SUMMARY
How commensal microbiota contributes to immune cell homeostasis at barrier surfaces is poorly understood. Lamina propria (LP) T helper 17 (Th17) cells participate in mucosal protection and are induced by commensal segmented filamentous bacteria (SFB). Here we show that MHCII-dependent antigen presentation of SFB antigens by intestinal dendritic cells (DCs) is crucial for Th17 cell induction. Expression of MHCII on CD11c+ cells was necessary and sufficient for SFB-induced Th17 cell differentiation. Most SFB-induced Th17 cells recognized SFB in an MHCII-dependent manner. SFB primed and induced Th17 cells locally in the LP and Th17 cell induction occurred normally in mice lacking secondary lymphoid organs. The importance of other innate cells was unveiled by the finding that MHCII deficiency in group 3 innate lymphoid cells (ILCs) resulted in an increase in SFB independent Th17 cell differentiation. Our results outline the complex role of DCs and ILCs in the regulation of intestinal Th17 cell homeostasis
INTRODUCTION
Commensal bacteria control mucosal and systemic immune responses (Macpherson and Harris, 2004). It is increasingly becoming appreciated that the composition of gut microbiota affects the homeostasis or function of most immune subsets in the intestinal lamina propria (LP) as well as systemically (Hill and Artis, 2010; Hooper et al., 2012). In particular, the homeostasis of steady state mucosal T cell subsets is controlled by signals from various components of the microbiota (Honda and Littman, 2012; Ivanov and Honda, 2012). T helper 17 (Th17) and regulatory T (Tregs) cells are the most abundant lamina propria CD4 T cell subsets at steady state. Treg cells are crucial for establishment of oral tolerance and for curbing excessive inflammatory responses toward the large numbers of resident commensal bacteria ((Josefowicz et al., 2012; Nutsch and Hsieh, 2012). Th17 cells are characterized by the production of the cytokine interleukin-17 (IL-17), but may also produce a number of other effector cytokines, e.g. IL-17F and IL-22. Th17 cell cytokines function as important activators of innate immune mechanisms, such as recruitment of neutrophils and induction of anti-microbial peptide production from epithelial cells and Th17 cells play key roles in mucosal defense against bacteria and fungi (Korn et al., 2009). In general, Treg cells and Th17 cells have antagonistic functions and the balance between these two subsets is an important determinant of how the mucosal immune system will respond to external challenges (Honda and Littman, 2012).
Treg and Th17 cell differentiation is controlled by the expression of the lineage-specific transcription factors forkhead box P3 (Foxp3) and RAR-related orphan receptor γt (RORγt) respectively, which are differentially induced during T cell activation by a specific combination of T cell receptor (TCR) and cytokine signals ((Josefowicz et al., 2012; Korn et al., 2009). Cytokines responsible for the differentiation of Th17 cells have been well defined in vitro (Korn et al., 2009). In contrast, the role of individual cytokines in controlling Th17 cell numbers or fine-tuning Th17 cell differentiation in vivo is not clearly understood and the role and nature of the TCR signals, including the context of antigen presentation, the participating antigens, the strength and location of antigen priming, and the receptor specificities of naturally-occurring Th17 cells are unknown.
At steady state, both Th17 and Treg cells are enriched in the intestinal LP. This is most likely due to their unique roles in mucosal protection and the immune requirements of the gut microenvironment. Treg and Th17 cell numbers in the gut are controlled by signals from different components of the commensal microbiota. Colonic Treg cells are induced by a combination of group IV and XIVa Clostridia, and small intestinal (SI) Th17 cells are induced by segmented filamentous bacteria (SFB) (Atarashi et al., 2013; Atarashi et al., 2011; Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009). Indeed, the increase in the Treg:Th17 cell ratio in the colon versus small intestine closely reflects the increase in relative abundance of group IV and XIVa Clostridia and decrease in SFB epithelial colonization between these two locations. Although both Treg and Th17 cells can be generated in the absence of the inducing bacteria, these commensals specifically increase the corresponding T cell subset, which profoundly influences intestinal immune responses (Atarashi et al., 2011; Ivanov et al., 2009). Moreover, in both cases, systemic effects on Th17 or Treg responses have also been demonstrated (Atarashi et al., 2011; Berer et al., 2011; Lee et al., 2011; Wu et al., 2010).
How Clostridia and SFB respectively modulate Treg and Th17 cell homeostasis is currently unknown. Both groups of commensals reside in the lumen and do not normally cross the epithelial barrier. It is generally thought that commensal-derived metabolites gain access to the LP and act on LP immune cells to generate a cytokine environment that promotes Treg or Th17 cell differentiation. In support of such mechanism commensal-derived short-chain fatty acids induce epigenetic changes to stabilize the Treg cell differentiation program (Arpaia et al., 2013; Furusawa et al., 2013; Smith et al., 2013). In the case of SFB-induced Th17 cell differentiation, Th17 cell-inducing microbiota modifies LP dendritic cell (DC) cytokine production and SFB induce secretion of cytokines, such as serum amyloid A, from IECs that may affect DC cytokine production (Ivanov et al., 2009). However, whether these SFB-induced changes in the cytokine environment are sufficient for Th17 cell induction is not known.
Here we focused on characterizing the mechanisms by which SFB induce Th17 cells in the SI LP, the innate immune cells involved, and the geography of Th17 cell induction. We find that although SFB-induced cytokine environment is important, it is not sufficient to promote Th17 cell differentiation of activated CD4 T cells. We show that, in vivo, SFB-mediated Th17 cell induction in the gut involves SFB-derived antigens and generation of SFB-specific Th17 cells. SFB-induced Th17 cells, but not intestinal Th17 cells in general, required antigen presentation by MHCII in the periphery. In contrast, SFB-independent Th17 cells could be generated even in the absence of MHCII. We also show that most, if not all, SFB-induced intestinal Th17 cells recognize SFB antigens, in contrast to non-Th17 CD4 T cells and to the small number of Th17 cells present in SFB-negative animals. DC restricted expression of MHCII was necessary and sufficient for induction of Th17 cells by SFB, demonstrating that CD11c+ intestinal DCs (iDCs) are the cells that acquire and present SFB antigens for Th17 cell induction. At the same time, ablation of MHCII-expression in group 3 RORγt+ innate lymphoid cells (ILC3) did not preclude induction of Th17 cells by SFB, but led to an increase in SFB-independent Th17 cell numbers in the gut, demonstrating that ILCs curb Th17 cell differentiation in the absence of SFB in an MHCII-dependent manner. Furthermore, priming of microbiota-induced Th17 cells was detected only in the gut mucosa and Th17 cell induction was unaffected in mice lacking peripheral lymph nodes (LN) and organized gut-associated lymphoid tissues (GALT), suggesting that antigen presentation, T cell priming, and Th17 cell differentiation occur locally in the LP.
