, A’ green arrowheads). As the actin caps expand during prophase, we continued to observe filaments in the interior of the cap using TIRF (Fig 2D, D’ red arrowheads), and filaments at the edge of the cap with both confocal and TIRF microscopy (Fig 2C, C’ and 2D, green arrowheads). Because of the inability of TIRF microscopy to resolve individual 6 nm thick filaments, we cannot determine whether the actin filaments are branched or unbranched.
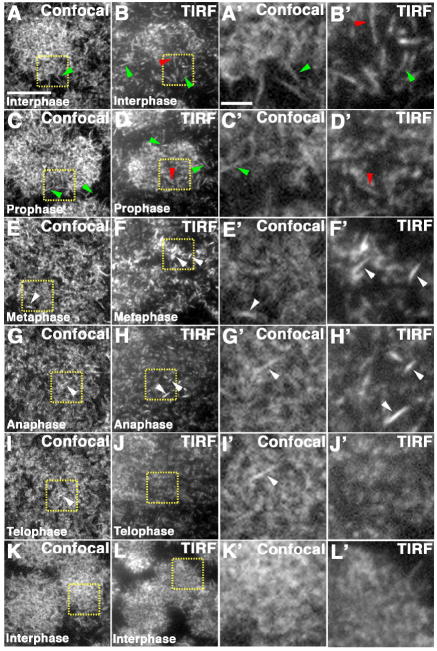
Figure 2. Cortical actin visualized with confocal and TIRF microscopy.
(A-L) Cortical actin in wild type cycle 10 syncytial embryos expressing MoesinGFP visualized with confocal (A, C, E, G, I, K) and TIRF microscopy (B, D, F, H, J, L). Yellow boxes indicate the area shown in higher magnification in A’-L’. Actin filaments can be seen in the interior of the actin cap with TIRF (B, B’, D, D’, red arrowheads), and at the edge of the actin cap with TIRF and confocal (A, A’, B, B’, C, C’, D, D’, green arrowheads) during interphase and prophase. In metaphase, anaphase, and telophase actin filaments can also be seen at the cortex (E-I’, white arrowheads). During anaphase and telophase, the actin is more evenly distributed (G, G’, H, H’, I, I’, J, J’). Invaginated furrows cannot be visualized with confocal at this focal plane and are never observed with TIRF microscopy as they extend outside of the TIRF field. Scale bar is 10 μm for A-L and 2 μm for A’-L’.
During metaphase of cycle 10, the actin cap is completely expanded and a thin, relatively uniform actin layer is maintained at the cortex ((Karr and Alberts, 1986), Fig 2E, F). More cortical actin filaments were resolved using TIRF microscopy, as compared to confocal (Fig 2E, E’, F, F’, white arrowheads). As expected, when actin rings and pseudocleavage furrows formed during metaphase, some of the actin remained at the cortex (Fig 2E, F). The invagination of the membrane in the pseudocleavage furrows beyond the axial resolution of TIRF microscopy resulted in the absence of actin rings, the tops of the pseudocleavage furrows, from the TIRF field (Fig 2F). This was clearly seen in cycle 12 metaphase embryos that have more robust furrows (Fig S2). In the TIRF images, we observed actin voids in the areas where the actin rings and pseudocleavage furrows had formed, but the actin rings themselves were not detected (Fig S2C, C’ C”, arrowheads). During anaphase and telophase, the cortical actin appears more evenly dispersed before the formation of new caps in the next interphase (Fig 2G-L).
Our initial findings demonstrate the utility of TIRF microscopy for resolving cortical actin structures in the syncytial blastoderm embryo (Fig 2 and Fig S2). In the future, this imaging approach will allow for a direct assessment of cortical actin dynamics during the cyclic rearrangements of cortical actin.
Cortical microtubule orientation and dynamics visualized with confocal and TIRF microscopy
During interphase when actin caps organize cortical to each nucleus, microtubules form a basket-like structure around each nucleus (Fig 1B). As the embryo progresses to metaphase, the microtubules reorganize into mitotic spindles. It has been shown that loss of proteins known to affect microtubules and spindles, including Centrosomin, a core centrosome component (Vaizel-Ohayon and Schejter, 1999), and EB1 (Webb et al., 2009), affect cortical actin organization. In addition, anaphase microtubules appear to play a role in actin pseudocleavage furrow organization in the subsequent cell cycle (Riggs et al., 2007). Thus, understanding microtubule organization and dynamics (activity of microtubules over time) in the syncytial embryo is necessary to our understanding of how microtubule and actin rearrangements are coordinated.
The mitotic spindle is composed of three populations of microtubules. The central spindle contains kinetochore and interpolar microtubules that extend from the spindle poles toward the chromosomes (Sharp et al., 2000b; Kwon and Scholey, 2004). The majority of these microtubules make up the spindle proper and extend into the area of the chromosomes, while some of the interpolar microtubules extend outside the area of the chromosomes. In the syncytial embryo, these interpolar microtubules extend toward the plasma membrane (cortical), away from the plasma membrane, and into the lateral cytoplasm, in an outer sphere surrounding the area of the chromosomes. The third population of microtubules are astral, non-central spindle microtubules, which extend from the spindle poles away from the chromosomes.
In cell culture and other systems, such as C. elegans, the astral and cortical interpolar microtubules are involved in establishing the cytokinetic furrow during anaphase (reviewed in (Glotzer, 2003; Rodriguez et al., 2003)). While the actin pseudocleavage furrow in the syncytial embryo forms during prophase and metaphase and not anaphase like conventional cytokinesis, it appears that microtubules in anaphase are necessary for the establishment of the actin furrows in the subsequent nuclear division cycle (Riggs et al., 2007). Thus, we hypothesized that astral and cortical interpolar microtubules are involved in organizing the actin cytoskeleton in the syncytial embryo. Studies that have examined microtubule organization and dynamics in the syncytial embryo have focused primarily on the dense population of microtubules in the spindle proper (Sharp et al., 2000a; Rogers et al., 2002; Kwon and Scholey, 2004). These microtubules are critical for proper nuclear division and it seems unlikely that they function in the organization of the actin cytoskeleton. The density of the microtubules in the central spindle makes it difficult to visualize astral and cortical interpolar microtubules. However, we have recently used confocal microscopy coupled with deconvolution to visualize these microtubules in fixed embryos (Webb et al., 2009).
