Abstract
The phenomenon of catabolite repression enables microorganisms to use their favourite carbon source first. New work reveals α-ketoacids as key effectors of this process, with their levels regulating gene expression.
Nutrients in the environment are a primary determinant of microbial physiology. When preferred nutrients are abundant, microbes grow fast. When they are scarce, growth slows down. This change in growth rate is accompanied by a change in cellular composition, with fast-growing cells being loaded with ribosomes (which are needed for rapid protein production), and slower-growing cells containing more metabolic enzymes for nutrient assimilation (catabolism)1,2. In a study published on Nature's website today, You et al.3 identify a striking linear relationship between the total protein composition (the proteome) of a cell and its growth rate, which extends beyond ribosomes to metabolic enzymes. They further demonstrate how such a relationship can arise, in part, from a new regulatory connection, in which a particular class of carbon catabolite called α-ketoacids, which form the carbon skeletons of amino acids, serves as a master transcriptional regulator by inhibiting the production of cyclic AMP — the primary inducer of carbon-catabolic genes.
Perhaps the most intensively studied example of gene regulation involves the enzymes of the bacterium Escherichia coli that mediate lactose catabolism. These enzymes are expressed only when lactose is present and glucose (the preferred carbon source) is not4. Escherichia coli detects the presence of lactose through binding of this sugar to, and inactivation of, the lac repressor protein5, and it senses the absence of glucose from elevated levels of cAMP6, which binds to and activates the transcription factor Crp (refs 7, 8). Identification of this classic regulatory loop involved seminal contributions from three Nobel laureates — François Jacob and Jacques Monod, who won the 1965 physiology prize for their pioneering studies of gene regulation5, and Earl Sutherland, winner of the 1971 prize for discoveries related to cAMP, who later identified cAMP in E. coli and showed that its levels rapidly fall in response to glucose6.
What controls the activity of adenylate cyclase (the enzyme that makes cAMP) and so cAMP levels? The phosphorylated form of the enzyme EIIAGlc can activate adenylate cyclase. EIIAGlc is a component of the bacterial phospho transferase system, which takes the phosphate group from phosphoenolpyruvate — the last intermediate in the biochemical process of glycolysis — and passes it through a series of enzymes, eventually leading to glucose import, phosphorylation and metabolism. When glucose is absent, the phosphorelay activity of EIIAGlc ceases, and the phosphorylated enzyme induces cAMP production9.
Although elegant, this is not the full story. Carbon sources that are not imported by way of the phosphotransferase system also tend to counteract cAMP production. In 1961, Boris Magasanik hypothesized that the general ability of carbon sources to repress the expression of catabolic enzymes reflects the fact that all carbon sources converge to produce a key signalling metabolite — a process he termed catabolite repression10. The relevant catabolites, however, were never identified.
You et al. now show that the key catabolites are α-ketoacids, which inhibit adenylate cyclase independently of the phosphotransferase system. Although several α-ketoacids can inhibit adenylate cyclase, α-ketoglutarate is the most abundant11 and therefore likely to be physiologically dominant. As both an intermediate of the energy-producing tricarboxylic acid cycle and the carbonaceous substrate for nitrogen assimilation, α-ketoglutarate reflects the balance of carbon to nitrogen in available nutrients. Inhibition of adenylate cyclase by α-ketoglutarate explains the long-standing observations12–14 that low nitrogen availability blocks expression of what we now know are cAMP-induced genes. Fascinatingly, the role of α-ketoglutarate as both a metabolite and a master regulator is evolutionarily conserved; in humans it serves as a cofactor to enzymes that covalently modify transcription factors, histone proteins and DNA.
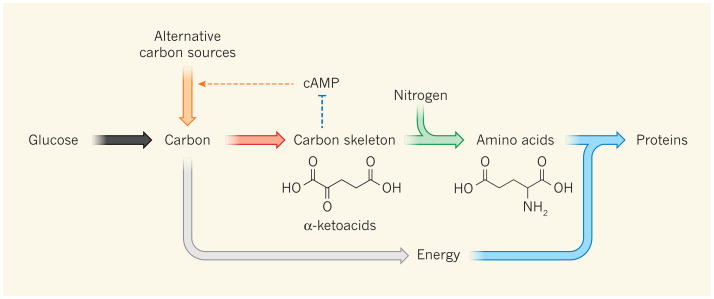
How and why does this regulatory connection lead to linear relationships between gene expression and cellular growth rate in microbes? When carbon limitation slows cellular growth, there is an increasing need for carbon-catabolic enzymes and a decreasing need for anabolic enzymes, which promote biosynthesis and use carbon as a building block (Fig. 1). Quantitatively, in the absence of futile cycling (in which two metabolic pathways operating in opposite directions cancel out each other's effects, wastefully using up energy), the cell's anabolic metabolism is directly proportional to growth rate. So, as growth slows, the required amount of anabolic enzymes, assuming their activities are constant, decreases linearly. Accordingly, the resulting ‘empty space’ in the proteome — which will be filled with enzymes required for coping with the carbon limitation, such as cAMP-regulated gene products — increases linearly with decreasing cellular growth.