RESULTS
Microbiota-induced intestinal Th17 cells are selected on MHCII
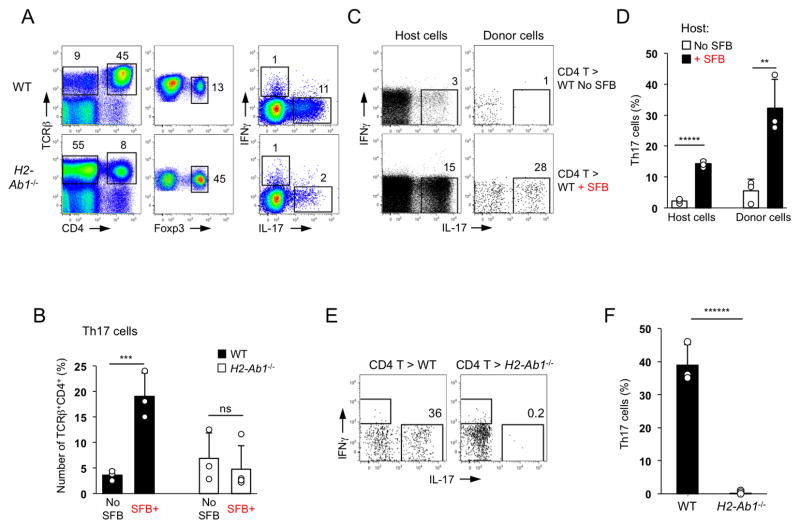
SFB preferentially increase Th17 cell proportions in the SI LP ((Ivanov et al., 2009) and Figure S1A). To examine the dependence of intestinal microbiota-induced Th17 cells on MHCII we analyzed intestinal CD4 T cell homeostasis in MHCII-deficient mice (IAb−/−). IAb−/− animals have greatly decreased numbers of CD4 T cells due to lack of selection on MHCII. However, a small subset of CD4 T cells is still present in spleen and lymph nodes (Cosgrove et al., 1991; Grusby et al., 1991). The small and large intestinal lamina propria of IAb−/− animals contained a substantial CD4 T cells population (Figure 1A). The CD4 T cell population in IAb−/− animals contained an expanded proportion of Foxp3+ Treg cells in all tissues examined (Figure 1A and S1B). In contrast, SI LP Th17 cells were decreased in SFB-positive IAb−/− animals compared to WT controls (Figure 1A). To examine whether Th17 cells in IAb−/− mice are controlled by SFB we compared T cell subsets in the presence and absence of SFB. Th17 cell numbers varied greatly in IAb−/− mice (Figure 1B). However, in contrast to WT mice, SFB-positive IAb−/− mice did not have significantly increased Th17 cell percentages in the gut (Figure 1B). Therefore, even though Th17 cells can be generated in the absence of MHCII, SFB do not induce further Th17 cell differentiation in MHCII-deficient animals.
Figure 1. Induction of intestinal Th17 cells by SFB requires MHCII expression in the periphery.
A. Th17 and Treg cell proportions in SI LP of SFB-positive WT and MHCII-deficient (IAb−/−) mice. Foxp3 and cytokine staining plots are gated on TCRβ+CD4+ cells
B. Th17 and Treg cell proportions in SI LP of SFB-negative (Jackson microbiota) and SFB-positive (Taconic microbiota) WT and IAb−/− mice. Plots gated on TCRβ+CD4+ cells
C–D. WT CD45.1+ CD4 T cells were adoptively transferred into WT CD45.2 mice before or 12 days after SFB colonization. Cytokine expression in host (CD45.2+) and donor (CD45.1+) SI LP TCRβ+CD4+ cells 2 weeks after transfer. Data from one of multiple experiments
E–F. Th17 cell induction in WT CD4 T cells two weeks after transfer into SFB-positive WT and MHCII-deficient recipients. Plots gated on TCRβ+CD4+ cells. Data from one of multiple experiments
IAb−/− mice possess diverse, but quite distinct CD4 T cell repertoire (Cardell et al., 1995). In order to examine if MHCII expression is required for SFB-mediated induction of WT Th17 cells, we performed adoptive transfer experiments. We first established that adoptively transferred WT CD4 T cells develop into Th17 cells in the SI LP in an SFB-dependent manner. Indeed, donor WT CD4 T cells were easily detected in the SI LP of SFB-negative and SFB-positive recipients, but acquired IL-17 expression only in SFB-positive recipients, similarly to endogenous CD4 T cells (Figure 1C, D). Purified naïve CD4 T cells also generated Th17 cells only in the SI LP of SFB-positive hosts (Figure S1C, D). We next transferred WT CD4 T cells into SFB-positive WT and IAb−/− recipients. WT CD4 T cells were detected in large numbers in the SI LP of IAb−/− recipients but did not generate any Th17 cells even in the presence of SFB (Figure 1E, F). Identical results were obtained after transfer of purified naïve CD4 T cells (Figure S1C, D). Collectively, these data demonstrate that the induction of LP Th17 cells by SFB requires MHCII expression in the periphery.
SFB-induced intestinal Th17 cells recognize SFB antigens
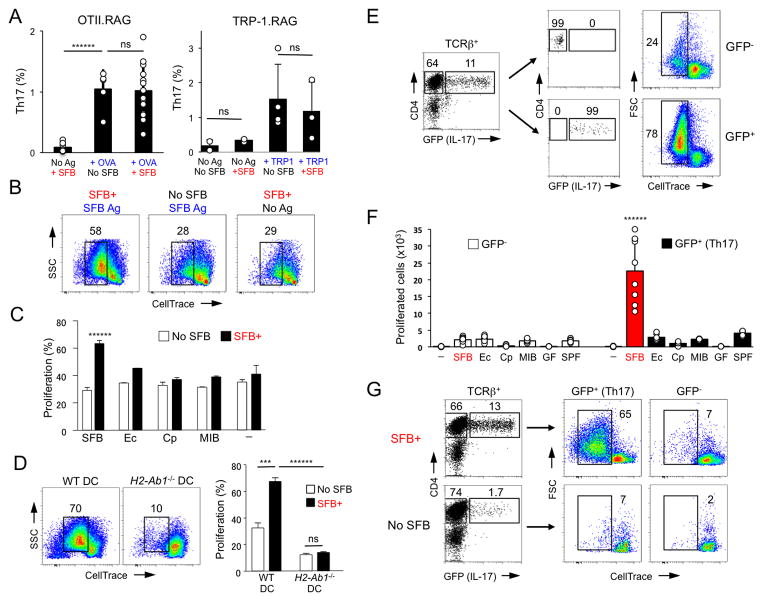
SFB presence does not promote Th17 cell differentiation in MHCII-deficient CD4 T cells, suggesting that SFB provide more than just specific cytokine environment. We, therefore, examined SFB effects on Th17 cell induction in two TCR Tg models – OTII and TRP-1, which recognize peptides from chicken ovalbumin (OVA) and mouse tyrosinase related protein 1 (TRP-1) respectively (Muranski et al., 2008). We first examined OTII mice on a RAG-sufficient background. These mice contained large number of CD4 T cells in the LP and a high proportion of these cells expressed IL-17 in SFB-positive mice, and even in the absence of OVA (Figure S2A). In contrast to spleen and MLNs, CD4 T cells in the gut contained a large fraction of non-Tg TCRs as demonstrated by the low proportion of Vα2hiVβ5hi Tg CD4 T cells (Figure S2B). We therefore used this selection against transgenic TCRs in the LP to compare the re-programming of these cells to Th17 cells in comparison to cells expressing alternative TCRs. As shown on Figure S2A, Vα2hiVβ5hi Tg CD4 T cells expressed very little IL-17, compared to the remaining CD4 T cells, even after activation with the cognate antigen (OVA). In agreement with this result, virtually all IL-17 expressing cells in the LP, expressed alternative TCRs, demonstrating an exclusion of Tg TCRs at the expense of endogenously formed TCRs with broad antigenic specificities in the Th17 cell subset (Figure S2C). As a result, purified intestinal IL-17+ CD4 T cells from OTII.B6 Tg mice responded equally well to OVA and to SFB antigens, in sharp contrast to lymph node CD4 T cells, which responded to OVA only (Figure S2D). These results demonstrate that the intestinal Th17 cell population in OTII.B6 Tg mice is enriched for non-Tg specificities, due to favorable co-expression of non-transgenic TCRs. They also suggest that non-SFB Tg T cells, e.g. OTII cells, are not efficiently induced into the Th17 cell lineage by SFB.
