Abstract
Miz1 is a zinc finger protein that regulates expression of cell cycle inhibitors as part of a complex with Myc. Cell cycle-independent functions of Miz1 are poorly understood. Here, we use a Nestin-Cre transgene to delete an essential domain of Miz1 in the central nervous system (Miz1ΔPOZNes). Miz1ΔPOZNes mice display cerebellar neurodegeneration characterized by the progressive loss of Purkinje cells. Chromatin immunoprecipitation (ChIP)-sequencing and biochemical analyses show that Miz1 activates transcription upon binding to a non-palindromic sequence present in core promoters. Target genes of Miz1 encode regulators of autophagy and proteins involved in vesicular transport that are required for autophagy. Miz1ΔPOZ neuronal progenitors and fibroblasts show reduced autophagic flux. Consistently, polyubiquitinated proteins and p62/Sqtm1 accumulate in cerebella of Miz1ΔPOZNes mice, characteristic features of defective autophagy. Our data suggest that Miz1 may link cell growth and ribosome biogenesis to the transcriptional regulation of vesicular transport and autophagy.
Introduction
The POZ (Pox virus and Zinc finger)-domain/zinc finger protein Miz1 was first identified as a partner protein of Myc and forms a ternary complex with Myc and Max on DNA. Myc/Miz1 complexes block transcription of their target genes in response to specific anti-mitogenic signals: for example, Myc and Miz1 inhibit activation of the cell cycle inhibitor p15Ink4b (Cdkn2b) in response to TGFbeta in tissue culture and in vivo 1, 2. Miz1 also forms repressive complexes with Bcl6 and Gfi1; the Bcl6/Miz1 complex represses expression of Cdkn1a and Bcl2 to control proliferation and survival of germinal center B-cells 3, 4, 5.
To test the role of Miz1 in vivo, we previously generated mice, in which exons that encode the POZ-domain of Miz1 are flanked by loxP sites (generating Miz1ΔPOZ mice) 6, 7. The Miz1ΔPOZ protein fails to stably associate with chromatin and lacks the ability to regulate gene expression 7, 8. Consistent with the biochemical model outlined above, some of the phenotypes observed in Miz1ΔPOZ mice can be explained by complex formation of Myc with Miz1. For example, both Myc and Miz1 are dispensable during normal skin development, but have critical functions during development of skin papillomas as repressors of Cdkn1a 9, 10.
In addition, Miz1 has essential functions during early B- and T-cell development that are independent of Myc 7, 11, 12. Specifically, Miz1 is required for the expression of the T-cell receptor β-chain at the cell surface and for IL-7-dependent signal transduction 7, 12. Since Miz1 itself is a DNA-binding protein, one model to explain these findings is that Miz1 binds and directly regulates target genes involved in signal transduction; accordingly, Myc-independent phenotypes might be due to deregulated expression of these genes. Currently, however, neither the DNA sequence to which Miz1 binds nor its direct target genes are known.
Miz1 is also highly expressed in many postmitotic cells, including neurons of the peripheral and central nervous systems (CNS) 13, arguing that Miz1 function extends beyond regulation of cell proliferation. This prompted us to use a Nestin-Cre strain to delete the POZ-domain in the CNS (Miz1ΔPOZNes). We report here a genome-wide analysis of Miz1 target genes by ChIP-sequencing, gel shift analyses and microarrays that identify Miz1 as a transcriptional regulator of multiple genes involved in vesicular transport, endocytosis and autophagy.
Results
Loss of Miz1 function causes cerebellar neurodegeneration
In order to identify roles for Miz1 in neuronal cells, we crossed Miz1flox/flox mice with mice expressing Cre-recombinase under the control of the Nestin promoter 14, generating mice that express an amino-terminally truncated protein lacking the POZ-domain of Miz1 in the central nervous system (Miz1ΔPOZNes) (Supplementary Figures S1a,b). Mice of the different genotypes were born at the expected Mendelian frequencies (Supplementary Figure S1c). Analysis of genomic DNA and qRT-PCR experiments confirmed that deletion of exons 3 and 4, which encode the POZ-domain, occurred efficiently in the cerebrum, cerebellum, olfactory bulb and, with a lower efficiency, in the eye, which are derived from Nestin-expressing progenitor cells (Supplementary Figure S1d,e).
Miz1ΔPOZNes mice initially appeared normal, but were smaller than either wild type or heterozygous littermates at five weeks after birth and this difference in size increased with age (Supplementary Figure S2a). In addition, Miz1ΔPOZNes mice progressively developed neurological symptoms characterized by slow movement, ataxia and tremor (Figure 1a). As a result, most Miz1ΔPOZNes mice had to be sacrificed at around 18 months of age. Macroscopic analysis revealed that the overall structure of the brain was largely normal in Miz1ΔPOZNes mice except for the cerebellum. The cerebellum was of normal size in newborn animals, but its relative size decreased over time compared to wild type and heterozygous animals (Figures 1b, Supplementary Figure S2b). Within the cerebellum, both the white matter and all three layers of the grey matter (Purkinje cells, molecular layer and granular layer) were reduced in size and appeared disorganized (Supplementary Figure S2c).
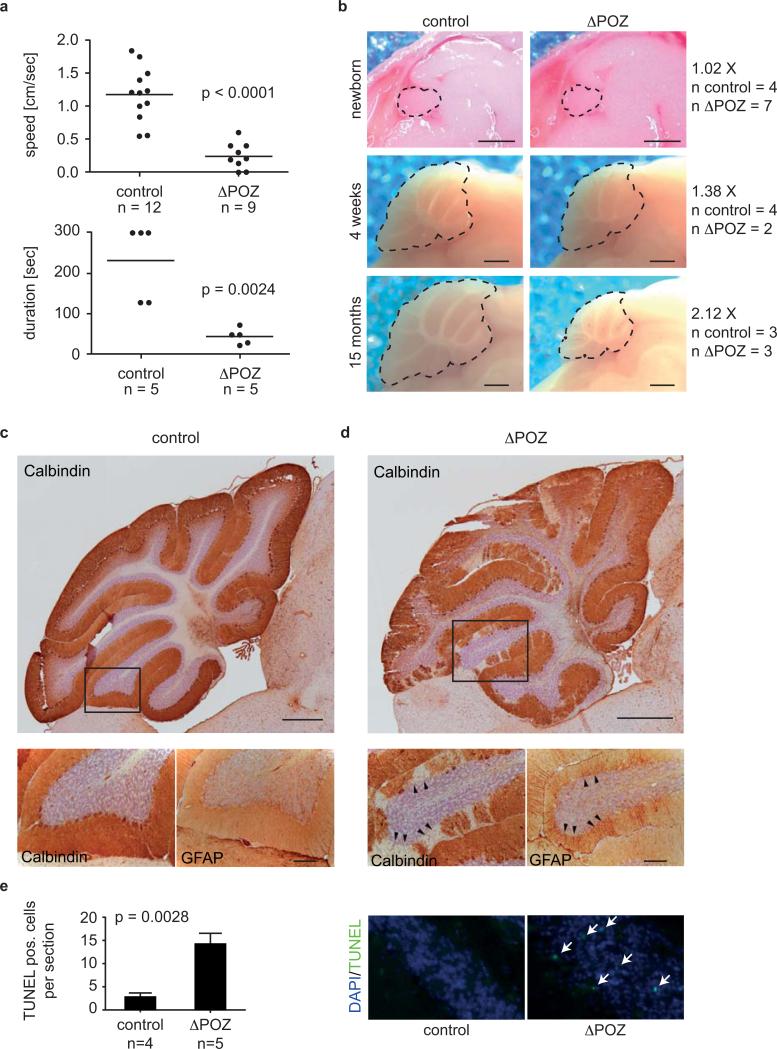
Figure 1. Phenotype of Miz1ΔPOZNes mice.
(a) Miz1ΔPOZNes mice move slowly and lack motor co-ordination. The panel shows the average speed of movement and time on a rotarod (maximum: 300 sec) maintained at constant speed of control and age-matched Miz1ΔPOZNes mice. The average age of mice tested was 16 months. p-values were calculated using an unpaired two-tailed Student's t-test (The number of animals tested is shown below each graph).
(b) Defect of cerebellar growth in Miz1ΔPOZNes mice. The panels show sagittal sections of brains of control and Miz1ΔPOZNes mice at the indicated age. Dotted lines show the circumference of the cerebellum. For each mouse, the cerebellar surface area was determined; numbers show the ratio between control and Miz1ΔPOZNes mice. The number of mice analyzed for each genotype is indicated on the right. Scale bar: 1mm.
(c,d): Histology of the cerebellum. Sections of the cerebellum of 11 month-old control (c) and Miz1ΔPOZNes (d, please note different scale) mice were stained with antibodies directed against calbindin, a marker for Purkinje cells, and GFAP, a marker for Bergman glial cells. Boxes indicate the area of a detail picture in the overviews. GFAP staining is shown for adjacent section (arrowheads: regions of missing Purkinje cells). Scale bar: Upper panels: 500 μm, lower panels: 100 μm.