To examine astral and cortical interpolar microtubule dynamics in live embryos, we employed embryos expressing a GFP-tagged microtubule binding protein, ZeusGFP (Morin et al., 2001). We observed a significant peak of cortical microtubules in late anaphase of cycles 10-13, as visualized with both confocal and TIRF microscopy (Fig. 3C, G, I, Fig S3, Movie S1 and data not shown). With TIRF microscopy, the cortical microtubules often appeared as individual spots during this time period (Fig. 3G, arrowhead). These spots were interpreted as microtubule ends that are oriented perpendicular to the cortex (Fig 3J, arrowhead). Time-lapse imaging revealed that these spots can extend into lines, indicating that the microtubules can extend parallel to the cortex (Fig. 3G, 3J arrow, Fig S3, Movie S1). These microtubule spots and lines were also visualized with confocal microscopy (Fig 3C, arrowhead, arrow), but most of the parallel microtubules appear to be part of the anaphase spindle proper (Fig. 3C, circle, as determined by correlation to subcortical images of the spindle not shown). Due to the 1 μm axial resolution of spinning disc confocal microscopy, we were unable to discern which microtubules were truly cortical from those that are subcortical. In comparison, the 100 nm axial resolution of TIRF microscopy ensures that the microtubules visualized are indeed cortical. These microtubules may be both astral and cortical interpolar microtubules.
Figure 3. Cortical microtubules visualized with confocal and TIRF microscopy.
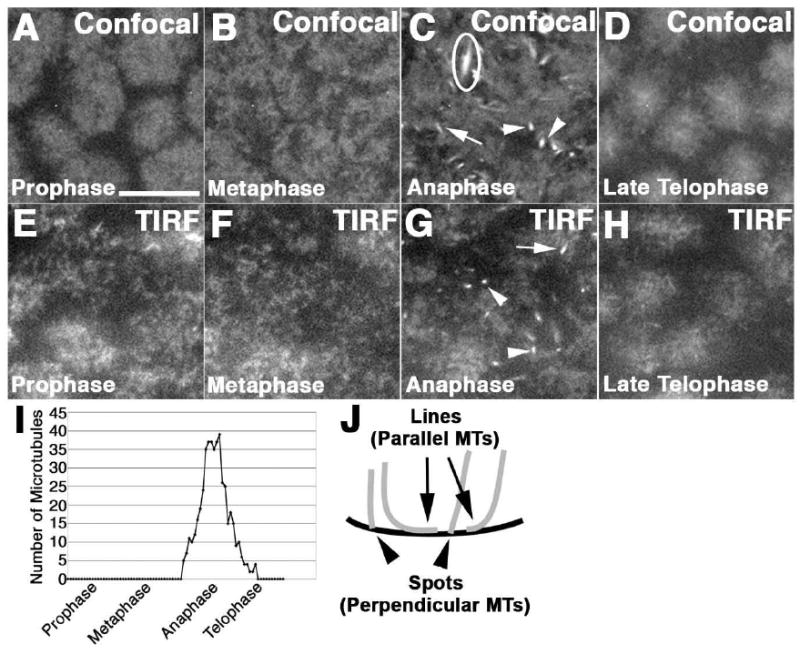
Wild type cycle 12 syncytial embryos expressing ZeusGFP visualized with confocal (A-D) and TIRF (E-H) microscopy, display a peak of cortical microtubules during anaphase (C,G). (C) Circle indicates parallel microtubules that are part of the spindle proper. (I) Quantification of the total number of microtubules visualized every 5 seconds in a 30 μm2 area. This is a representative example of the peak of cortical microtubules in a single cycle 12 embryo. (J) The microtubules oriented parallel to the cortex appear as lines (arrows), and those perpendicular to the cortex appear as spots (arrowheads), also indicated in C and G. Scale bar is 10μm.
Differences in microtubule orientation and dynamics can be assessed using TIRF microscopy
To begin to understand the changes in the cytoskeleton in the syncytial embryo, we asked whether TIRF microscopy could be used to evaluate microtubule orientation and dynamics in wild type embryos. We assessed microtubule orientation, pattern of behavior, cortical lifetime and persistence of cortical position relative to other microtubules (relative persistence).
Microtubule orientation was assayed at two time points within the peak of cortical microtubules in wild type embryos. We determined that at two time points during the peak of cortical microtubules in cycle 12 (Fig 4A), the majority (71%) of microtubules were oriented perpendicular to the cortex (spots) (Fig 4A, D, arrowheads), while only 29% were oriented parallel to the cortex (lines) (Fig 4A, D, arrows). Tracking the behavior of these microtubules during their cortical lifetime (residence in the TIRF field), showed three different microtubule behavior patterns: microtubules remained as spots (spot-only microtubules), transitioned from spots to lines (spot-line microtubules), or were only lines (line-only microtubules). We did not observe microtubules transitioning from lines to spots. In wild type embryos, 65% of the microtubules were spot-only microtubules, and 29% of the microtubules were spot-line microtubules (Fig 4E, Fig S3, Movie S1). It was rare for microtubules to behave as line-only microtubules (Fig 4E, 6%). The average cortical lifetime was 44 seconds in wild type embryos (Fig 4F, Fig S4A, black bars). When we analyzed the cortical lifetime of the microtubules based on their behavior pattern, we found no difference in the cortical lifetime between spot-only and spot-line microtubules (Fig 4G, black bars). The frequency of line-only microtubules in wild type embryos was too low to compare using statistical measures.
Figure 4. Cortical microtubule dynamics assessed with TIRF microscopy.