To more directly examine the effects of SFB on non-SFB Tg T cells, we examined TCR Tg animals on a RAG-deficient background, which lack alternative endogenous TCRs. In both, OTII.RAG and TRP-1.RAG Tg mice small numbers of Tg T cells were present in the SI LP in the absence of the cognate antigen, but none of these cells expressed IL-17 even after SFB colonization (Figure 2A and S2E, F). LP Tg T cells were activated and expanded following administration of cognate antigen, which also led to induction of effector Th1 and Th17 cells (Figure 2A and S2E, F). However, even in the presence of the cognate antigen, SFB colonization did not induce further conversion of Tg T cells into Th17 cells (Figure 2A). Moreover, SFB colonization did not induce Th17 cell differentiation of TRP-1.RAG Tg CD4 T cells transferred into WT mice separately or together with WT CD4 T cells, despite considerable expansion and activation of the Tg T cells and despite the presence of endogenous SFB-induced Th17 cells and induction of Th17 cell differentiation in co-transferred WT cells (Figure S3A, B). Combined, these experiments demonstrate that SFB-conditioned intestinal environment is not sufficient to induce IL-17 expression in all activated CD4 T cells.
Figure 2. SFB-induced intestinal Th17 cells preferentially respond to SFB antigens.
A. Th17 cell proportions in the SI LP of OTII.RAG and TRP-1.RAG TCR Tg mice before and after SFB colonization in the absence or presence of cognate antigen. Representative data from 5 independent experiments
B–C. Proliferation response of sorted SI LP TCRβ+CD4+ cells from SFB-negative (Jax) and SFB-positive (Tac) WT B6 mice to SFB (B, C) or other bacterial antigens (C). T cell proliferation was scored by dye dilution on Day 3. Ec, E. coli, Cp, Clostridium perfringens; MIB, mouse intestinal bacteria (cultured isolates from feces of SFB-negative (Jackson) mice); “-” – no antigen. Representative data from 5 independent experiments
D. SI LP TCRβ+CD4+ cells were purified from SFB-negative (No SFB) and SFB-positive (SFB+) WT mice and co-cultured with SFB antigens as in (B) and WT or IAb−/− DCs. Data from 2 independent experiments
E–F. SI LP GFP+ (Th17) and GFP− (non-Th17) TCRβ+CD4+ cells from SFB-positive Il17GFP mice were stimulated in vitro with SFB (E, F) or various bacterial antigens (F) as in (B) or with lysates from germ-free (GF) or SFB-negative SPF (SPF) animals. Representative data from multiple experiments
G. SI LP GFP+ (Th17) and GFP− (non-Th17) TCRβ+CD4+ cells from SFB-positive (SFB+) or SFB-negative (No SFB) Il17GFP mice were stimulated in vitro with SFB antigens as in (B). Representative data from 2 independent experiments
We next examined whether SFB-induced Th17 cells preferentially respond to SFB. We isolated CD4 T cells from SI LP of SFB-positive and SFB-negative WT mice and compared their response to SFB antigens ex vivo. Purified SI LP CD4 T cells from SFB-positive WT mice responded to SFB antigens, while SI LP CD4 T cells isolated from SFB-negative mice, did not (Figure 2B). In contrast, SI LP CD4 T cells from SFB-positive and SFB-negative mice did not respond significantly to a number of non-SFB bacteria, including Gram-negative E. coli, Gram-positive Clostridium perfringens, and cultured murine intestinal isolates (Figure 2C), demonstrating that LP CD4 T cells from SFB-positive animals are specifically enriched for SFB reactivities. The SFB-specific response required antigen presenting cells and MHCII expression, because purified WT SI LP CD4 T cells from SFB-positive mice did not respond to SFB antigens in the absence of DCs or when co-cultured with MHCII-deficient DCs as antigen presenting cells (Figure 2D and data not shown).
To investigate directly the response of gut Th17 cells, we purified GFP+ (Th17) and GFP− (non-Th17) CD4 T cells from the SI LP of SFB-colonized Il17GFP reporter mice (Figure S3C) and stimulated them in vitro with various bacterial lysates. Th17 cells responded strongly to SFB, while non-Th17 cells did not respond to SFB above background (Figure 2E, F). The response of purified Th17 cells was specific to SFB, because the same cells did not respond significantly to cultured bacteria (Figure 2F). In response to SFB, all proliferated GFP+ cells continued to express IL-17 (GFP) (Figure S3G), in contrast to the small proportion of proliferated GFP− cells, which remained mostly IL-17− (Figure S3G). Furthermore, LP Th17 cells did not respond to lysates prepared from feces of germ-free (GF) or SFB-negative conventionally raised mice (SPF) that contained similar numbers of total bacteria (Figure 2F) confirming that the response is SFB-specific. To examine whether the response is directed towards an antigen from SFB, as opposed to an SFB-induced host protein, we prepared lysates from SFB filaments purified by cell sorting (Figure S3D). LP Th17 cells responded to sorted SFB filaments, while non-Th17 cells did not (Figure S3E). We conclude that LP Th17 cells respond to SFB-derived protein antigens. To examine whether the SFB-specific response in intestinal Th17 cells is directed by the presence of SFB, we purified GFP+ (Th17) cells from SI LP of Il17GFP reporter mice before and after SFB colonization and examined their response to SFB. In contrast to Th17 cells isolated from SFB-positive mice, Th17 cells from SFB-negative mice did not proliferate in response to the same SFB antigen preparation (Figure 2G). Similar results were obtained when Th17 cells were isolated on the basis of RORγt expression from SFB-positive and SFB-negative RorcGFP reporter mice (Figure S3F). These results demonstrate that Th17 cells from SFB-positive, but not from SFB-negative, mice preferentially recognize SFB antigens and are, therefore, enriched for SFB specificities.