(e) Enhanced apoptosis in the granular layer of cerebella in Miz1ΔPOZNes mice. The upper panel shows representative TUNEL staining of cerebella of age-matched control and Miz1ΔPOZNes mice. The lower panel shows a quantification of staining observed in four control and five Miz1ΔPOZNes mice. Error bars show standard error of the mean (SEM). p-value was calculated using an unpaired two-tailed Student's t-test.
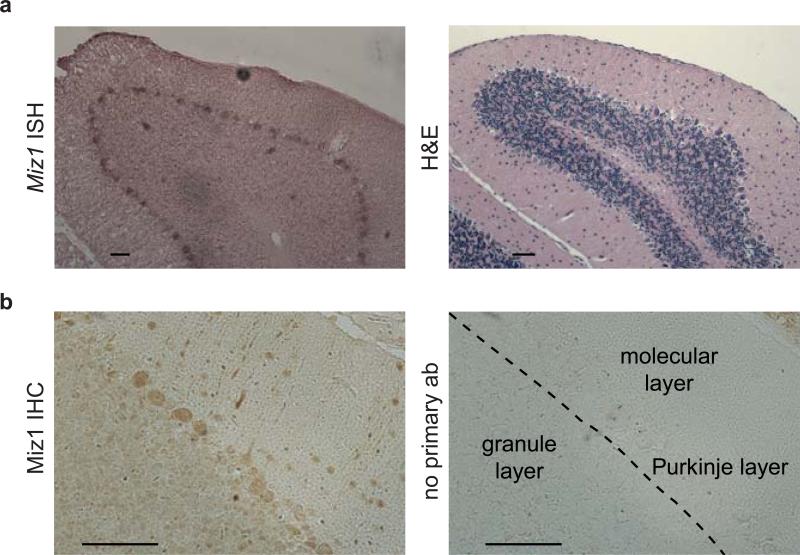
Staining of calbindin, a marker for Purkinje cells, revealed disruption of the Purkinje cell layer at 10 months of age and a corresponding increase in Bergmann glia cells as documented by staining with antibodies directed against glial fibrillary acid protein (GFAP) (Figure 1c,d). The Purkinje cell layer appeared normal at four weeks of age, but was largely disrupted at 15 months of age (Supplementary Figure S2d). We also observed increased apoptosis in the granular layer (Figure 1e). This is consistent with observations that the survival of granular neurons depends on synaptic connections with Purkinje cells 15. In order to understand why Miz1 loss has the strongest effects on Purkinje cells, we performed in situ hybridization and immunohistochemistry (Figure 2a,b). Both techniques revealed elevated expression of Miz1 mRNA and protein in Purkinje cells, suggesting that these cells are particularly sensitive to the lack of a functional Miz1 protein.
Figure 2. Miz1 is highly expressed in Purkinje cells.
(a) In-situ hybridization (ISH) documenting expression of Miz1 mRNA in Purkinje cells of the cerebellum in wild type mice. Left panel shows in situ hybridization, right panel shows H&E staining. Scale bar: 50 μm.
(b) Immunohistochemistry (IHC) documenting the presence of Miz1 in the nuclei of Purkinje cells. The panel on the left shows staining with (H190) antibody, staining on the right is a control staining lacking primary antibody. Scale bar: 50 μm.
Identification of Miz1 target genes and DNA binding sequence
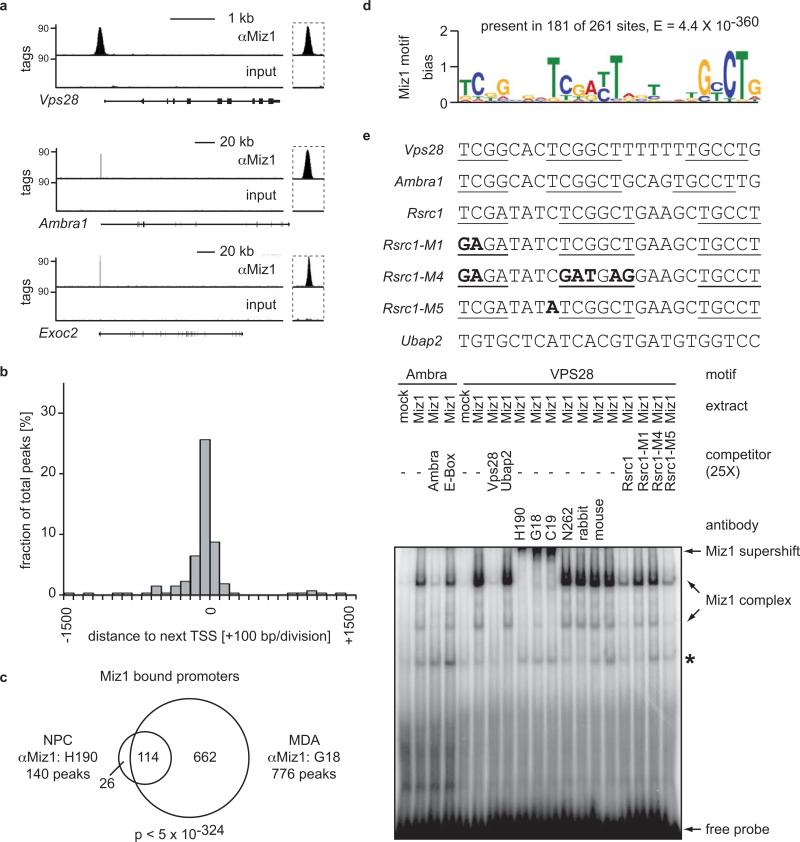
To understand the molecular basis of this phenotype, we identified Miz1 target genes in neuronal cells. We isolated neural progenitor cells from E13.5 brain. After expansion as neurospheres in culture, we performed chromatin immunoprecipitation experiments coupled with high-throughput sequencing to systematically map Miz1 binding sites on chromatin. Statistics for all sequencing experiments are shown in Supplementary Table S1. This analysis identified 261 sites to which Miz1 bound with high occupancy (Figure 3a; see Supplementary Figure S3a for more examples and Supplementary Table S2 for a complete list). Control chromatin immunoprecipitation experiments using independent preparations of chromatin confirmed the specificity of binding (Supplementary Figure S3b). Furthermore, we used a different α-Miz1 antibody and confirmed specific binding of Miz1 to 10/10 sites identified in the genome-wide analysis relative to control regions (Supplementary Figure S3c). The majority of Miz1 binding sites were localized in core promoters close to the transcription start site, unequivocally identifying 140 Miz1-bound promoters (binding site +/− 1.5 kb of the transcription start site, Figure 3b). Comparison with ChIP-sequencing datasets from human mammary epithelial cells (MDA-MB231) showed that binding of Miz1 to these promoters is largely conserved between different cell types and species (Figure 3c; p<5×10−324 using a hypergeometric test).
Figure 3. Identification of Miz1 target genes and DNA binding sequence.
(a) Representative examples for ChIP-sequencing traces obtained from neuronal progenitor cells using an α-Miz1 antibody. Input traces are shown as negative control. For each gene, the intron/exon structure is shown below the traces with exons indicated as vertical bars and the transcriptional start site at the left. An enlarged picture of the peak region is shown on the right.
(b) Miz1 binds predominantly to core promoters. The panel shows a histogram of the percentage of promoters with the indicated distances between the peak of the Miz1 binding site and the next transcriptional start site (TSS). 140/261 (54 %) of all peaks are located in core promoters (+/− 1.5 kb).
(c) Miz1 binding is conserved across different cell types and species. The plot shows a Venn diagram documenting the overlap in Miz1-bound promoters between murine neuronal progenitor cells (NPCs) and human MDA-MB231 cells (a mammary epithelial tumor cell line). The p-value was calculated using a hypergeometric test. Note that different α-Miz1 antibodies (H190, G18) were used for the analysis of mouse and human Miz1 binding sites.
(d) Miz1 binding sites contain a conserved sequence element. Sequences (+/−50bp) around all 261 Miz1 binding sites observed in NPCs were analyzed by MEME-ChIP 16.
(e) Electrophoretic-shift mobility assay documenting binding of Miz1 to the conserved sequence element (“Miz1-binding motif”). Nuclear extracts from control HeLa cells (“mock”) or cells ectopically expressing Miz1 were incubated with radioactive oligonucleotides spanning the Miz1 binding motif of the Ambra1- and Vps28-promoters. Where indicated, binding reactions included a 25-fold excess of non-labeled oligonucleotides (“competitor”), the indicated mutants thereof or of an oligonucleotide containing an E-box sequence from the Ubap2-promoter. Supershift assays were performed using α-Miz1 (H190, G18, C19), α-Myc (N262), or control antibodies. * designates a non-specific band. The sequences of the oligonucleotides used are shown above the panel; mutated nucleotides are shown in bold.