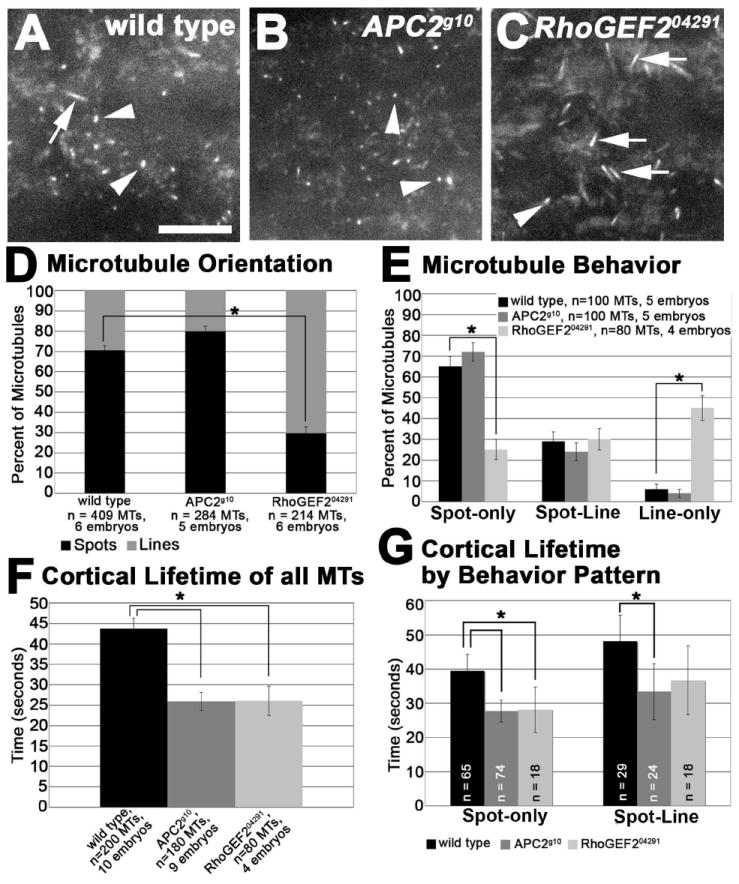
(A-C) Microtubules oriented perpendicular to the cortex (spots, arrowheads) and parallel to the cortex (lines, arrows) in cycle 12 anaphase wild type (A), APC2g10 (B), and RhoGEF204291 (C) embryos expressing ZeusGFP. (D) Quantification of the orientation of the microtubules. (E) Quantification of the behavior of the microtubules. (F) Quantification of the average cortical lifetime of all the microtubules analyzed for each genotype. (G) Quantification of the cortical lifetime for the spot-only and spot-line microtubule behavior patterns. Scale bar is 10 μm. * is p<0.001.
Changes in cortical microtubule orientation, behavior, and cortical lifetime may reflect a modulation of microtubule dynamic instability. Microtubules may rapidly grow thus appearing as lines, or may have stopped growing, thus appearing as spots throughout their cortical lifetime. Alternatively, spindle pole movement and the consequent movement of the cortical microtubules extending from those poles may result in changes in microtubule orientation and dynamics.
Syncytial embryos lack post-translationally modified, stabilized microtubules (Wolf et al., 1988; Warn et al., 1990; Webb et al., 2009). However, as they reside in the TIRF field during anaphase, microtubules may be relatively stable at the cortex. This could be due to a physical association with the cortex, or through other microtubule end binding proteins that prevent their growth and shrinkage. To begin to understand whether microtubules in the TIRF field are stabilized, we asked whether spot-only microtubules move in relation to each other. We termed this property relative persistence. We predicted that if spot-only microtubules in the TIRF field are stabilized, they will retain their position relative to neighboring spot-only microtubules in the X-Y field. Because the embryonic cortex and the entire embryo can move over time, we predicted that neighboring stabilized microtubules would translate en masse. Three to five spot-only microtubules within 55 μm2 circular areas (patches) in wild type embryos were analyzed and their positions were tracked relative to each other and to the edges of the field. Spot-only microtubules persist in the same configuration relative to each other during their cortical lifetimes (Fig 5A, 90% of the RMS dispersion values are less than measurement error of 2 pixels, gray box). The coordinated movement of microtubules within a patch suggests that these microtubules are stabilized at or near the cortex.
Figure 5. Relative persistence of spot-only microtubules.
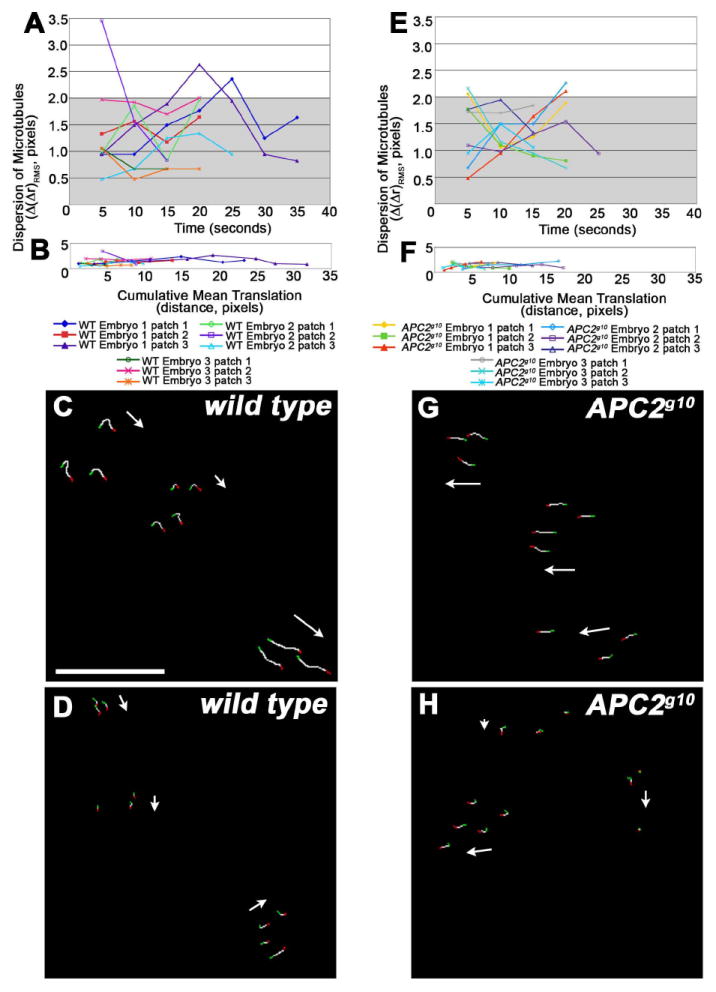
(A, E) Quantification of the dispersion of wild type (A) and APC2g10 (E) spot-only microtubules over time in three patches for each of three embryos. This is a measure of the relative persistence of microtubules. Dispersion values ≤ 2 pixels were considered to be within the measurement error (gray box in A,E, see Methods). (B, F) Quantification of the dispersion of wild type (B) and APC2g10 (F) spot-only microtubules over cumulative mean translation, as a measure of the patch distance moved. (C, D, G, H) Tracking patterns for microtubule movement in wild type (C, D) and APC2g10 (G, H). Green pixels indicate the point at which the microtubules enter the TIRF field, white pixels the path of microtubule movement, and red pixels the point at which the microtubules leave the TIRF field. White arrows indicate the direction of patch movement. Patches can translate in the same direction (C, G) or in different directions (G, H).