Most lamina propria Th17 cells recognize SFB antigens
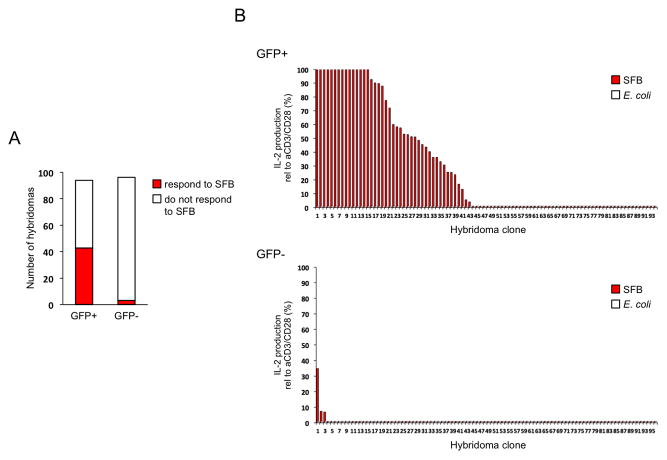
The in vitro co-culture experiments showed that LP Th17 cells from SFB-positive mice respond to SFB antigens. However, the strong proliferative response in these experiments can be due to expansion of a small subset of clones within the starting Th17 population. To more directly quantify the proportion of LP Th17 cells that recognize SFB antigens we decided to query the TCR specificities of individual cells in the total LP Th17 population. To this goal we generated a collection of T cell hybridomas from GFP+ (Th17) and GFP− (non-Th17) SI LP CD4 T cells, isolated from SFB-positive mice, and examined the response of individual clones to SFB and control bacteria. As shown on Figure 3A, 43 out of 94 hybridomas from LP Th17 cells, or 46%, responded to SFB. In contrast, only 3% of the non-Th17 cell hybridomas (3 out of 96) responded to SFB (Figure 3A). Most Th17 cell hybridomas responded strongly to SFB (50–100% of the maximum anti-CD3/anti-CD28 stimulation) and did not respond to E. coli or C. perfringens antigens (Figure 3B and data not shown). In comparison, the three positive hybridomas from non-Th17 cells responded weakly (5–35%), though specifically, to SFB antigens (Figure 3B). Taking into account that Th17 cells from SFB-negative mice do not respond to SFB antigens and that there is 5–7 fold Th17 cell increase upon SFB colonization, the hybridoma results show the presence of at least 55–57% SFB-reactive cells in the SFB-induced Th17 cell population. This response was diverse and polyclonal, as demonstrated by the sequences of SFB-responsive hybridomas (Table S1). 14 out of 15 sequenced SFB-responsive hybridomas had unique TCRβ CDR3 junctions with diverse lengths (Table S1). SFB-induced Th17 cells in vivo, as well as Th17 cell hybridomas used a wide range of Vβ gene segments, though we did note a relative abundance of Vβ14 TCRs (Figure S4 and Table S1). We, therefore, conclude that most SFB-induced Th17 cells recognize SFB antigens and this SFB-specific Th17 cell response is diverse and polyclonal.
Figure 3. Most intestinal SFB-induced Th17 cells recognize SFB.
T cell hybridomas were generated from SI LP GFP+ (Th17) and GFP− (non-Th17) CD4 T cells from SFB-positive Il17GFP mice. Data combined from 2 independent experiments
A. Number of hybridomas responding to SFB
B. Response of individual hybridomas (percentage of maximum anti-CD3 and anti-CD28 stimulation) to SFB or E. coli antigens as assessed by IL-2 production. Clones were ordered in decreasing amounts of IL-2 production
MHCII expression on DC is necessary and sufficient for induction of Th17 cells by SFB
MHCII expression in the periphery is required for SFB-mediated Th17 cell induction. We, therefore, next investigated the nature of the participating MHCII-expressing cells that present SFB antigens. Several types of MHCII-expressing cells exist in the LP. These include professional APCs, DCs and B cells, as well as other cell types, such as intestinal epithelial cells (IECs) and ILCs. To investigate the role of professional APCs, we first examined Th17 cell induction by SFB in the absence of B cells. Th17 cell induction after SFB colonization occurred normally in Cμ-deficient mice (Kitamura et al., 1991), which lack mature B cells (Figure S5H), demonstrating that B cells are not required for SFB-mediated Th17 cell induction.
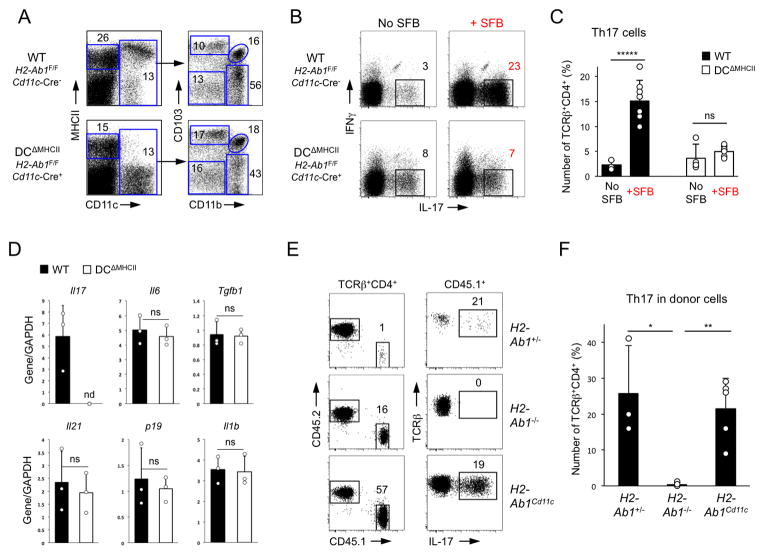
To investigate whether MHCII-dependent antigen presentation by intestinal DCs is required, we generated mice with DC-specific ablation of MHCII (DCΔMHCII mice), by inter-crossing IAbflox mice (Hashimoto et al., 2002) with CD11c-Cre mice (Caton et al., 2007) (Figure 4). All major LP CD11c+ subsets were present in DCΔMHCII mice in similar numbers, however they completely lacked MHCII expression (Figure 4A and Figure S5A). In contrast, MHCII expression was present on CD11c-negative cells (Figure 4A), including IgA+ plasma cells and B220+ B cells, though it was reduced on the latter (Figure S5B).
Figure 4. DC expression of MHCII is necessary and sufficient for SFB-mediated Th17 cell induction.