The binding site for Miz1 on DNA is unknown. To determine whether Miz1 specifically recognizes its target promoters, we used MEME-ChIP (“Motif Analysis of Large DNA Datasets“) algorithms to search the ChIP-sequencing dataset obtained from neural progenitor cells for conserved sequence motifs in Miz1-bound chromatin. This analysis yielded an extended non-palindromic sequence that is present in the center of 181 of the 261 Miz1 bound sites (Figure 3d) (E=4.4×10−360; for definition of E-value, see 16). Gel-shift experiments using extracts of HeLa cells ectopically expressing Miz1 identified two complexes that were super-shifted with several independent Miz1 antibodies, but not with control or α-Myc antibodies; a weaker, co-migrating complex was observed in control cells (Figure 3e). Competition experiments showed that mutation of conserved nucleotides abolished binding to Miz1 to this site, whereas mutation of non-conserved nucleotides did not (Figure 3e). Virtually identical results were obtained with a recombinant Miz1 protein (Supplementary Figure S4). We concluded that this sequence constitutes a direct binding site for Miz1.
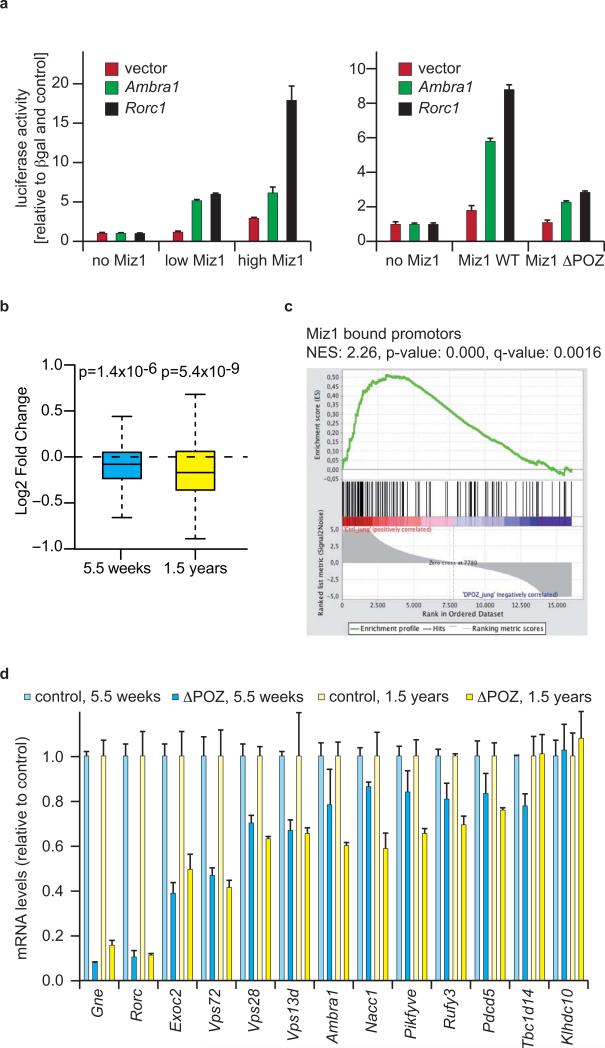
To test whether Miz1 activates transcription of it's target genes, we cloned a 1kb fragment of the Rorc promoter and oligonucleotides (71bps) spanning the Miz1 binding site from the Ambra1 promoter, both covering the respective start sites of transcription, in front of a luciferase open reading frame and performed transient reporter assays. Co-expression of Miz1 stimulated expression of both constructs relative to a control vector, whereas the Miz1ΔPOZ protein was impaired in transactivation (Figure 4a). Similar results had been obtained previously in reporter assays using the Cdkn2b promoter 8, 17. Consistent with these results, microarray analyses of RNA isolated from cerebella of wild type and Miz1ΔPOZNes mice showed that the expression of the 140 genes that are direct targets of Miz1 (see Figure 3c) was significantly downregulated in cerebella of Miz1ΔPOZNes mice (Figure 4b). To extend these results, we spiked a gene set enrichment analysis (GSEA) with a gene set of these Miz1-bound promoters and found that this was the most strongly downregulated and only statistically significant set of genes in this analysis (Figure 4c; p=0.000; FDR q=0.002. In this analysis, the nominal P value is calculated using an empirical phenotype-based permutation test procedure. The q value (FDR) is corrected for gene set size and multiple hypothesis testing)18. qRT-PCR analyses confirmed that multiple direct target genes of Miz1 were expressed at lower levels in cerebella of Miz1ΔPOZNes mice and that down-regulation of Miz1 target genes preceded the appearance of overt pathology since it was already apparent in phenotypically normal young animals (Figure 4d). In addition, expression of multiple genes specifically expressed in Purkinje cells, such as Calbindin, Car8 and Cbln1 was strongly downregulated in cerebella of 1.5 years old Miz1ΔPOZNes animals although these genes were not direct target genes of Miz1, suggesting that their expression was reduced as an indirect consequence due to the loss of Purkinje cells. Reduced expression of direct Miz1 target genes was also observed in mouse embryonic fibroblasts isolated from Miz1ΔPOZ mice, demonstrating that Miz1-dependent regulation is not restricted to the cerebellum (Supplementary Figure S5a). Ectopic expression of wild type Miz1 restored expression of Miz1 target genes (Supplementary Figure S5b). Furthermore, siRNA-mediated depletion of Miz1 decreased expression of Miz1 target genes in mouse embryo fibroblasts (Supplementary Figure S8b). Finally, immunohistochemistry as well as immunoblotting confirmed that the protein encoded by one of the Miz1 target genes, Ambra1, is expressed at lower levels in cerebella and brains of Miz1ΔPOZNes mice relative to wild type and heterozygous mice (Supplementary Figure S5c,d) and in Miz1ΔPOZ mouse embryo fibroblasts (Supplementary Figure S5e). Taken together, the data show that Miz1 activates a set of direct target genes in vivo and in tissue culture.
Figure 4. Miz1 is a transcriptional activator of genes involved in vesicular transport.
(a) Miz1 activates the Rorc and Ambra1 promoters and the POZ domain is required for activation. HEK293 cells were transiently transfected with plasmids containing promoter sequences of Ambra1 or Rorc in front of luciferase and with expression vectors encoding Miz1 or MizΔPOZ. Empty vectors were used as control. Values were normalized with CMV-βGal. Error bars are standard deviation (SD) of biological triplicates.
(b) Direct target genes of Miz1 are downregulated in cerebella of Miz1ΔPOZNes mice. Microarray analysis was performed from cerebella of mice at the indicated ages. The panels show box plots of relative gene expression (plotted as log2 fold change) of the 140 Miz1 direct target genes comparing control and Miz1ΔPOZNes mice. The p-value was calculated using a Mann–Whitney–Wilcoxon-test. Microarrays were performed as technical duplicates from RNA pooled from two animals for each genotype.
(c) Gene set enrichment analysis 18 documenting significant downregulation of Miz1 target genes in cerebella of Miz1ΔPOZNes mice. NES: normalized enrichment score (NES). Target genes of Miz1 were the only significant gene set in this analysis.
(d) qRT-PCR documenting expression of selected Miz1 target genes in cerebella of control and of Miz1ΔPOZNes mice at the indicated ages. Data represent the mean of two independent biological experiments each with three technical replicates. Error bars are SEM.
Miz1 target genes are involved in transport and autophagy
Neither Ingenuity (www.ingenuity.com), gene ontology (GO)-term, nor GSEA analysis identified statistically enriched functions or previously identified expression profiles involving Miz1 target genes that could explain the observed phenotype. Functional annotation revealed that multiple target genes of Miz1 encode proteins involved in vesicular transport pathways, endocytosis and lysosomal biogenesis (Table 1). Examples include Vps28, which is involved in sorting ubiquitinated proteins in the endosomal compartment as part of the ESCRT-I complex (Endosomal sorting complex required for transport-I) 19, Exoc2, which is part of the exocyst complex, a multi-protein complex involved in exocytosis and transport from recycling endosomes 20 as well as Pikfyve (Fab1), a phosphatidylinositol kinase that regulates vesicular transport and endocytosis 21. Endosomal transport and lysosomal biogenesis have been implicated in late steps of autophagy; hence several target genes of Miz1 involved in vesicular transport have been linked to autophagy (Table 1 and Supplementary Table S3). In addition, several Miz1 target genes encode proteins that have been directly linked to autophagy; examples for this group include Ambra1, an activating cofactor of Beclin1, Gne and Tbc1d14 (Table 1) 22.
Table 1.