Individual patches of microtubules in wild type embryos exhibit translation ranging from 4.5 to 32 pixels (0.48 to 3.39 μm) from the initial location (Fig 5B). Examination of individual patches within the same embryo revealed that patches move independently of each other; patches may translate together (Fig 5C) or in different directions (Fig 5D). There are two possible explanations for the movement of the patches of microtubules. Patch movement may be due to the movement of the cortex to which these microtubules are physically attached. We have visualized general cortical actin movement in syncytial embryos using confocal microscopy (data not shown). Alternatively, the movement of the cortical microtubules may be due to the movement of the subcortical spindle poles, and the consequent movement of the cortical microtubule ends. In the latter model, patches that consist of microtubules originating from different poles should not exhibit relative persistence. If this is the case, it suggests that the microtubules in the patches we examined originate from a single pole. While our data cannot distinguish between these models for patch translation, it is consistent with the hypothesis that spot-only microtubules in the TIRF field are relatively stabilized and are neither growing nor shrinking.
Microtubule dynamics in syncytial embryos with different genotypes can be evaluated with TIRF imaging
To demonstrate that differences in microtubule orientation and dynamics can be quantitatively assessed using TIRF microscopy, we examined and analyzed microtubules in embryos null for APC2 and RhoGEF2. Both the APC family proteins and the Rho specific guanine nucleotide exchange factor, RhoGEF2, have been shown to associate with microtubules. RhoGEF2 interacts with microtubules via the microtubule end-binding protein EB1 (Rogers et al., 2004), while APC family proteins can interact with microtubules through a variety of mechanisms, including an interaction with EB1 (Nathke, 2004; Rogers et al., 2004; Wen et al., 2004; Kita et al., 2006). Both APC2 and RhoGEF2 are present in the syncytial embryo (McCartney et al., 1999; Padash Barmchi et al., 2005). However, APC2 does not have the EB1 binding domain and does not bind EB1 directly (McCartney et al., 1999; Webb et al., 2009). APC2g10 null mutant embryos have defects in actin pseudocleavage furrow extension, but no apparent defects in microtubule spindle dynamics, or astral microtubule density or distribution (Webb et al., 2009). Loss of function of RhoGEF2, in RhoGEF204291 null mutant embryos, results in areas of reduced actin and areas of increased actin accumulation in the actin ring (Webb et al., 2009). Microtubules have not been analyzed in RhoGEF2 mutant syncytial embryos.
We observed a peak of cortical microtubules during anaphase in both APC2 and RhoGEF2 mutants as in wild type (data not shown). Within this peak, we examined microtubule orientation, behavior, cortical lifetime and relative persistence in the mutant embryos. Embryos lacking RhoGEF2 exhibited a striking increase in the proportion of microtubule lines (Fig 4C, arrows, Movie S2) compared to wild type (Fig 4A, arrow, Movie S1) and APC2g10 (Fig 4B, Movie S3) embryos. Approximately 70% of the microtubules were lines in RhoGEF2 mutants, in comparison to 30% in wild type and 20% in APC2 mutant embryos (Fig 4D, p<0.001). The microtubule behavior also differs in RhoGEF2 embryos. Over 60% of the cortical microtubules analyzed in wild type and APC2g10 embryos were spot-only microtubules, while only 25% of the microtubules in the RhoGEF2 mutant embryos were spot-only microtubules (Fig. 4A-C arrowheads, E, p<0.001). Greater than 40% of the microtubules in the RhoGEF2 mutants were line-only microtubules, in comparison to 6% in wild type and 4% in APC2 mutants (Fig 4E, p<0.001). Interestingly, approximately 25% of microtubules were spot-line microtubules in all three genotypes (Fig. 4E).
We evaluated whether these differences in microtubule orientation and behavior in RhoGEF2 mutants were associated with a change in the cortical lifetime of the microtubules. Both RhoGEF2 and APC2 mutants have an average cortical lifetime of approximately 26 seconds (s.d. ± 3.6 and ± 2.2 seconds, respectively), compared to 44 seconds (s.d. ± 2.6 seconds) in wild type embryos (Fig 4F, p<0.001, Fig S4A). To understand whether the difference in cortical lifetime is correlated with the behavior of the microtubules, we analyzed the cortical lifetime for the spot-only and spot-line microtubules. We did not determine the cortical lifetime for the line-only microtubules because this behavior is very rare in both wild type and APC2g10 embryos, and thus the sample size in these genotypes was very small. This analysis revealed that the difference in the cortical lifetime between wild type and the two mutants is not due to differences in behavior pattern (i.e. spot-only and spot-line microtubules, Fig 4G). The cortical lifetime of wild type spot-only microtubules is significantly longer than that of the mutants (Fig 4G, p<0.001, Fig S4B). The cortical lifetime of wild type spot-line microtubules is significantly longer than that of APC2g10 mutants (p<0.001), but similar to that of RhoGEF204291 mutants (p=0.06) (Fig 4G, Fig S4C). The increased variability in cortical lifetimes of RhoGEF204291 mutant spot-line microtubules resulted in no statistically significant change from wild type (Fig 4G). Similar to wild type, the cortical lifetime of microtubules in APC2 (Fig 4G, dark gray bars) or RhoGEF2 (Fig 4G, light gray bars) mutant embryos is constant regardless of behavior pattern.
Finally, we assessed spot-only relative persistence in APC2g10 mutant embryos. We did not observe enough spot-only microtubules in RhoGEF2 mutant embryos to assess this property. In APC2 null embryos, spot-only microtubules within a patch persist in the same configuration relative to each other, similar to wild type (Fig 5E, 88% of RMS dispersion values are less than measurement error of 2 pixels (gray box)). It appears that the maximum translation of the patches is less in APC2 mutants as compared to wild type, with translation ranging from 4 to 17 pixels (0.42 to 1.80 μm) (Fig 5F). Similar to wild type, individual patches of microtubules within the same APC2 mutant embryo translate independently (Fig 5G, H).
These assays demonstrate the utility of TIRF microscopy for evaluating cortical microtubule orientation, behavior, cortical lifetime, and relative persistence. The differences in microtubule orientation and dynamics that we described for APC2 and RhoGEF2 mutants are intriguing and further study will reveal the biological significance of these differences.
Summary
The use of TIRF microscopy in the Drosophila embryo is a powerful tool for understanding cortical dynamics with improved axial resolution, resulting in improved spatial and temporal resolution. Using this technique, we have begun to evaluate the organization of cortical actin during the actin cycle in the syncytial blastoderm embryo. We have also identified a peak of cortical microtubules during anaphase. In addition, our analysis of wild type, APC2 mutant and RhoGEF2 mutant embryos, indicates that TIRF imaging can be used to evaluate changes in microtubule orientation and dynamics.