A. SI LP lymphocytes from DCΔMHCII and control littermates. Left panels, gated on TCRβ−CD4− cells. Right panels, gated on CD11c+ cells
B–C. Th17 cell induction in DCΔMHCII mice and control littermates 2 weeks after SFB colonization. Plots gated on TCRβ+CD4+ cells. Representative data from 4 independent experiments
D. Relative cytokine expression (RT-PCR) in terminal ileum of DCΔMHCII and control littermates (WT) 2 weeks after colonization with SFB. nd – below threshold of detection
E–F. Th17 cell differentiation of WT CD45.1+ CD4 T cells in the SI LP 2 weeks after transfer into SFB-positive IAb+/−, IAb −/−, and IAbCD11c CD45.2+ recipient littermates. Data from 2 independent experiments
In the absence of SFB, DCΔMHCII mice had slightly increased Th17 cell percentage over control littermates (Figure 4B, C). SFB colonization induced robust Th17 cell differentiation in control animals. In contrast, SFB colonization did not lead to a statistical increase in Th17 cells in DCΔMHCII mice, even though SFB numbers and attachment were similar to the littermate controls (Figure 4B, C and Figure S5F, G). Ablation of MHCII on DCs did not affect the expression of major Th17-inducing cytokines [e.g. IL-6, IL-23, transforming growth factor-β (TGF-β), IL-1β], or the induction of a number of other cytokines and enzymes, such as SAA1, SAA3, and iNOS, by SFB (Figure 4D and S5C), demonstrating lack of major changes in the cytokine milieu. We conclude that MHCII expression on intestinal DCs is required for induction of Th17 cells by SFB. Consistent with our previous studies (Ivanov et al., 2009), SFB colonization did not affect Th1 or Treg cell proportions in WT or DCΔMHCII mice (Figure S5D, E). Treg cell proportions were increased in the intestinal LP of DCΔMHCII mice, though, as previously reported (Darrasse-Jeze et al., 2009), they were decreased in spleen and MLN, suggesting that MHCII expression on DCs may have different roles in Treg cell control in gut versus secondary lymphoid organs (Figure S5E).
To examine whether MHCII expression on DCs is sufficient for induction of Th17 cells by SFB, we examined Th17 cell induction in mice in which MHCII expression is restricted to CD11c+ cells (MHCIICD11c mice) (Lemos et al., 2003). Because MHCIICD11c mice lack MHCII expression on thymic epithelium and are deficient in CD4 thymic positive selection, we performed adoptive transfers. As in previous experiments, transfer of WT CD4 T cells led to an SFB-dependent Th17 cell induction in the SI LP, which was abrogated in the absence of peripheral MHCII expression in MHCII-deficient recipients (Figure 4E, F). Transfer of WT CD4 T cells into littermate MHCIICD11c mice led to considerable induction of Th17 cells in the transferred cells, demonstrating that recovery of MHCII expression only on CD11c+ cells is sufficient to promote Th17 cell induction (Figure 4E, F).
Combined, these results show that MHCII expression on intestinal DCs is necessary and sufficient for SFB-mediated induction of Th17 cells.
MHCII expression on ILCs controls intestinal Th17 cells
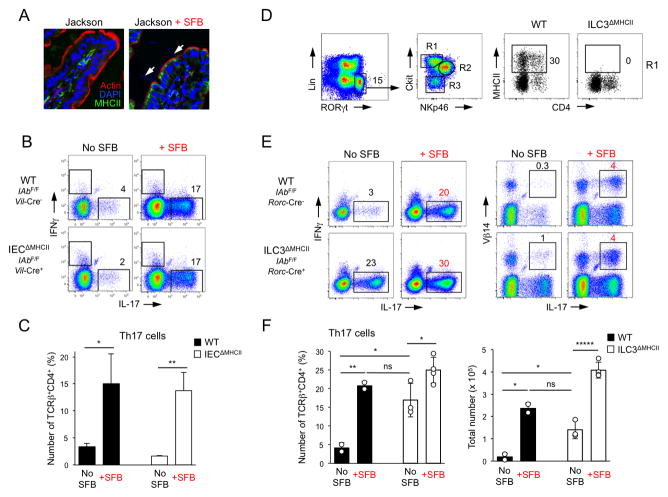
Several other non-conventional antigen-presenting cell subsets express MHCII in the intestine. These include IECs and ILCs (Hepworth et al., 2013; Hershberg and Mayer, 2000). MHCII expression on IECs has an unknown function and is controlled by commensal bacteria (Umesaki et al., 1995). Notably, MHCII expression on IECs was induced very specifically by SFB only in the terminal ileum (Figure 5A and data not shown). To investigate the role of IEC MHCII expression in SFB-mediated Th17 cell induction we generated IECΔMHCII mice, in which MHCII was deleted only on IECs by crossing IAbflox mice with Villin-Cre mice (Madison et al., 2002) (Figure S6A, C). Colonization of SFB-free IECΔMHCII mice with SFB (Figure S6A, B) led to induction of Th17 cells, similar to that in littermate controls (Figure 5B, C), demonstrating that MHCII expression on IECs is not required for this process.
Figure 5. RORγt+ ILCs inhibit differentiation of SFB-independent intestinal Th17 cells through MHCII.
A. MHCII expression on IECs in Jackson B6 mice before and 2 weeks after SFB colonization. Arrows point to SFB filaments attaching to IECs
B–C. Th17 cell induction in IECΔMHCII mice and control littermates 2 weeks after SFB colonization. Plots gated on TCRβ+CD4+ cells. Data from one of 2 independent experiments
D. Expression of MHCII on SI LP c-kit+NKp46−RORγt+ group 3 ILCs in ILC3ΔMHCII mice and control littermates
E–F. Th17 cell induction in ILC3ΔMHCII and control littermates 2 weeks after SFB colonization. Plots gated on TCRβ+CD4+ cells. Data from 2 independent experiments
We also discovered that a subset of intestinal ILCs express MHCII (Figure 5D), as also reported recently (Hepworth et al., 2013). A large proportion of Lin−RORγt+ ILC3 cells express MHCII (Figure 5D). MHCII expression was most prevalent in c-kit+NKp46−RORγt+ ILCs, and on only a small fraction of NKp46+ or c-kit− ILCs (Figure 5D and S6D). To examine the role of MHCII expression on ILCs in SFB-mediated Th17 cell induction we generated ILC3ΔMHCII mice, in which MHCII was deleted only on RORγt+ ILCs by inter-crossing MHCII-floxed mice with RORγt-Cre mice (Eberl and Littman, 2004) (Figure 5D and S6D, E). All three ILC3 subsets were present in ILC3ΔMHCII mice (data not shown). MHCII expression was completely ablated on Lin−NKp46−c-kit+RORγt+ LTi-like cells and significantly decreased in the small subset of MHCII+ cells within the remaining two ILC3 subsets (Figure 5D and S6D). ILC3ΔMHCII mice did not demonstrate any signs of rectal prolapse or intestinal inflammation in our colony and MHCII deletion on ILC3s did not affect the percentage of SI LP Tregs (Figure S6F, G). Surprisingly, in contrast to SFB-free control littermates, which had low numbers of Th17 cells, ILC3ΔMHCII animals contained high percentage and numbers of SI LP Th17 cells even in the absence of SFB (Figure 5E, F). SFB-free ILC3ΔMHCII mice contained as many Th17 cells as SFB-colonized WT littermates (Figure 5F). Colonization of WT mice with fecal bacteria from SFB-negative ILC3ΔMHCII animals did not induce Th17 cells, arguing against an outgrowth of other Th17 cell-inducing bacteria (Figure S6H). In contrast to Th17 cells in SFB-positive WT animals, Th17 cells in SFB-negative ILC3ΔMHCII mice did not respond to SFB antigens in vitro (Figure S6I). Colonization with SFB induced further increase in both percentages and total numbers of Th17 cells in 9-week old ILC3ΔMHCII mice (Figure 5F and Figure S6J, K). In agreement with our observation that SFB induce SFB-specific Th17 cells, SFB colonization induced Th17 TCR repertoire changes in ILC3ΔMHCII mice, such as the induction of Vβ14+IL-17+ CD4 T cells (Figure 5E and S6L), and a response to SFB antigens in vitro (Figure S6I). Collectively, these results demonstrate that Th17 cells are increased in SFB-negative ILC3ΔMHCII mice, but SFB are still capable of inducing Th17 cells in these animals.