Functional annotation of selected Miz1 target genes. Microarray results of the listed genes show the comparison of cerebella samples between old control and Miz1ΔPOZNes mice. A (+) in the column Miz1 bound promoters indicates that Miz1 binding was detected in the ChIP-Seq experiment. See Supplementary Table S3 for all microarray results of the listed genes, additional functional annotation, and full references.
| Gene symbol | gene name | control/ΔPOZ [logFC] | Miz1 bound promoter | documented role in vesicular transport | documented role in autophagy |
|---|---|---|---|---|---|
| Rorc | RAR-related orphan receptor gamma | −2.70 | + | + | + |
| Gne | glucosamine (UDP-N-acetyl)-2-epimerase/N- acetylmannosamine kinase | −1.70 | + | + | |
| Trpml1 | mucolipin 1 | −1.34 | + | + | |
| Vps72 | vacuolar protein sorting 72 | −0.64 | + | + | |
| Vps13d | vacuolar protein sorting 13 D | −0.62 | + | + | |
| Mlst8 | MTOR associated protein, LST8 homolog | −0.60 | + | + | |
| Pikfyve | phosphoinositide kinase, FYVE finger containing | −0.58 | + | + | + |
| Rufy3 | RUN and FYVE domain containing 3 | −0.55 | + | + | + |
| Exoc2 | exocyst complex component 2 | −0.50 | + | + | + |
| Ambra1 | autophagy/beclin 1 regulator 1 | −0.49 | + | + | |
| Nacc1 | nucleus accumbens associated 1, BEN and BTB (POZ) domain containing | −0.49 | + | + | |
| Vps28 | vacuolar protein sorting 28 | −0.38 | + | + | + |
| Klhdc10 | kelch domain containing 10 | −0.19 | + | + | |
| Tbc1d14 | TBC1 domain family, member 14 | −0.07 | + | + | + |
| Pdcd5 | programmed cell death 5 | −0.04 | + | + | |
| Vamp4 | vesicle-associated membrane protein 4 | −0.03 | + | + | + |
| Pex14 | peroxisomal biogenesis factor 14 | 0.00 | + | + | + |
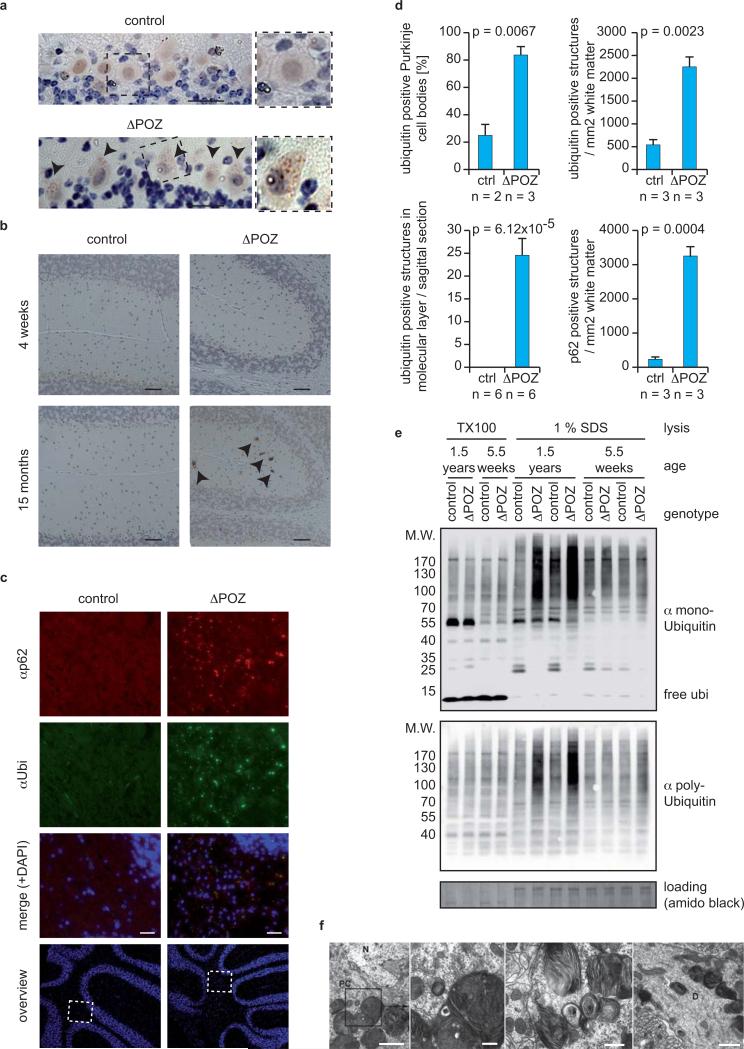
Since the phenotype of knock-out mice that carry brain-specific deletions of the Atg5 or Atg7 genes, which encode E1- and E3-like enzymes for Atg12 conjugation during autophagy, is reminiscent to that of Miz1ΔPOZNes mice, we tested whether Miz1ΔPOZNes mice display phenotypes that might indicate a defective autophagy 23, 24. During autophagy, both monoand polyubiquitinated proteins are sequestered in specific vesicles (“autophagosomes“) that subsequently fuse with lysosomes leading to proteasome-independent degradation of long-lived proteins 25. We therefore performed immunohistochemistry with an antibody against ubiquitin and observed a punctate staining in the body of Purkinje cells of Miz1ΔPOZNes, but not control mice, arguing that ubiquitinated proteins accumulate in cytoplasmic vesicles or aggregates in these neurons (Figure 5a; for a quantification of staining see Figure 5d). In addition, strongly ubiquitin-positive cellular fragments were found in the molecular layer and the white matter of 15month-old Miz1ΔPOZNes mice, but neither in age-matched control nor in young Miz1ΔPOZNes mice, correlating with the observed phenotypes (Figure 5b,c,d). Such cellular fragments most likely corresponded to protrusions (dendrites or axons) of cerebellar neurons (Figure 5f). Biochemical fractionation showed that both poly- and monoubiquitinated proteins accumulated in an insoluble form in the cerebella of Miz1ΔPOZNes mice, but not in brains of control mice, in an age-dependent manner (Figure 5e). Importantly, proteasome activity was identical between cerebella of wild type and Miz1ΔPOZNes mice, arguing that the accumulation of ubiquitinated proteins was not due to a decrease in proteasome activity (Supplementary Figure S6a).
Figure 5. Ubiquitinated proteins and p62/Sqstm1 accumulate in an age-dependent manner in cerebella of Miz1ΔPOZNes mice.
(a,b) Immunostaining using an α-ubiquitin antibody of (a) Purkinje cells and (b) the molecular layer of Miz1ΔPOZNes and age-matched control mice (dotted area: higher magnification). Arrowheads designate ubiquitin-positive structures. Scale bar: (a) 25 μm; (b) 50 μm.
(c) Immunofluorescence staining using an α-p62 and an α-ubiquitin antibody of the white matter of cerebella from Miz1ΔPOZNes and age-matched control mice. Nuclei were counterstained with DAPI and overviews of the cerebella clarifying the selected areas are shown. Scale bar: 25 μm.
(d) Quantification of a, b and c. Error bars represent SEM derived from the indicated numbers of animals. p-values were calculated using an unpaired two-tailed Student's t-test.
(e) Immunoblots of Triton X-100 soluble (“TX100”) and insoluble (“1% SDS”) fractions of cerebella obtained from control and Miz1ΔPOZNes mice of the indicated ages. Blots were probed with antibodies recognizing either mono- or polyubiquitinated proteins. Staining of the membrane with amido black was used as loading control.
(f) Transmission electron microscopy documenting the accumulation of large multilamellar bodies in Purkinje cell bodies (PC) and Purkinje dendrites (D) of Miz1ΔPOZNes mice (N: nucleus). Such structures were not observed in age-matched control mice (as quantified in Supplementary Figure S7b). Mean age of mice for all electron microscopy pictures is 13 months for Miz1ΔPOZNes and 12.5 months for control mice. Morphologically similar structures accumulate in neurons of mice deficient in late steps of autophagy 28. Scale bars: 2, 0.2, 0.5 and 1 μm, respectively.
Defects in autophagy also cause accumulation of p62/SQSTM1 26. p62 acts as a receptor for ubiquitinated cargoes and delivers them to the autophagosome; p62 itself is incorporated into the autophagosome and subsequently degraded by autophagy 26. Cerebella of Miz1ΔPOZNes mice also displayed strongly elevated levels of p62 aggregates, (Figure 5c,d). Notably, ubiquitin-positive structures in the white matter co-localize with p62; virtually identical observations have been reported for other autophagy-deficient strains of mice 27. Furthermore, p62, like the ubiquitinated proteins, accumulated in a detergent-insoluble form (Supplementary Figure S6b). Finally, transmission electron microscopy revealed the accumulation of large polymorphic multilamellar bodies occasionally containing multivesicular or amorphous material in the Purkinje cell bodies and dendrites of Miz1ΔPOZNes mice (Figure 5f and Supplementary Figure S6c-e; for a quantitation of organelles, see Supplementary Figure S7b). These multilamellar bodies are morphologically very similar to membranous cytoplasmic bodies accumulating in autophagy-deficient neurons of Trpml1 (transient receptor potential cation channel/mucolipin1)-deficient animals, expression of which is strongly downregulated in Miz1ΔPOZNes cerebella (Table 1) 28.