Microtubules cycle between periods of slow growth and rapid disassembly at their plus ends, which is referred to as dynamic instability (Desai and Mitchison, 1997). This property provides for the spatial and temporal flexibility that allows for the rapid remodeling of the microtubule cytoskeleton. Dynamic instability can be modulated by a host of microtubule associated proteins that can promote catastrophe (rapid shrinkage) or stabilize plus ends and prevent disassembly, for example. The cortical microtubules in the syncytial embryo appear to be a diverse population; the majority of these microtubules enter the cortical region perpendicular to the cortex (spot-only microtubules) and become relatively stabilized during their cortical lifetimes. Others appear to slide along the cortex over time (spot-line microtubules). The transitioning from a spot to a line suggests microtubule growth at the plus end. However, using TIRF microscopy we are unable to visualize an entire microtubule from the pole to the cortical tip. Consequently, we cannot distinguish between growth and movement of the microtubules due to pole separation. Because established dynamic parameters of microtubules measure the rates of growth and shrinkage, we are unable to apply these quantitative analyses to this system. Fluorescent speckle microscopy (Kwon and Scholey, 2004) in combination with TIRF may help to distinguish between growth and movement, and could be used to assess more established dynamic parameters in this system.
In other systems, astral and interpolar microtubules have been implicated in a number of processes during mitosis including spindle positioning, delivery and localization of proteins, and in the formation and progression of the cytokinetic furrow (Goode et al., 2000; Rodriguez et al., 2003; D’Avino et al., 2005). In the syncytial embryo, anaphase microtubules are implicated in the formation of pseudocleavage furrows in the next nuclear cycle (Riggs et al., 2007). Interestingly, the timing of this microtubule requirement is the same as for conventional cytokinesis (Riggs et al., 2007). It has been postulated that overlapping plus ends of astral microtubules originating from neighboring centrosomes during anaphase establish the site of the future pseudocleavage furrows (Riggs et al., 2007). One mechanism by which microtubules appear to contribute to furrow formation is through the localization of components of the recycling endosome that are required for membrane recruitment and actin remodeling in the early stages of furrow formation (Riggs et al., 2003). Taken together this suggests that the astral and cortical interpolar microtubules we find in the TIRF field may be involved in the establishment of pseudocleavage furrows. Further study using multicolor TIRF microscopy will help to elucidate the interdependence of the actin and microtubule cytoskeletons, and the role of other associated proteins in their organization and dynamics. Defining the normal behavior and dynamic properties of these cortical microtubules as assessed by their orientation, behavior, cortical lifetime and persistence is a first step toward a comprehensive understanding of the interrelationships between microtubules and the factors required for furrow formation.
Experimental Procedures
Fly stocks
The following stocks were used: MoesinGFP (Edwards et al., 1997), ZeusGFP (Morin et al., 2001), ZeusGFP APC2g10/TM6 Tb (generated from APC2g10, (McCartney et al., 2006)) and FRTG13 RhoGEF204291/CyO; ZeusGFP (generated from FRTG13 RhoGEF204291, (Hacker and Perrimon, 1998)). Mutant embryos maternally FRTG13 RhoGEF204291; ZeusGFP were generated using the FLP/FRT/DFS technique (Chou and Perrimon, 1996). Genotypes indicated are maternal, as zygotic transcription has not significantly begun in the syncytial embryo.
Embryo Preparation
To visualize actin and microtubules using TIRF microscopy, 0-2 hour embryos (25-27°C) were collected on apple juice agar plates and dechorionated in a homemade mesh basket using standard procedures (Fig S1A, B). Dechorionated embryos were arranged width-wise in a line on the edge of a piece of agar, with approximately one embryo length in between each embryo (Fig S1C-E). Embryo glue (double stick tape adhesive from Scotch® 3M dissolved in heptane) was applied to half the surface of a coverslip, creating a straight line of glue down the center of the coverslip. The coverslip was inverted on the agar, such that approximately one third to one half of the lengths of the dechorionated embryos were adhered to the coverslip by the glue (Fig S1F-H). The embryos were mounted in approximately 50 μl of phosphate buffered saline with 1% normal goat serum. The coverslip was then placed on a dish with a gas-permeable membrane (PetriPERM dish, Sigma) to allow gas exchange during imaging (Fig S1I). This sandwiched the embryos between the coverslip to which they are adhered and the gas permeable membrane. The embryos were imaged through the coverslip using an inverted microscope, where the surface tension generated by a minimal amount of media held the coverslip to the membrane (Fig S1J). The same conditions were used for confocal imaging.
Image acquisition and processing
Images were acquired every 5 seconds with TIRF and every 30 seconds an epifluorescent image was taken 0.8 μm into the embryo. The epifluorescent images were used to properly stage the embryos. Confocal single plane images were acquired every 5 seconds.
TIRF images were acquired on a Nikon 2000TE microscope (Melville, NY) with an argon laser (laser bench provided by Prairie Technologies, Madison, WI) and a 60X, 1.45 NA oil immersion objective capable of both epifluorescence and TIRF illumination, using Metamorph6.1 imaging software (Molecular Devices, Downingtown, PA) and a Retiga-SRV camera (Qimaging) or a Hamamatsu EM CCD C9100 camera. Confocal images were acquired with a spinning-disc confocal microscope (Solamere Technology Group) with a Yokogawa scanhead on a Zeiss Axiovert 200M using QED InVivo software and a QICAM fast 1394 camera (QImaging). ImageJ and Adobe Photoshop were used for image analysis.
Assays of microtubule orientation and dynamics
The total number of microtubules during the peak of cortical activity (Fig 3I) was determined by counting all the microtubules in a 30 μm2 area, for every time point (every 5 seconds) from cycle 12 prophase to cycle 13 interphase.
To analyze the orientation of the microtubules (Fig 3J, 4D), the proportion of the total microtubules perpendicular to the cortex (spots) or parallel to the cortex (lines), was calculated based on the microtubules in a 30 μm2 area in two different time points within the peak of cortical microtubule activity for 5 or more cycle 12 embryos per genotype. Lines were defined as objects with a linear shape and a length ≥ 5 pixels. Statistics employed the binomial approximation of the normal distribution.
Twenty microtubules per embryo for 4 or 5 cycle 12 embryos per genotype were tracked to determine their behavior pattern (spots-only, spot-line, or line-only (Fig 4E)), and their average cortical lifetime (Fig 4F, G). We did not observe any microtubules that transitioned from lines to spots. Statistics employed the binomial approximation of the normal distribution for behavior and the t-test assuming unequal variance for cortical lifetimes.