Th17 cell induction by SFB does not require LN or organized GALT
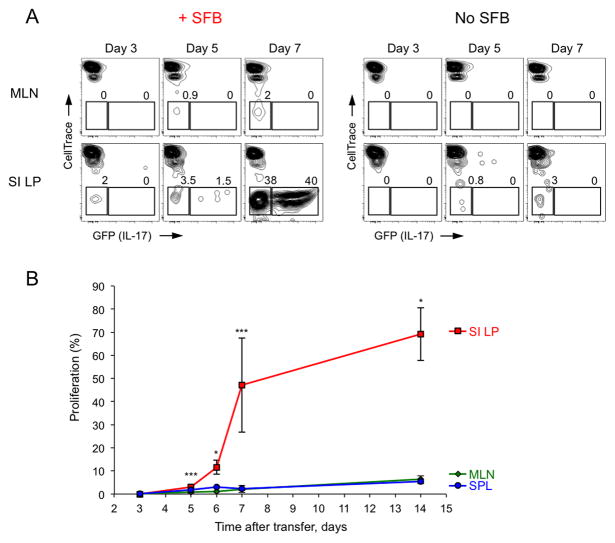
Our results show that SFB antigens are presented by iDCs in the context of MHCII to induce SFB-specific Th17 cells. To examine the site of Th17 cell priming in WT mice we analyzed the kinetics of SFB-mediated CD4 T cell proliferation and Th17 cell differentiation in different tissues following adoptive transfer (Figure 6). CD4 T cells were purified from spleens and LNs of Il17GFP reporter mice, labeled with proliferation dye and transferred into congenic WT recipients before or after SFB colonization. Proliferation was scored by dye dilution and Th17 cell differentiation by induction of GFP (IL-17) expression at different time points post transfer. A small number of proliferating transfer cells were first detected in the SI LP at Day 3 after transfer (Figure 6A). The number of proliferating cells increased by Day 5 and some of those produced IL-17, again only in the SI LP, but not in spleen or MLN. By Day 7, transferred cells in SFB-colonized animals proliferated robustly and differentiated into Th17 cells in the SI LP. T cell proliferation and Th17 cell induction was dependent on the presence of SFB and was very low in SFB− animals (Figure 6A). In contrast, despite being present in larger numbers, very few transferred cells proliferated in the MLN at Day 7 and none expressed IL-17 (Figure 6A). Albeit lower than the SI LP, proliferation and Th17 cell induction was also observed in Peyer’s Patches (PPs) starting at Day 6 (Figure S7). Further increase in proliferation and Th17 cell differentiation of transferred cells in the SI LP was observed at 2 weeks post transfer (Figure 6B and data not shown). However, we did not detect significant proliferation or IL-17 expression in MLNs, iLNs, or spleen of SFB-colonized animals at any time-point, suggesting that SFB priming and induction of Th17 cells occurs in the small intestine itself (Figure 6B and data not shown).
Figure 6. Priming and induction of Th17 cells by SFB occurs in the small intestine.
CellTrace Violet labeled CD45.2+ CD4 T cells from Il17GFP mice were transferred into WT CD45.1+ recipients before (No SFB) or after (+SFB) SFB colonization
A–B. Proliferation (A, B) and Th17 cell induction (A) at indicated time points. Plots are gated on CD45.2+TCRβ+CD4+ transferred cells. SI LP, small intestinal LP; MLN, mesenteric lymph nodes; SPL, spleen. Combined data from 3 independent experiments
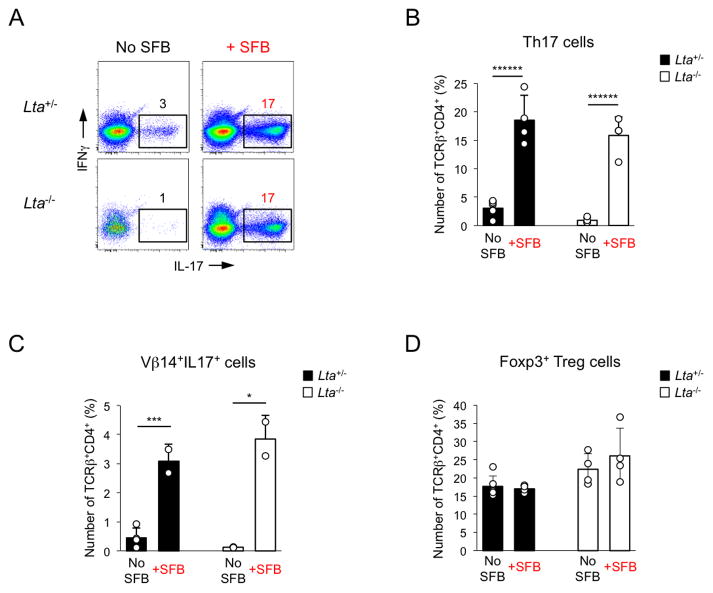
To investigate directly whether organized GALT is required, we examined the induction of Th17 cells by SFB in lymphotoxin-α (LTα)-deficient mice. Lta−/− mice possess a defect in generation of secondary lymphoid organs and lack PPs and isolated lymphoid follicles (ILFs) in the intestine, as well as peripheral lymph nodes, including MLNs. Lta−/− animals also lack LP B cells (Newberry et al., 2002). Despite these defects, induction of Th17 cells by SFB, including induction of Vβ14+IL-17+ cells, was unimpeded in Lta−/− mice (Figure 7), demonstrating that organized GALT is not required for this process and confirming that LP B cells are also dispensable. Therefore, sampling of SFB antigens can occur outside Peyer’s Patches or MLNs and Th17 cell induction does not require priming in peripheral lymph nodes, suggesting that iDCs acquire SFB antigens and prime CD4 T cells locally in the LP.