Collectively, the findings suggested that Purkinje cells in Miz1ΔPOZNes mice had a defect in autophagy that could account for the phenotype of Miz1ΔPOZNes mice. Surprisingly, however, steady state levels of Lc3/Atg8 that is conjugated to phosphatidylethanolamine (Lc3-II), a marker of autophagy, were unaltered in the cerebella of Miz1ΔPOZNes mice (Supplementary Figure S7a). Furthermore, transmission electron microscopic analysis revealed strong decreases in the number of lysosomes and lipofuscin-containing vesicles, which are thought to be end-stage lysosomes, but did not detect significant changes in the number of autophagosomes (Supplementary Figure S7b). These results either indicate that autophagy is unaltered or that autophagic flux is inhibited at several steps, such that flux is low but no specific intermediates accumulate 29. Conjugated Lc3 is degraded upon fusion of autophagosomes with lysosomes. Therefore, accumulation of Lc3 in the presence of lysosomal inhibitors can be taken as a measure of autophagic flux allowing us to distinguish between both alternatives 29.
Miz1-dependent autophagic flux
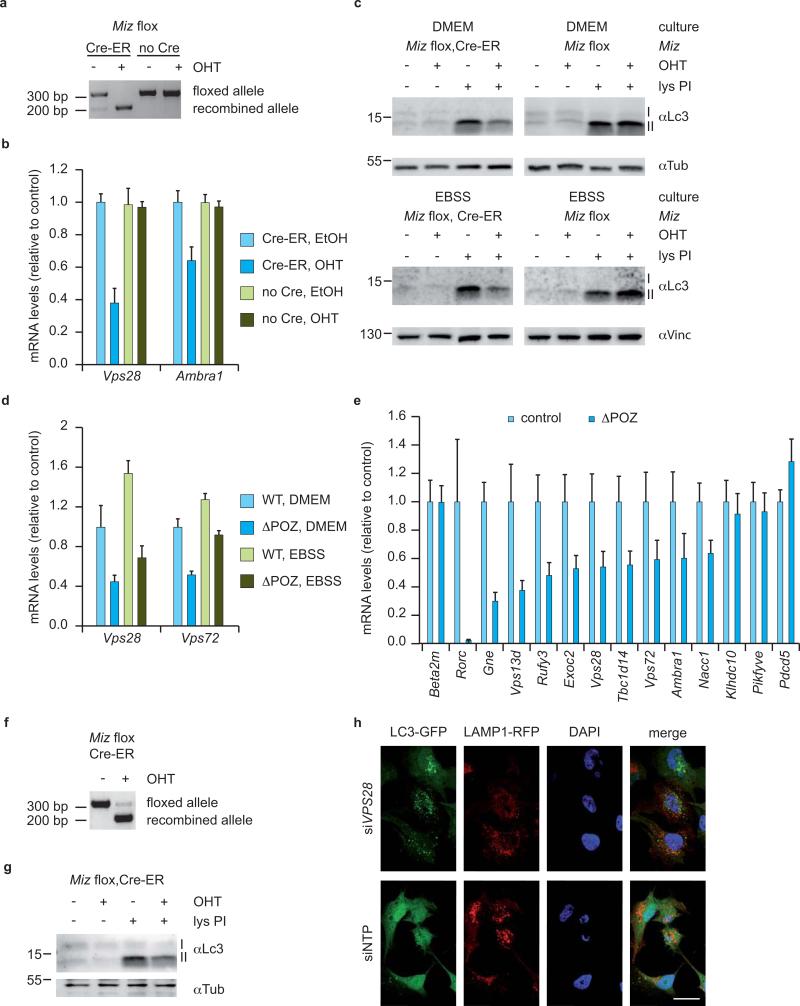
To test whether Miz1 is required for maintaining autophagic flux, we isolated primary embryonic fibroblasts from Miz1flox/flox mice expressing a Cre-ER transgene. Exposure of Miz1flox/floxCre-ER+ cells to 4-OHT induced acute deletion of exons 3 and 4 and decreased expression of Miz1 target genes (Figure 6a,b). Consistent with the results obtained in vivo, deletion of Miz1 had little effect on Lc3 conjugation in unperturbed cells (Figure 6c). In contrast, the accumulation of conjugated Lc3 that occurs in response to inhibition of lysosomal protein degradation was significantly impaired in Miz1ΔPOZ cells “lys PI” lanes in Figure 6c). Addition of 4-OHT had no effect on the expression of Miz1 target genes and autophagic flux in control (Miz1flox/flox) cells (Figure 6b,c), indicating that loss of functional Miz1 impairs autophagic flux. This conclusion is supported by the observation that Miz1ΔPOZ mouse embryo fibroblasts have a reduced capacity for degradation of long-lived proteins upon amino acid starvation (Supplementary Figure S8a). Notably, deletion of the POZ domain of Miz1 diminished autophagic flux both in normal medium (DMEM) and in medium depleted for amino acids (EBSS), demonstrating that Miz1 is not required for the induction of autophagy in response to amino acid starvation (Figure 6c). Consistent with this interpretation, expression of Miz1 target genes was decreased in both media (Figure 6d). Furthermore, treatment of mouse embryo fibroblasts with siRNA targeting Miz1 strongly reduced expression of multiple target genes of Miz1 involved in membrane traffic and autophagy (Supplementary Figure S8b) and diminished autophagic flux in response to inhibition of lysosomal protein degradation (Supplementary Figure S8c). These observations confirm that the reduction of autophagic flux observed in Miz1ΔPOZ cells is due to loss of Miz1 function.
Figure 6. Miz1 is required for maintaining autophagic flux.
(a) Genomic PCR documenting recombination of the floxed Miz1 alleles in Cre-ER positive and negative MEFs. 4-OHT was added to the media for 4 days.
(b) qRT-PCR assays of Miz1 target genes in Miz1flox/floxCre-ER- and Miz1flox/floxCre-ER+ MEFs. The experiment was performed independently from four embryos with identical results. Error bars depict SD of technical triplicates from one experiment.
(c) Immunoblots using an antibody detecting Lc3-I and conjugated Lc3-II. 4-OHT-treated and control-treated MEFs of the indicated genotype were incubated with leupeptin and ammonium chloride (lys PI) for two hours in DMEM (+ amino acids) or in EBSS (- amino acids) medium. The expression of α-tubulin (upper panels) or α-vinculin (lower panels) was used as loading control.
(d) qRT-PCRs of Miz1 target genes in control and Miz1ΔPOZ MEFs after incubation in DMEM or in EBSS. Error bars depict SD of technical triplicates from one experiment.
(e) qRT-PCR assays documenting expression of the indicated Miz1 target genes in control and Miz1ΔPOZ neurospheres; assays were performed at passage three. SEM values are derived from six biological replicates, each with technical triplicates.
(f) Recombination of Miz1flox/floxCre-ER+ neurospheres was tested by genomic PCR. 4-OHT was added to the media for three days.
(g) Immunoblots using an α-Lc3 antibody detecting Lc3-I and Lc3-II. 4-OHT treated and non-treated Miz1flox/floxCre-ER+ neurospheres were incubated with leupeptin and ammonium chloride (lys PI) for 30 minutes. The expression of α-tubulin was used as loading control.
(h) Fluorescent microscopy of SHEP cells stably infected with lentiviruses expressing Lc3-GFP and Lamp1-RFP (to mark lysosomes). Vps28 was depleted with siRNAs and a non-targeting pool (siNTP) was used as a control. Note the accumulation of Lc3 in vesicles that do not overlap with lysosomes upon depletion of Vps28. Scale bar: 25 μm.