Quantification of relative persistence for spot-only microtubules
A group of spot-only microtubules within a patch, a circular area with a radius of 40 pixels (4.2 μm, total area = 55 μm2), were analyzed for relative persistence. Using the tracking function in ImageJ, 2D vector coordinates for 3 to 5 microtubules in each patch were marked and tracked between consecutive time points in serial TIRF images in cycle 12 embryos. Three patches were analyzed in each of three embryos per genotype. The pixel coordinate system origin was located at the upper left corner of each image. Tracking data therefore consisted of vectors representing N time-points (N images) for spot-only microtubules 1 through M in each patch. For one patch, the data consisted of a location vector for each spot-only microtubule at each time-point:
For each time interval (i.e., 1→2, 2→3, etc.), we compute the displacement vector, Δrm(n, n+1) for each spot-only microtubule in the patch. For example, in interval (1, 2):
For these data sets, the time interval was 5 sec. In a patch undergoing pure translation, Δrm would be the same for all microtubules in that group. If there is another type of motion superposed on the translation (random movement of microtubules, patch rotation, patch elongation or shear, etc.) then the M displacement vectors will differ to some degree.
To quantify relative persistence, we first compute the average displacement vector for the microtubules in the patch:
and then the root-mean-square difference of the displaced microtubules from the average displacement:
which is a scalar measure of the ‘splay’ or dispersion among the displacements in one patch. It was found that individual spot-only microtubules could be located with a precision of ±1 pixel, so any displacements or RMS distances ≤ 2 pixels were considered to be zero within measurement error (Fig 5A, E, grayed area). For each cortical patch, RMS dispersion, Δ[Δr(t1,t2)]RMS was plotted versus cumulative elapsed time (Fig 5A, E), or against cumulative average scalar displacement, XCUM(n), without regard to the direction of the displacement (Fig 5B, F);
where n = 1, 2, …, N−1. From these plots (Fig 5A, B, E, F), it can be seen that spot-only microtubules in a patch undergo mainly coherent translation, since the RMS difference rarely exceeds positional measurement error, whether over short or long time intervals.
Supplementary Material
Prophase to telophase of a wild type cycle 12 syncytial embryo expressing ZeusGFP visualized with TIRF microscopy. Starting at approximately 4 minutes, during anaphase, we observe a peak of cortical microtubules. Most of the microtubules have a spot-only, or spot-line behavior pattern.
Prophase to telophase of a RhoGEF204291 cycle 12 syncytial embryo expressing ZeusGFP visualized with TIRF microscopy. Similar to wild type, embryos of this genotype display a peak of cortical microtubules during anaphase (starting at approximately 4 minutes). Most of the microtubules have a line-only, or spot-line behavior pattern.
Prophase to telophase of an APC2g10 cycle 12 syncytial embryo expressing ZeusGFP visualized with TIRF microscopy. Similar to the wild type peak and behavior of microtubules, embryos of this genotype display a peak of cortical microtubules during anaphase (starting at approximately 4 minutes) and have primarily a spot-only, or spot-line behavior pattern.
(A) Embryos are laid on an applejuice agar plate. (B) To dechorionate the embryos, they are collected into a homemade mesh basket. (C-E) Embryos arranged width-wise in a line on a block of agar. (F) A coverslip (the edge of which is indicated with an asterisk) with a line of embryo glue is inverted onto the embryos. (G, H) Approximately one third of the length of embryo is glued to the coverslip. (I) A phosphate buffered saline solution containing normal goat serum is applied to the embryos (see Methods), and the coverslip is inverted onto a dish with a gas-permeable membrane. In this way the embryos are sandwiched between the membrane and the coverslip. This constitutes the imaging chamber. (J) The entire chamber is inverted for imaging on an inverted microscope.
Cortical actin in a wild type cycle 12 metaphase embryo expressing MoesinGFP visualized with TIRF microscopy (A-F, red) and with epifluorescence approximately 0.8 μm into the embryo (A’-F’, green). Actin voids (arrowheads in C, C”) exist where the actin rings and pseudocleavage furrows have formed (arrowheads in C’, C”). Scale bar is 10 μm.
(A-X) Prophase to telophase of a wild type cycle 12 syncytial embryo expressing ZeusGFP visualized with TIRF microscopy. We observed a peak of cortical microtubules during anaphase (starting at approximately 4 minutes). Epifluorescent images, taken every 30 seconds, were used to stage the embryos (insets). Most of the microtubules have a spot orientation (K, L, arrowheads), and a spot-only (L-O, red arrowhead), or spot-line (L-O, white arrowhead, P-U, arrow) behavior pattern. Scale bar is 10 μm.
(A) Distribution of cortical lifetimes of all microtubules analyzed for each genotype. (B) Distribution of cortical lifetimes for microtubules with the spot-only behavior pattern for each genotype. (C) Distribution of cortical lifetimes for microtubules with the spot-line behavior pattern for each genotype. Wild type microtubules exhibited fewer, short durations, and more, longer durations compared to mutants in all cases (A-C).
Acknowledgments
We would like to thank Frederick Lanni for his essential contributions to the analysis of relative persistence, James Fitzpatrick for discussions of TIRF microscopy, the members of the McCartney and Minden laboratories for their critical reading of the manuscript, the Center for Biologic Imaging for the use of the Nikon 2000TE microscope, and U. Hacker for the RhoGEF204291 stock. This work was supported through the Pittsburgh Technology Center for Networks and Pathways (5U54-RR022241-03) funded under the NIH Roadmap for Biomedical Research for S.W. and B.M, and the Howard Hughes Medical Institute Undergraduate Research Grant (52005865) for O.R.