Figure 7. SFB induce Th17 cells in the absence of secondary lymphoid organs.
SI LP lymphocytes were isolated from Lta−/− and control littermates 2 weeks after colonization with SFB
A–C. Th17 and Vβ14+IL-17+ cells induction in TCRβ+CD4+ cells. Representative data from one of 2 independent experiments
D. Foxp3+ Treg cell proportions in TCRβ+CD4+ cells. Combined data from 2 independent experiments
DISCUSSION
Intestinal T cell homeostasis is a major factor in preventing chronic intestinal inflammation, while maintaining mucosal immunity and response to intestinal infections. The mechanisms by which commensal microbes modulate T cell homeostasis are, therefore, of considerable interest, but have remained poorly understood due to the complexity and diversity of commensal-host interactions. SFB are the only currently known individual commensal capable of specifically inducing Th17 cells. However, the mechanisms of this induction have not been investigated in detail. SFB can induce changes in the gut cytokine environment by affecting cytokine expression from both iDCs and IECs (Ivanov et al., 2009; Shima et al., 2008), but whether these cytokines are required or sufficient to drive Th17 cell induction is not known. Here, we find that simply presence of SFB-induced cytokine environment is not sufficient to induce Th17 cells in vivo. However, presentation of SFB-derived antigens by iDCs is crucial and drives generation of SFB-specific Th17 cells.
Th17 cells were present in the intestines of MHCII-deficient mice and therefore Th17 cells can be selected on alternative ligands. However, SFB colonization did not induce Th17 cells in IAb−/− animals or in transferred WT CD4 T cells in IAb−/− recipients. Therefore, even though Th17 cells can be generated in the absence of MHCII, induction of Th17 cells by SFB requires MHCII expression in the periphery. These results also suggest that antigen presentation is crucial for Th17 cell induction by SFB and that SFB-induced Th17 cells may have skewed TCR specificities. Indeed, we found that most intestinal Th17 cells recognize SFB antigens in an MHCII-dependent manner. The enrichment for SFB specificities is quite substantial as demonstrated by our hybridoma experiments, in which close to 60% of independent intestinal Th17 cell TCRs responded to SFB, mostly with strong IL-2 production. At the same time only 3% of non-Th17 hybridomas from the same animals responded only weakly to SFB, demonstrating that most, if not all, SFB-specific T cells become Th17 cells. A strong MHCII-dependent response to bacterial antigens may be mediated by the expression of a superantigen. However, SFB-induced Th17 cells had a diverse Vβ-chain usage and there was a high diversity in Vβ usage among SFB-recognizing hybridomas generated from intestinal Th17 cells. Therefore, Th17 cells are not likely to be induced by an SFB superantigen, but more likely by conventional presentation of SFB antigens in the context of MHCII. The importance of Th17 cell priming by SFB antigens was also illustrated by the fact that SFB colonization did not induce Th17 cell differentiation in two types of non-SFB Tg T cells even after activation of the Tg T cells by providing the cognate antigen and even after placing them in an SFB environment that induced Th17 cells in WT cells. Therefore, in addition to providing important cytokine environment, SFB also provide antigens for Th17 cell differentiation. Our results differ from a previous study that showed microbiota-dependent induction of LP Th17 cells in the absence of cognate antigen (Lochner et al., 2011). The presence of SFB was not specifically examined in this study and therefore the effects observed might be due to other microbial entities, which are strongly affecting the local cytokine environment and are antigen and/or MHCII-independent. Similarly, we show here that Th17 cells can also be present in the gut even in the absence of antigen presentation in the context of MHCII. However, our results clearly show that SFB induce Th17 cells by a unique mechanism that requires antigen presentation by iDCs. Indeed, our results show that SFB-induced cytokine environment is not sufficient to induce Th17 cell differentiation of non-SFB Tg T cells in the absence, or even presence, of antigen. It is unclear why SFB-specific CD4 T cells differentiate preferentially into Th17 cells. Our results show that most Th17 cells in SFB-positive mice are SFB-specific and combined with our hybridoma experiments suggest that most, if not all, SFB-specific CD4 T cells in the gut are Th17 cells. To preferentially induce Th17 cells, SFB may provide antigens that induce an appropriate amount of TCR stimulation or are presented in the appropriate context by certain APCs. This may explain why we were unable to skew Th17 cell differentiation of Tg T cells in SFB-positive mice by providing the antigen orally or intravenously, which may differ from the context, way, or location these antigens are delivered in vivo by a commensal (Iqbal et al., 2002). In this respect, all our efforts to colonize SFB-positive mice containing OTII cells with OVA-expressing E. coli were unsuccessful.
The number and type of SFB antigens contributing to Th17 cell induction remain to be determined. Despite an increase in Vβ14+IL-17+ cells we did not find evidence of oligoclonality. SFB-induced Th17 cells utilized a diverse range of Vβ segments and SFB-reacting hybridomas showed a diverse and polyclonal CDR3 repertoire. This suggests that SFB provide multiple antigens for Th17 cell induction, although we cannot exclude a highly polyclonal response to an individual antigen. At the same time SFB seem to secrete only a handful of proteins as the highly reduced SFB genome lacks any additional protein secretion systems, as well as sortase-dependent secretory LPXTG-motif containing proteins (Sczesnak et al., 2011). Combined, these observations suggest that Th17 cell-inducing SFB antigens may be acquired by direct interaction and sampling of the bacteria by host cells. The geography and mechanisms of this sampling, as well as all of the participating host cell types, will be important to examine in detail in future studies. The experiments described here show that SFB antigens are ultimately acquired by iDCs. Ablation of MHCII expression on CD11c+ cells resulted in abrogation of the effect of SFB on promoting Th17 cell expansion. Of note, Th17 cells were still present in the LP of SFB-free DCΔMHCII mice in similar proportions, or even increased, compared to WT littermates, but were not further induced by SFB. Moreover, when MHCII expression was restricted only to DCs, in MHCIICD11c mice, transferred WT cells differentiated into Th17 cells in an SFB-dependent matter. Therefore, MHCII expression on intestinal DCs is both required and sufficient for the induction of Th17 cells by SFB. The CD11c+ subset that we term DCs here represents at least three developmentally and functionally distinct subsets that include CD103+ conventional DCs and CX3CR1+ monocytes (Bogunovic et al., 2012; Farache et al., 2013). Both CD103+ and CX3CR1+ cells have been implicated in mucosal Th17 cell differentiation, generally through production of Th17 cell-inducing cytokines (Bogunovic et al., 2012; Farache et al., 2013). In contrast, here we show that one or several of these subsets must also present SFB antigens in order for SFB to induce Th17 cells. Further studies will be needed to dissect the specific role of each of the three CD11c+ subsets in the process.