Consistent with the results obtained in cerebella of Miz1ΔPOZNes mice, expression of multiple target genes of Miz1 was decreased in cultured neurospheres from E13 embryos of Miz1ΔPOZ and of homozygous Miz1ΔPOZNes mice (Figures 6e, S8d). In contrast, effects on target gene expression were much weaker in heterozygous (Mizflox/wt Cre+) neurospheres, correlating with the lack of phenotype of these animals (Supplementary Figure S8d). Exposure of neurospheres isolated from Miz1flox/floxCre-ER+ mice to 4-OHT induced the acute deletion of exons 3 and 4 and diminished autophagic flux as documented by reduced accumulation of conjugated Lc3 in the presence of inhibitors of lysosomal protein degradation (Figure 6f,g). In contrast to the in vivo situation, we failed to detect accumulation of polyubiquitinated proteins in cultured neurospheres (Supplementary Figure S8e). We propose that apoptosis and subsequent cell lysis prevents the detection of poly-ubiquitinated proteins in cultured Miz1ΔPOZ neurospheres. Consistent with this interpretation, microarray analyses from neurospheres at different passages indicated a progressive reduction in expression of genes characteristic of neuronal progenitors Miz1ΔPOZ neurospheres in addition to downregulation of Miz1 target genes, suggesting that accumulation of ubiquitinated proteins might lead to the elimination of neuronal progenitors via apoptosis (Supplementary Table S4). Furthermore, transmission electron microscopy revealed the presence of multilamellar bodies in early apoptotic cells found in Miz1ΔPOZ neurospheres (Supplementary Figure S8f).
The findings suggested that the lack of changes in steady state levels of Lc3 conjugation is observed due to blockade of autophagic flux at several steps. Of the Miz1 target genes, Ambra1 is required for autophagosome formation and Lc3-conjugation in mammalian cells 30. Several members of the ESCRT complex have been implicated in mammalian autophagy, but none of the Miz1 target genes listed in Supplementary Table S2. We therefore used siRNA to deplete Vps28 in neuroblastoma cells that stably expressed Lc3-GFP and Lamp1-RFP, which marks lysosomes. Upon depletion of Vps28, Lc3 accumulated in vesicles (Figure 6h). Importantly, Lc3 staining did not overlap with staining for the lysosomal marker Lamp1, indicative of a late block in autophagy (Figure 6h). These findings are consistent with observations in Drosophila cells 31. We concluded that target genes of Miz1 are required at early and late steps of autophagy.
Discussion
We showed here that Miz1 has a critical function in maintaining the viability of Purkinje cells in the cerebellum in vivo. In the absence of functional Miz1, cerebella undergo progressive neurodegeneration, which is characterized by an age-dependent accumulation of ubiquitinated proteins and of p62, indicative of defective autophagy.
In order to explain these phenotypes, we used ChIP-sequencing to show that Miz1 binds to approximately 260 target sites in neuronal progenitor cells. Target sites of Miz1 had an extended non-palindromic consensus sequence as might be expected for a protein with multiple (13) zinc fingers. Biochemical analysis confirmed that Miz1 directly bound to this sequence. Comparison with data obtained in human mammary epithelial cells (MDA-MB231) showed that these direct target genes were largely conserved between different tissues and species. Miz1 binding sites were predominantly found in core promoters, close to the transcription start site. Reporter assays and comparison with gene expression data showed that Miz1 activates transcription upon binding to its cognate DNA sequence, arguing that the phenotype of Miz1ΔPOZNes mice was due to reduced expression of Miz1 target genes. Comparison with published microarray data showed that the direct Miz1 target genes described here are not subject to regulation by Myc, arguing that their expression may reflect Myc-independent functions of Miz1 (Supplementary Table S5).
Defects in distinct cellular processes can lead to accumulation of polyubiquitinated proteins 32, 33. One is autophagy and one of the Miz1 target genes described here, Ambra1, is a bona-fide autophagy gene 30; other target genes encode regulators of autophagy (e.g. Tbc1d14 and Pdcd5) (see Supplementary Table S3 for references). Late steps of autophagy involve the fusion of autophagosomes with endosomal vesicles and ultimately lysosomes 22, 34. As a result, defects in the progression of endosomes along the endosomal-lysosomal pathway, such as dysfunction of ESCRT complex proteins, indirectly cause defects in autophagy and accumulation of polyubiquitinated proteins 35. Multiple proteins encoded by target genes of Miz1 (e.g. Vps28, Pikfyve/Fab1, Rorc) were involved in vesicular transport, late steps of autophagosome processing and in fusion of endosomes with lysosomes. We confirmed that depletion of mammalian Vps28, a direct target gene of Miz1, caused a late block in autophagy. We propose that autophagic flux is reduced in Miz1ΔPOZ cells due to the decreased expression of proteins that act at both early and late stages of the autophagic pathway. This model can explain why steady-state levels of Lc3 conjugation were unaffected by loss of Miz1 in vivo and in cell culture 29. In post-mitotic cells, autophagy is critical for removal of damaged organelles and toxic protein aggregates; notably, deletion of Atg5 or Atg7 in the CNS has the strongest effect on Purkinje cells, similar to what is observed in Miz1ΔPOZNes mice, arguing that survival of these cells is particularly dependent on autophagy 23, 24, 36, 37. The reduction of autophagic flux observed in Miz1ΔPOZNes mice could therefore account for their progressive neurodegeneration. Notably, the neurodegenerative phenotype of Miz1ΔPOZNes mice is weaker than that of Atg5 or Atg7 knockout mice, most likely due to residual autophagic flux in Miz1ΔPOZNes cells and, by inference, in cerebella of Miz1ΔPOZNes mice. Since several target genes of Miz1 encode proteins that are required for intracellular trafficking of multiple cell surface receptors and since regulated transport to the cell surface and endocytosis are critical for the function of the TCR and IL-7 receptors that are functionally deficient in Miz1ΔPOZ lymphocytes, we propose that the target genes identified here can also account for other phenotypes observed in Miz1ΔPOZ animals 38, 39, 40, 41, 42. Several other transcription factors, such as p53, TFEB and the FoxO family of factors have been implicated in the transcriptional control of autophagy 43, 44, 45 . Notably, there is little overlap in the direct target genes of these factors with those bound by Miz1, suggesting that the precise process within the overall autophagic process that is enhanced by each factor may differ functionally and reflect the specific stimulus or stress response that activates each factor.
Previous work had shown that the ribosomal assembly factor, nucleophosmin, is a co-activator of Miz1 46. In growing cells, nucleophosmin is retained in the nucleolus by the ribosomal protein, Rpl23. Nucleophosmin is released into the nucleolus when Rpl23 levels fall, linking transactivation by Miz1 to defects in ribosome biogenesis. Together with the findings reported here, the data argue that Miz1 is a stress-responsive factor that balances biosynthetic capacity and cell growth with nutrient and energy supply in addition to promoting cell cycle arrest.
Methods
Mice
Miz1flox/flox and Miz1+/ΔPOZ mice were crossed to each other and to Nestin-Cre mice 14 to obtain Miz1ΔPOZNes mice. Mice were genotyped by standard PCRs using three different primers listed in Supplementary Table S4 (genomic PCRs). Mice of both sexes between 0 and 18 months (as indicated in figure legends) were used and all mice were included in the analysis. Unless indicated otherwise, mice that carry one or two wild type alleles of Miz1 and Miz1flox mice that do not express Nestin-Cre were collectively used as control mice. Speed of movement was measured by placing mice on a grid and determining distance covered per time. To measure motor coordination, mice were trained for three days to the rotarod (Ugo Basile). Subsequently, the time each animal was able to run at a constant speed of 21 rpm was determined. The circumference of the cerebellum was measured and used to estimate surface area. All experiments were carried out according to German law and are approved by an institutional panel (Tierschutzkommission der Regierung von Unterfranken).
Immunofluorescence, immunohistochemistry and histology
All staining were performed on tissue sections collected from OCT- or paraffin-embedded tissues. The following primary antibodies were used: α-Calbindin (1:100, Sigma, C9848), α-glial acidic fibrillary protein (GFAP) (OPA1-06100, Pierce), α-Miz1 (1:100, H190, Santa Cruz), α-Ubiquitin (1:100, MBL, MK 11-3), α-p62 (1:1000, MBL, pm045). Detection of primary antibodies was carried out with appropriate peroxidase- or fluorochrome-coupled secondary antibodies. Sections were counterstained with hematoxylin. TUNEL staining was performed using an in situ cell death detection kit (Roche). Klüver-Barrera and H&E staining was performed according to standard procedures. Pictures were taken with a Leica DM 6000B microscope.
Cell culture
To isolate neuronal progenitor cells (NPCs), forebrains of E13.5 Miz1+/+, Miz1ΔPOZ or Miz1flox/floxCre-ER+ embryos were cut in small pieces, digested with trypsin and filtered through sterile gauze. Cells were cultivated in 2:1 DMEM/F12 supplemented with 1×B27 (Life technologies), 20 ng/μl EGF (Biomol), 20 ng/μl basic FGF (Biomol), 1 μg/ml fungizone (Gibco) and Penicillin/Streptomycin (PAA). NPCs were passaged every seven days. Mouse embryo fibroblasts (MEFs) were isolated from E13.5 Miz1flox/floxCre-ER+ and Miz1flox/floxCre-ER− embryos. To induce recombination in Miz1flox/floxCre-ER+ NPCs or MEFs, cells were incubated with 200 nM 4-OHT for 3-4 days. Cells were passaged and treated with 20 μM Leupeptin and 20 mM NH4Cl for 30 minutes (NPCs) or 2 hours (MEFs) in NPC medium or DMEM and EBSS (Earle's balanced salt solution) (MEFs). To measure degradation of long-lived proteins, cells were labelled with 5 μCi of 35S-Met/Cys (Hartmann) for 3 h, washed and incubated for 17 h to release short-lived proteins. To block autophagy, cells were incubated with 10 mM 3-methyl-adenine (Tocris Bioscince) for 3 h. After washing, cells were incubated with EBSS or DMEM in the presence/absence of 3-methyl-adenine for 4 h. TCA-cleared supernatant and TCA-washed cells were measured with a scintillation counter. Long-lived protein degradation was calculated by dividing the released radioactivity by total radioactivity.