References
- Afshar K, Stuart B, Wasserman SA. Functional analysis of the Drosophila diaphanous FH protein in early embryonic development. Development. 2000;127:1887–1897. doi: 10.1242/dev.127.9.1887. [DOI] [PubMed] [Google Scholar]
- Albertson R, Cao J, Hsieh TS, Sullivan W. Vesicles and actin are targeted to the cleavage furrow via furrow microtubules and the central spindle. J Cell Biol. 2008;181:777–790. doi: 10.1083/jcb.200803096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann KJ, Pollard TD. The Arp2/3 complex nucleates actin filament branches from the sides of pre-existing filaments. Nat Cell Biol. 2001;3:306–310. doi: 10.1038/35060104. [DOI] [PubMed] [Google Scholar]
- Axelrod D. Total internal reflection fluorescence microscopy in cell biology. Traffic. 2001;2:764–774. doi: 10.1034/j.1600-0854.2001.21104.x. [DOI] [PubMed] [Google Scholar]
- Cao J, Albertson R, Riggs B, Field CM, Sullivan W. Nuf, a Rab11 effector, maintains cytokinetic furrow integrity by promoting local actin polymerization. J Cell Biol. 2008;182:301–313. doi: 10.1083/jcb.200712036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino PP, Savoian MS, Glover DM. Cleavage furrow formation and ingression during animal cytokinesis: a microtubule legacy. J Cell Sci. 2005;118:1549–1558. doi: 10.1242/jcs.02335. [DOI] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Edwards KA, Demsky M, Montague RA, Weymouth N, Kiehart DP. GFP-moesin illuminates actin cytoskeleton dynamics in living tissue and demonstrates cell shape changes during morphogenesis in Drosophila. Dev Biol. 1997;191:103–117. doi: 10.1006/dbio.1997.8707. [DOI] [PubMed] [Google Scholar]
- Egner A, Andresen V, Hell SW. Comparison of the axial resolution of practical Nipkow-disk confocal fluorescence microscopy with that of multifocal multiphoton microscopy: theory and experiment. J Microsc. 2002;206:24–32. doi: 10.1046/j.1365-2818.2002.01001.x. [DOI] [PubMed] [Google Scholar]
- Foe VE, Field CM, Odell GM. Microtubules and mitotic cycle phase modulate spatiotemporal distributions of F-actin and myosin II in Drosophila syncytial blastoderm embryos. Development. 2000;127:1767–1787. doi: 10.1242/dev.127.9.1767. [DOI] [PubMed] [Google Scholar]
- Gates J, Mahaffey JP, Rogers SL, Emerson M, Rogers EM, Sottile SL, Van Vactor D, Gertler FB, Peifer M. Enabled plays key roles in embryonic epithelial morphogenesis in Drosophila. Development. 2007;134:2027–2039. doi: 10.1242/dev.02849. [DOI] [PubMed] [Google Scholar]
- Glotzer M. Cytokinesis: progress on all fronts. Curr Opin Cell Biol. 2003;15:684–690. doi: 10.1016/j.ceb.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Goode BL, Drubin DG, Barnes G. Functional cooperation between the microtubule and actin cytoskeletons. Curr Opin Cell Biol. 2000;12:63–71. doi: 10.1016/s0955-0674(99)00058-7. [DOI] [PubMed] [Google Scholar]
- Hacker U, Perrimon N. DRhoGEF2 encodes a member of the Dbl family of oncogenes and controls cell shape changes during gastrulation in Drosophila. Genes Dev. 1998;12:274–284. doi: 10.1101/gad.12.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelrigg T. GFP and Other Reporters. In: Sullivan W, Ashburner M, Scott Hawley R, editors. Drosophila Protocols. Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Jankovics F, Brunner D. Transiently reorganized microtubules are essential for zippering during dorsal closure in Drosophila melanogaster. Dev Cell. 2006;11:375–385. doi: 10.1016/j.devcel.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Karr TL, Alberts BM. Organization of the cytoskeleton in early Drosophila embryos. J Cell Biol. 1986;102:1494–1509. doi: 10.1083/jcb.102.4.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart DP, Galbraith CG, Edwards KA, Rickoll WL, Montague RA. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J Cell Biol. 2000;149:471–490. doi: 10.1083/jcb.149.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita K, Wittmann T, Nathke IS, Waterman-Storer CM. Adenomatous polyposis coli on microtubule plus ends in cell extensions can promote microtubule net growth with or without EB1. Mol Biol Cell. 2006;17:2331–2345. doi: 10.1091/mbc.E05-06-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M, Scholey JM. Spindle mechanics and dynamics during mitosis in Drosophila. Trends Cell Biol. 2004;14:194–205. doi: 10.1016/j.tcb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- Mavrakis M, Rikhy R, Lippincott-Schwartz J. Plasma membrane polarity and compartmentalization are established before cellularization in the fly embryo. Dev Cell. 2009;16:93–104. doi: 10.1016/j.devcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney BM, Dierick HA, Kirkpatrick C, Moline MM, Baas A, Peifer M, Bejsovec A. Drosophila APC2 is a cytoskeletally-associated protein that regulates wingless signaling in the embryonic epidermis. J Cell Biol. 1999;146:1303–1318. doi: 10.1083/jcb.146.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney BM, Price MH, Webb RL, Hayden MA, Holot LM, Zhou M, Bejsovec A, Peifer M. Testing hypotheses for the functions of APC family proteins using null and truncation alleles in Drosophila. Development. 2006;133:2407–2418. doi: 10.1242/dev.02398. [DOI] [PubMed] [Google Scholar]
- Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci U S A. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Goode BL. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol Mol Biol Rev. 2006;70:605–645. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathke IS. The Adenomatous Polyposis Coli Protein: The Achilles Heel of the Gut Epithelium. Annu Rev Cell Dev Biol. 2004 doi: 10.1146/annurev.cellbio.20.012103.094541. [DOI] [PubMed] [Google Scholar]
- Padash Barmchi M, Rogers S, Hacker U. DRhoGEF2 regulates actin organization and contractility in the Drosophila blastoderm embryo. J Cell Biol. 2005;168:575–585. doi: 10.1083/jcb.200407124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Bloom K. Dynamic microtubules lead the way for spindle positioning. Nat Rev Mol Cell Biol. 2004;5:481–492. doi: 10.1038/nrm1402. [DOI] [PubMed] [Google Scholar]
- Riggs B, Fasulo B, Royou A, Mische S, Cao J, Hays TS, Sullivan W. The concentration of Nuf, a Rab11 effector, at the microtubule-organizing center is cell cycle regulated, dynein-dependent, and coincides with furrow formation. Mol Biol Cell. 2007;18:3313–3322. doi: 10.1091/mbc.E07-02-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs B, Rothwell W, Mische S, Hickson GR, Matheson J, Hays TS, Gould GW, Sullivan W. Actin cytoskeleton remodeling during early Drosophila furrow formation requires recycling endosomal components Nuclear-fallout and Rab11. J Cell Biol. 2003;163:143–154. doi: 10.1083/jcb.200305115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat Cell Biol. 2003;5:599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC, Sharp DJ, Vale RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Wiedemann U, Hacker U, Turck C, Vale RD. Drosophila RhoGEF2 associates with microtubule plus ends in an EB1-dependent manner. Curr Biol. 2004;14:1827–1833. doi: 10.1016/j.cub.2004.09.078. [DOI] [PubMed] [Google Scholar]