Figure 1. Regulation of cAMP levels by carbon and nitrogen availability.
The main biosynthetic task of the bacterium Escherichia coli is protein production. This requires energy generation (grey arrow), carbon-skeleton synthesis, nitrogen incorporation to make amino acids, and protein synthesis. The anabolic fluxes (red, green and blue arrows) increase linearly with growth rate, as does the required anabolic proteome fraction except for processes that are directly slowed by nutrient limitation (such as amino-acid synthesis during nitrogen limitation). Carbon catabolism provides energy and building blocks for anabolism, and cAMP allocates the proper fraction of the proteome to carbon-catabolic enzymes. You et al.3 find that this is achieved by a new regulatory loop, wherein α-ketoacids inhibit cAMP production: when favoured carbon sources such as glucose are present or nitrogen is limiting, carbon influx exceeds anabolic capability and α-ketoacid accumulation inhibits cAMP. Conversely, when favoured carbon sources are depleted, α-ketoacid levels fall, and cAMP increases to stimulate production of the required carbon-catabolic machinery (orange arrows).
When growth slows because of nitrogen limitation, there is less need for carbon-catabolic enzymes and more demand for those involved in nitrogen assimilation. This time, the decrease in the requirement for carbon-catabolic flux is linear with decreasing growth rate. Thus, the situation is flipped, but the optimal responses still remain linear.
Inhibition of cAMP production by α-ketoglutarate naturally produces the desired responses. Whenever carbon-catabolic machinery is in excess relative to anabolic machinery, α-ketoglutarate accumulates, cAMP levels fall and carbon-catabolic enzymes are repressed. Conversely, when anabolic machinery is in excess, α-ketoglutarate is depleted, cAMP levels rise and carbon-catabolic enzymes increase. The steady-state concentration of cAMP is therefore the factor that ensures that the proper amount of the proteome is devoted to carbon-catabolic enzymes. Consequently, the physiological function of cAMP signalling goes beyond simply enabling hierarchical utilization of carbon sources, which is of unclear significance for fitness and can also be achieved through other mechanisms (including inducer exclusion15). Instead, cAMP controls the fraction of the proteome devoted to carbon catabolism.
In a broader historical context, during the half-century between the coining of the term catabolite repression and the present work, physiology has largely taken a back seat to molecular genetics and, more recently, to genomics. Therefore, You and colleagues' use of quantitative physiology to elucidate the molecular mechanism of catabolite repression is particularly noteworthy. One hopes that it is the beginning of a rebalancing, in which physiology-driven systems biology emerges as a full equal to research driven by molecular biology. As the present paper shows, the strength of the physiology-driven approach is not in finding the full scope of molecular events occurring in biological systems, but in identifying the most functionally important ones.
Contributor Information
Joshua D. Rabinowitz, Email: joshr@genomics.princeton.edu, the Department of Chemistry and at the Lewis-Sigler Institute for Integrative Genomics, Princeton University New Jersey 08544, USA.
Thomas J. Silhavy, the Department of Molecular Biology, Princeton University, Princeton, New Jersey 08544, USA
References
- 1.Maaløe O. In: Biological Regulation and Development. Goldberger RF, editor. 1979. pp. 487–542. Plenum. [Google Scholar]
- 2.Klumpp S, Zhang Z, Hwa T. Cell. 2009;139:1366–1375. doi: 10.1016/j.cell.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.You C, et al. Nature. 2013 http://dx.doi.org/10.1038/nature12446.
- 4.Monod J. PhD thesis. Univ. Paris; 1942. Recherches sur la croissance des cultures bactériennes. [Google Scholar]
- 5.Jacob F, Monod J. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 6.Makman RS, Sutherland EW. J Biol Chem. 1965;240:1309–1314. [PubMed] [Google Scholar]
- 7.Emmer M, deCrombrugghe B, Pastan I, Perlman R. Proc Natl Acad Sci USA. 1970;66:480–487. doi: 10.1073/pnas.66.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zubay G, Schwartz D, Beckwith J. Proc Natl Acad Sci USA. 1970;66:104–110. doi: 10.1073/pnas.66.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deutscher J, Francke C, Postma PW. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magasanik B. Cold Spring Harb Symp Quant Biol. 1961;26:249–256. doi: 10.1101/sqb.1961.026.01.031. [DOI] [PubMed] [Google Scholar]
- 11.Bennett BD, et al. Nature Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandelstam J. Nature. 1957;179:1179–1181. doi: 10.1038/1791179a0. [DOI] [PubMed] [Google Scholar]
- 13.Daniel J, Danchin A. Biochimie. 1986;68:303–310. doi: 10.1016/s0300-9084(86)80027-x. [DOI] [PubMed] [Google Scholar]
- 14.Doucette CD, Schwab DJ, Wingreen NS, Rabinowitz JD. Nature Chem Biol. 2011;7:894–901. doi: 10.1038/nchembio.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inada T, Kimata K, Aiba H. Genes Cells. 1996;1:293–301. doi: 10.1046/j.1365-2443.1996.24025.x. [DOI] [PubMed] [Google Scholar]