Chromatin immunoprecipitation and parallel sequencing
For chromatin immunoprecipitation (ChIP), cells were cross-linked with 1 % formaldehyde at 37°C for 10 min. Sonication was carried out after swelling in a Bransson sonifier until the majority of fragments showed nucleosomal size. α-Miz1 antibodies (neuronal progenitors: H190; MDA cells: G18 (Santa Cruz)) were coupled to ProteinA/G-Dynabeads (Invitrogen) and used for immunoprecipitation. DNA was purified with Qiagen PCR purification kit after elution of the bound chromatin with 1% SDS and reversion of the crosslink. ChIP DNA was end repaired and A-tailed. Illumina adaptors were ligated to the ChIP DNA fragments. 175 -225bp size fractions were cut out from a 1% agarose gel, extracted by Qiagen gel extraction kit and enriched by 18 cycles of PCR amplification. Library size was controlled with the Experion-system (BioRad) and subsequently quantified by picogreen assay and sequenced on an Illumina GAIIx sequencer. Only reads passing the internal Illumina raw data-filter were considered. Resulting fastq files were used for alignment to a precompiled reference index (neuronal progenitor cells: mm9, MDA: hg19) with BOWTIE 47. Peaks were called by MACS 48 using data from the input sample as control and determining a p-value of 1010. GEO accession number for the ChIP-sequencing results is: GSE48602.
To compare the Miz1 peaks in neuronal progenitor and MDA-MB231 cells, a hypergeometric distribution was used: first the total number of base pairs covered by peaks in each dataset was calculated (NPC 41,500bp and MDA-MD231 4,47494bp), and second the number of overlapping base pairs of MDA-MD231 and NPCs peaks was counted (33,176bp). As population size all murine promoter regions (+/−1.5kb around a TSS; total 93,582,000bp) were used.
Gene Expression
Total cellular RNA was isolated from whole cerebella or from neurospheres using RNAeasy (Qiagen) including on-column DNA digestion (microarray-analysis) or extraction with peqGOLD (Peqlab) for qRT-PCR. For isolation of RNA from cerebellum, tissue samples were placed into peqGOLD and homogenized with an ultra-turrax. For microarray analyses, RNA was additionally treated with DNAase on an RNAeasy column. Agilent Mouse Genome Microarray 4×44 K v2 was used for the analysis of the gene expression. For qRT-PCR, the first-strand was synthesized with M-MLV Reverse Transcriptase (Invitrogen) and random hexamer primers (Roche). Primer sequences are shown in Supplementary Table S6. For in situ hybridizations, DNA corresponding to exons 3 and 4 of murine Miz1 was cloned into pBluescript KS+ vector and used for synthesis of sense and antisense probe. ArrayExpress accession numbers for the microarray results are: E-MEXP-3878 (cerebella) and E-MEXP-3883 (neurospheres).
Cell fractionation and immunoblotting
Detergent-soluble and -insoluble protein fractions from mouse brains were prepared as described 23. Briefly, after homogenization, cells were lysed with 0.5 % Triton X-100. After centrifugation the pellet was re-suspended in 1 % SDS. After SDS-PAGE proteins were transferred on PVDF-membrane and immunostained for ubiquitinated proteins (07-375, upstate; MK-11-3, MBL). For immunoblotting of Lc3 in MEFs and NPCs, cells were lysed in buffer containing 50 mM Hepes-KOH, pH 7.5, 150 mM NaCl, 100 mM NaF, 10 mM EDTA, 10 mM Na4P2O7. Immunoblots were probed with antibodies directed against Lc3 (1:1000, MBL, PM036), α-vinculin (1:10.000, Sigma) and α-tubulin (1:1000, Chemicon).
Electrophoretic mobility shift assay (EMSA)
GST-Miz1ΔPOZ protein was expressed in E. coli (BL-21) and affinity purified using a glutathione-sepharose (GE Healthcare) column. After elution with glutathione, purity was controlled with a Coomassie Brilliant Blue-stained SDS-PAGE. Dignam nuclear extracts were prepared from HeLa cells transiently transfected with pCDNA3-Miz1. For labeling, 25 pmol of annealed oligonucleotides were labeled using 0.15 mCi γ32P-ATP (Hartmann) and 25U of PNK (NEB) and purified on a G50 column (GE). 1 μl of purified probe was incubated with 2 μl of nuclear extract (~ 2 mg/ml) in the presence of 20 mM Hepes-KOH , pH 7.9, 10 % (v/v) glycerol, 105 mM NaCl, 2 mM MgCl2, 50 μM EDTA, 0.25 μg/μl salmon sperm DNA, 1.5 mM DTT, and protease- and phosphatase-inhibitors (Sigma) in a 20 μl reaction. For supershifts 1 μg of antibody was added (Miz1: H190, G18, C19 (all Santa Cruz) Control: rabbit/mouse IgG). After 10min incubation at room temperature, samples were separated on a 5 % PAA-/0.5× TBE-gel for 5h at 180V and exposed on phosphoimager storage screens.
Transmission electron microscopy
Female mice of different genotypes (3 Miz1ΔPOZNes mice and 3 control mice, 11-15 months of age) were perfusion-fixed in deep anesthesia via the left ventricle using, after a brief pre-rinse with heparinized saline, 4 % PFA and 2.5% glutaraldehyde (GA) in 0.1 M cacodylate buffer pH 7.3 - 7.4 (CB). After dissection of the brain and postfixation for at least 18 hours, small blocks of cerebellar tissue were taken from a cerebellar hemisphere of each mouse, washed in CB, osmicated in 2% osmium tetroxide in CB for 2 hours, dehydrated in acetone and embedded in Spurr's medium. Contralateral hemispheres were washed in PBS, cut into coronal 150 μm vibratome sections, osmicated in 1 % osmium tetroxide in PBS for 1 hour, dehydrated in ethanol and flat-embedded in Epon Ultrathin sections were contrasted with uranyl acetate and lead citrate and observed in a Leo 912 AB electron microscope (Zeiss NTS, Oberkochen, Germany). Both embedding methods yielded equivalent ultrastructural details for Miz1ΔPOZNes specimens.
In situ Hybridization (ISH)
Dissected brains were frozen in 2-methylbutane and stored at −80°C. Serial cryosections (15 μm) were thaw-mounted on microscopic slides, fixed in 4% paraformaldehyde in PBS and stored in 100% ethanol at 4°C. ISH was carried out using chromogenic detection.
Transient transfection assays
PCR fragments of the human Ambra1 and Rorc promotor were cloned in a pGL4 plasmid (Clontech). HEK293 cells were transiently transfected (PEI) with the reporter constructs, Miz-WT and MizΔPOZ expression plasmids and CMV-βgal plasmid. Cells were harvested after 48 h, luciferase and β-galactosidase activity was measured according to the manufacture's protocol. Endogenous Vps28 protein was depleted in SHEP neuroblastoma cells stably expressing Lc3-GFP 49 and Lamp1-RFP 50 by siRNA (Dharmacon).
Long-lived protein degradation assays
Degradation of long lived proteins was measured according to 51. Cells were labeled with 5 μCi of 35S-Met/Cys (Hartmann) for 3 h, washed and incubated for 17 h to release short-lived proteins. To block autophagy, cells were partially incubated with 10 mM 3-methyl-adenine (Tocris Bioscience) for 3 h. After another washing, cells were incubated with EBSS or DMEM in the presence/absence of 3-methyl-adenine for 4 h. TCA-cleared supernatant and TCA-washed cells were measured with a scintillation counter. Long-lived protein degradation was calculated by dividing the released radioactivity by total radioactivity.
Determination of proteasomal activity
Proteasomal activity in cerebella of ΔPOZ and control mice was measured with a Proteasome Activity Assay Kit (Abcam). Fluorescence was measured with a microtiter plate reader (Tecan) in the presence/absence of MG132 after 15 min at 37°C for 100 min. The MG132-sensitive increase of fluorescence at 350/440 nm was considered as proteasomal activity.