- Rothwell WF, Fogarty P, Field CM, Sullivan W. Nuclear-fallout, a Drosophila protein that cycles from the cytoplasm to the centrosomes, regulates cortical microfilament organization. Development. 1998;125:1295–1303. doi: 10.1242/dev.125.7.1295. [DOI] [PubMed] [Google Scholar]
- Schejter ED, Wieschaus E. Functional elements of the cytoskeleton in the early Drosophila embryo. Annu Rev Cell Biol. 1993;9:67–99. doi: 10.1146/annurev.cb.09.110193.000435. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Brown HM, Kwon M, Rogers GC, Holland G, Scholey JM. Functional coordination of three mitotic motors in Drosophila embryos. Mol Biol Cell. 2000a;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000b;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- Sokac AM, Wieschaus E. Local actin-dependent endocytosis is zygotically controlled to initiate Drosophila cellularization. Dev Cell. 2008;14:775–786. doi: 10.1016/j.devcel.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Croix C, Pitt B, Watkins S. Imaging of biological systems with fluorescent technologies. Science and Medicine. 2005;10:15–29. [Google Scholar]
- Stevenson V, Hudson A, Cooley L, Theurkauf WE. Arp2/3-dependent pseudocleavage [correction of psuedocleavage] furrow assembly in syncytial Drosophila embryos. Curr Biol. 2002;12:705–711. doi: 10.1016/s0960-9822(02)00807-2. [DOI] [PubMed] [Google Scholar]
- Stevenson VA, Kramer J, Kuhn J, Theurkauf WE. Centrosomes and the Scrambled protein coordinate microtubule-independent actin reorganization. Nature Cell Biology. 2001;3:68–75. doi: 10.1038/35050579. [DOI] [PubMed] [Google Scholar]
- Sullivan W, Fogarty P, Theurkauf W. Mutations affecting the cytoskeletal organization of syncytial Drosophila embryos. Development. 1993;118:1245–1254. doi: 10.1242/dev.118.4.1245. [DOI] [PubMed] [Google Scholar]
- Sullivan W, Theurkauf WE. The cytoskeleton and morphogenesis of the early Drosophila embryo. Curr Opin Cell Biol. 1995;7:18–22. doi: 10.1016/0955-0674(95)80040-9. [DOI] [PubMed] [Google Scholar]
- Thompson NL, Lagerholm BC. Total internal reflection fluorescence: applications in cellular biophysics. Curr Opin Biotechnol. 1997;8:58–64. doi: 10.1016/s0958-1669(97)80158-9. [DOI] [PubMed] [Google Scholar]
- Vaizel-Ohayon D, Schejter ED. Mutations in centrosomin reveal requirements for centrosomal function during early Drosophila embryogenesis. Curr Biol. 1999;9:889–898. doi: 10.1016/s0960-9822(99)80393-5. [DOI] [PubMed] [Google Scholar]
- Warn RM, Harrison A, Planques V, Robert-Nicoud N, Wehland J. Distribution of microtubules containing post-translationally modified alpha-tubulin during Drosophila embryogenesis. Cell Motil Cytoskeleton. 1990;17:34–45. doi: 10.1002/cm.970170106. [DOI] [PubMed] [Google Scholar]
- Webb RL, Zhou MN, McCartney BM. A novel role for an APC2-Diaphanous complex in regulating actin organization in Drosophila. Development. 2009;136:1283–1293. doi: 10.1242/dev.026963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrle-Haller B, Imhof BA. Actin, microtubules and focal adhesion dynamics during cell migration. Int J Biochem Cell Biol. 2003;35:39–50. doi: 10.1016/s1357-2725(02)00071-7. [DOI] [PubMed] [Google Scholar]
- Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Wallar BJ, Alberts AS, Gundersen GG. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- Wolf N, Regan CL, Fuller MT. Temporal and spatial pattern of differences in microtubule behaviour during Drosophila embryogenesis revealed by distribution of a tubulin isoform. Development. 1988;102:311–324. doi: 10.1242/dev.102.2.311. [DOI] [PubMed] [Google Scholar]
- Zallen JA, Cohen Y, Hudson AM, Cooley L, Wieschaus E, Schejter ED. SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J Cell Biol. 2002;156:689–701. doi: 10.1083/jcb.200109057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Prophase to telophase of a wild type cycle 12 syncytial embryo expressing ZeusGFP visualized with TIRF microscopy. Starting at approximately 4 minutes, during anaphase, we observe a peak of cortical microtubules. Most of the microtubules have a spot-only, or spot-line behavior pattern.
Prophase to telophase of a RhoGEF204291 cycle 12 syncytial embryo expressing ZeusGFP visualized with TIRF microscopy. Similar to wild type, embryos of this genotype display a peak of cortical microtubules during anaphase (starting at approximately 4 minutes). Most of the microtubules have a line-only, or spot-line behavior pattern.
Prophase to telophase of an APC2g10 cycle 12 syncytial embryo expressing ZeusGFP visualized with TIRF microscopy. Similar to the wild type peak and behavior of microtubules, embryos of this genotype display a peak of cortical microtubules during anaphase (starting at approximately 4 minutes) and have primarily a spot-only, or spot-line behavior pattern.
(A) Embryos are laid on an applejuice agar plate. (B) To dechorionate the embryos, they are collected into a homemade mesh basket. (C-E) Embryos arranged width-wise in a line on a block of agar. (F) A coverslip (the edge of which is indicated with an asterisk) with a line of embryo glue is inverted onto the embryos. (G, H) Approximately one third of the length of embryo is glued to the coverslip. (I) A phosphate buffered saline solution containing normal goat serum is applied to the embryos (see Methods), and the coverslip is inverted onto a dish with a gas-permeable membrane. In this way the embryos are sandwiched between the membrane and the coverslip. This constitutes the imaging chamber. (J) The entire chamber is inverted for imaging on an inverted microscope.
Cortical actin in a wild type cycle 12 metaphase embryo expressing MoesinGFP visualized with TIRF microscopy (A-F, red) and with epifluorescence approximately 0.8 μm into the embryo (A’-F’, green). Actin voids (arrowheads in C, C”) exist where the actin rings and pseudocleavage furrows have formed (arrowheads in C’, C”). Scale bar is 10 μm.
(A-X) Prophase to telophase of a wild type cycle 12 syncytial embryo expressing ZeusGFP visualized with TIRF microscopy. We observed a peak of cortical microtubules during anaphase (starting at approximately 4 minutes). Epifluorescent images, taken every 30 seconds, were used to stage the embryos (insets). Most of the microtubules have a spot orientation (K, L, arrowheads), and a spot-only (L-O, red arrowhead), or spot-line (L-O, white arrowhead, P-U, arrow) behavior pattern. Scale bar is 10 μm.
(A) Distribution of cortical lifetimes of all microtubules analyzed for each genotype. (B) Distribution of cortical lifetimes for microtubules with the spot-only behavior pattern for each genotype. (C) Distribution of cortical lifetimes for microtubules with the spot-line behavior pattern for each genotype. Wild type microtubules exhibited fewer, short durations, and more, longer durations compared to mutants in all cases (A-C).