Supplementary Material
Acknowledgements
We thank Michael Krause, Florian Finkernagel and Lukas Rycek for performing and analyzing the microarray analyses, Angela Grün, Renate Metz and Barbara Bauer for excellent technical assistance and members of the Eilers laboratory for comments on the manuscript. This work was supported the Deutsche Forschungsgemeinschaft via SFB 581 (M.E. and E.A.) and via grant 222/5-3 (to M.E.), by the NCI CA-096832 and CA-21765 (MFR), The Anderson Fellowship (DK), and the American Lebanese Syrian Charities of St Jude Children's Research Hospital.
Footnotes
Author contributions
A.G., E.W., D.K., S.W., B.v.E., N.W., C.R., and G.K. performed the experiments, A.G., E.A., E.W and M.E. designed experiments, M.F.R. provided critical advice and A.G., E.W. and M.E. wrote the paper.
Accession codes
ChIP-sequencing data have been deposited in the NCBI Gene Expression Omnibus under accession code GSE48602.
The authors declare no competing financial interests.
References
- 1.Gebhardt A, Frye M, Herold S, Benitah SA, Braun K, Samans B, et al. Myc regulates keratinocyte adhesion and differentiation via complex formation with Miz1. J Cell Biol. 2006;172(1):139–149. doi: 10.1083/jcb.200506057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Riggelen J, Muller J, Otto T, Beuger V, Yetil A, Choi PS, et al. The interaction between Myc and Miz1 is required to antagonize TGFbeta-dependent autocrine signaling during lymphoma formation and maintenance. Genes & development. 2010;24(12):1281–1294. doi: 10.1101/gad.585710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6(10):1054–1060. doi: 10.1038/ni1245. [DOI] [PubMed] [Google Scholar]
- 4.Saito M, Novak U, Piovan E, Basso K, Sumazin P, Schneider C, et al. BCL6 suppression of BCL2 via Miz1 and its disruption in diffuse large B cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(27):11294–11299. doi: 10.1073/pnas.0903854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basu S, Liu Q, Qiu Y, Dong F. Gfi-1 represses CDKN2B encoding p15INK4B through interaction with Miz-1. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(5):1433–1438. doi: 10.1073/pnas.0804863106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gebhardt A, Kosan C, Herkert B, Moroy T, Lutz W, Eilers M, et al. Journal of cell science. Vol. 120. Pt 15: 2007. Miz1 is required for hair follicle structure and hair morphogenesis. pp. 2586–2593. [DOI] [PubMed] [Google Scholar]
- 7.Kosan C, Saba I, Godmann M, Herold S, Herkert B, Eilers M, et al. Transcription factor miz-1 is required to regulate interleukin-7 receptor signaling at early commitment stages of B cell differentiation. Immunity. 2010;33(6):917–928. doi: 10.1016/j.immuni.2010.11.028. [DOI] [PubMed] [Google Scholar]
- 8.Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, et al. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell. 2002;10(3):509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 9.Oskarsson T, Essers MA, Dubois N, Offner S, Dubey C, Roger C, et al. Skin epidermis lacking the c-Myc gene is resistant to Ras-driven tumorigenesis but can reacquire sensitivity upon additional loss of the p21Cip1 gene. Genes & development. 2006;20(15):2024–2029. doi: 10.1101/gad.381206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honnemann J, Sanz-Moreno A, Wolf E, Eilers M, Elsasser HP. Miz1 is a critical repressor of cdkn1a during skin tumorigenesis. PLoS One. 2012;7(4):e34885. doi: 10.1371/journal.pone.0034885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saba I, Kosan C, Vassen L, Moroy T. IL-7R-dependent survival and differentiation of early T-lineage progenitors is regulated by the BTB/POZ domain transcription factor Miz-1. Blood. 2011;117(12):3370–3381. doi: 10.1182/blood-2010-09-310680. [DOI] [PubMed] [Google Scholar]
- 12.Saba I, Kosan C, Vassen L, Klein-Hitpass L, Moroy T. Miz-1 is required to coordinate the expression of TCRbeta and p53 effector genes at the pre-TCR “beta-selection” checkpoint. J Immunol. 2011;187(6):2982–2992. doi: 10.4049/jimmunol.1101451. [DOI] [PubMed] [Google Scholar]
- 13.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 14.Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nature genetics. 1999;23(1):99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 15.Lossi L, Mioletti S, Merighi A. Synapse-independent and synapse-dependent apoptosis of cerebellar granule cells in postnatal rabbits occur at two subsequent but partly overlapping developmental stages. Neuroscience. 2002;112(3):509–523. doi: 10.1016/s0306-4522(02)00112-4. [DOI] [PubMed] [Google Scholar]
- 16.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27(12):1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nature cell biology. 2001;3(4):392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106(2):145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 20.Langevin J, Morgan MJ, Sibarita JB, Aresta S, Murthy M, Schwarz T, et al. Drosophila exocyst components Sec5, Sec6, and Sec15 regulate DE-Cadherin trafficking from recycling endosomes to the plasma membrane. Dev Cell. 2005;9(3):365–376. doi: 10.1016/j.devcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 21.Ikonomov OC, Sbrissa D, Foti M, Carpentier JL, Shisheva A. PIKfyve controls fluid phase endocytosis but not recycling/degradation of endocytosed receptors or sorting of procathepsin D by regulating multivesicular body morphogenesis. Mol Biol Cell. 2003;14(11):4581–4591. doi: 10.1091/mbc.E03-04-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22(1):R29–34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 23.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 24.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 25.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34(3):259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 26.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 27.Liang CC, Wang C, Peng X, Gan B, Guan JL. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. The Journal of biological chemistry. 2010;285(5):3499–3509. doi: 10.1074/jbc.M109.072389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curcio-Morelli C, Charles FA, Micsenyi MC, Cao Y, Venugopal B, Browning MF, et al. Macroautophagy is defective in mucolipin-1-deficient mouse neurons. Neurobiology of disease. 2010;40(2):370–377. doi: 10.1016/j.nbd.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447(7148):1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 31.Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol. 200717(20):1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 32.Jaeger PA, Wyss-Coray T. All-you-can-eat: autophagy in neurodegeneration and neuroprotection. Molecular neurodegeneration. 2009;4:16. doi: 10.1186/1750-1326-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kroemer G, White E. Autophagy for the avoidance of degenerative, inflammatory, infectious, and neoplastic disease. Curr Opin Cell Biol. 2010;22(2):121–123. doi: 10.1016/j.ceb.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eskelinen EL. Maturation of autophagic vacuoles in Mammalian cells. Autophagy. 2005;1(1):1–10. doi: 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- 35.Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17(18):1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 36.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madeo F, Eisenberg T, Kroemer G. Autophagy for the avoidance of neurodegeneration. Genes & development. 2009;23(19):2253–2259. doi: 10.1101/gad.1858009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rusten TE, Rodahl LM, Pattni K, Englund C, Samakovlis C, Dove S, et al. Fab1 phosphatidylinositol 3-phosphate 5-kinase controls trafficking but not silencing of endocytosed receptors. Mol Biol Cell. 2006;17(9):3989–4001. doi: 10.1091/mbc.E06-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaccari T, Rusten TE, Menut L, Nezis IP, Brech A, Stenmark H, et al. Comparative analysis of ESCRT-I, ESCRT-II and ESCRT-III function in Drosophila by efficient isolation of ESCRT mutants. Journal of cell science. 2009;122(Pt 14):2413–2423. doi: 10.1242/jcs.046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11(6):859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 41.McLeod IX, Zhou X, Li QJ, Wang F, He YW. The class III kinase Vps34 promotes T lymphocyte survival through regulating IL-7Ralpha surface expression. J Immunol. 2011;187(10):5051–5061. doi: 10.4049/jimmunol.1100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geisler C. TCR trafficking in resting and stimulated T cells. Critical reviews in immunology. 2004;24(1):67–86. doi: 10.1615/critrevimmunol.v24.i1.30. [DOI] [PubMed] [Google Scholar]
- 43.Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126(1):121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 44.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell metabolism. 2007;6(6):472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 46.Wanzel M, Russ AC, Kleine-Kohlbrecher D, Colombo E, Pelicci PG, Eilers M. A ribosomal protein L23-nucleophosmin circuit coordinates Mizl function with cell growth. Nature cell biology. 2008;10(9):1051–1061. doi: 10.1038/ncb1764. [DOI] [PubMed] [Google Scholar]
- 47.Langmead B. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics. 2010 doi: 10.1002/0471250953.bi1107s32. Chapter 11: Unit 11.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7(9):1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19(3):797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zoncu R, Sabatini DM. Cell biology. The TASCC of secretion. Science. 2011;332(6032):923–925. doi: 10.1126/science.1207552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bauvy C, Meijer AJ, Codogno P. Assaying of autophagic protein degradation. Methods Enzymol. 2009;452:47–61. doi: 10.1016/S0076-6879(08)03604-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.