How and where SFB antigens are delivered or acquired by the DCs is also unknown. Most antigen specific responses in the LP originate in secondary lymphoid organs, such as PPs and MLNs. However, induction of commensal-specific IgA may also occur in the lamina propria itself (Fagarasan et al., 2001; Fritz et al., 2012). Whether antigen-specific mucosal T cell responses are exclusively initiated in MLNs and PPs is less clear. An earlier study, found normal numbers of Th17 cells in PP-null, but MLN-sufficient mice, suggesting that Th17 cell responses do not require PPs (Atarashi et al., 2008). To examine the niche for Th17 cell priming by SFB, we followed proliferation and IL-17 induction of transferred CD4 T cells. SFB-specific T cell proliferation, followed by IL-17 expression occurred shortly after transfer only in the small intestine, including SI LP and PPs, although proliferation was always higher in the SI LP. Proliferation and IL-17 expression were not observed in peripheral secondary lymphoid tissues, including MLNs. In addition, SFB-colonization induced normal numbers of Th17 cells in Lta−/− mice, which lack all peripheral LNs and PPs. Combined, these data suggest that SFB-mediated T cell priming and Th17 cell differentiation occur locally in the small intestine. This means that SFB antigens are acquired by iDCs and presented directly in the intestinal mucosa. The normal induction of Th17 cells in Lta−/− mice demonstrates that the process can take place in the SI LP in the absence of PPs. However in WT mice SFB antigens may be delivered in both PPs and LP and further studies will be needed to pinpoint the exact mechanism of this acquisition. SFB are closely associated with the intestinal epithelium in the terminal ileum. Therefore direct sampling of SFB may occur for example through interdigitating DCs or absorption of damaged or apoptotic IECs (Rescigno et al., 2001; Torchinsky et al., 2009).
In further support that Th17 cell homeostasis is controlled by signals locally in the LP, we found that lack of MHCII expression on RORγt+ ILCs leads to an increase in Th17 cell differentiation. ILC3ΔMHCII mice had an increase in Th17 cell proportions even in the absence of SFB. An earlier study found that break in T cell homeostasis in ILC3ΔMHCII mice leads to intestinal inflammation at 12 weeks of age, correlated with increased IL-17 and IL-22 cytokine production (Hepworth et al., 2013). Inflammation and cytokine production were alleviated after antibiotic treatment, demonstrating dependence on intestinal bacteria (Hepworth et al., 2013). In contrast, we did not observed any rectal prolapse or evidence of intestinal inflammation in ILC3ΔMHCII mice in our colony up to 30 weeks of age. Nevertheless, the increase in Th17 cells was present as early as 6 weeks of age (data not shown). Therefore ILCs inhibit Th17 cell differentiation in the gut in an MHCII-dependent manner. The increase in Th17 cells most likely leads to intestinal pathology in the presence of pro-inflammatory microbiota (Hepworth et al., 2013), however our results suggest that the effect on T cell homeostasis precedes that of the pro-inflammatory microbiota. Our results are in agreement with the conclusion of an earlier report that ILCs inhibit Th17 cell differentiation (Qiu et al., 2013). In this study, the effect was attributed to a negative control on SFB colonization. Increase in SFB numbers in ILC-deficient mice leads to more Th17 cells and increase in intestinal inflammation (Qiu et al., 2013). In contrast, the ILC-mediated inhibition of Th17 cell differentiation that we report here is clearly SFB-independent. Moreover, the presence of SFB, even in the presence of larger numbers of Th17 cells did not lead to an increase in intestinal inflammation, arguing that the increase in intestinal inflammation in models with decreased ILC function depends on the presence of other local intestinal pathogens (Hepworth et al., 2013; Qiu et al., 2013). SFB-negative ILC3ΔMHCII mice contained as many Th17 cells as SFB-positive WT mice. Nevertheless, SFB were still able to induce Th17 cells in these animals. Our experiments demonstrate the existence of SFB-dependent and SFB-independent intestinal Th17 cells, controlled by different homeostatic mechanisms and by different innate immune subsets. How this occurs requires further study. Our experiments show that Th17 cells present in the absence of SFB in ILC3ΔMHCII mice and Th17 cells induced by SFB have different specificities and are therefore induced by different mechanisms, e.g. in different priming context or by different types of antigen presenting cells. As we show here, acquisition and presentation of SFB antigens involves selective sampling by iDCs in the LP and skewed Th17 cell induction. In contrast, loss of epithelial barrier function and increased bacterial translocation that have been reported in ILC3ΔMHCII mice (Hepworth et al., 2013), will likely lead to Th17 cell induction by alternative mechanisms. More studies will be needed to establish whether SFB-dependent and SFB-independent intestinal Th17 cells differ functionally and whether the SFB-mediated Th17 cell induction is an evolutionary adaptation for bacterial colonization or an alternative mechanism of Th17 cell induction acquired by the host.
Our results shed further light into the complex interactions involved in controlling intestinal Th17 cell homeostasis. Antigen presentation by MHCII plays central role in the induction of Th17 cells by commensal microbiota. Intestinal DCs acquire antigens from Th17 cell-inducing bacteria to promote antigen-specific Th17 responses locally in the lamina propria. At the same time, RORγt+ ILCs control excessive Th17 cell responses by inhibiting Th17 cell differentiation also in an MHCII-dependent manner. Further characterization of this process, such as identification of the participating SFB antigens or the involvement of specific subsets of DCs or ILCs, will help in better understanding the molecular and cellular mechanisms involved in the host-commensal crosstalk regulating T cell homeostasis in the gut.
EXPERIMENTAL PROCEDURES
Mice
MHCII-floxed (IAbF/F), Cd11c-Cre, Vil-Cre, Cμ-deficient, Il17GFP, TRP-1.RAG1 and Lta−/− mice were obtained from Jackson Laboratory. Rorc-Cre mice (Eberl and Littman, 2004) were a gift from Dan Littman (NYU). MHCII-deficient (IAb−/−), OTII.B6 and OTII.RAG1 mice were obtained from Taconic Farms, the latter through the NIAID Exchange Program (Barnden et al., 1998). MHCIICD11c mice (also known as CD11c-Aβb mice) were previously described (Lemos et al., 2003). All mice were bred and housed under specific pathogen-free conditions at Columbia University Medical Center. To control for microbiota and caging effects all experiments were performed with littermate control and gene-deleted animals housed in the same cage.
SFB colonization and Th17 cell induction
Bacterial genomic DNA isolation from fecal pellets and quantitative PCR for the SFB 16S rRNA gene were performed as previously described (Ivanov et al., 2009). SFB colonization was performed by oral gavage with fecal pellets from SFB-monocolonized mice or with fecal pellets from SFB-negative Jackson B6 mice colonized with feces from SFB-monocolonized mice unless otherwise noted. Control mice were gavaged with fecal pellets from SFB-negative littermates. SI LP Th17 cell induction was assessed 2–3 weeks after colonization.
Activation of TCR Tg T cells
To activate TCR Tg T cells, OTII.B6 and OTII.RAG Tg mice were fed 1% OVA protein in the drinking water for 12 days. In addition the mice were orally gavaged with 50 mg of OVA protein (OVA, grade V; Sigma) on Day 1,3, and 5. TRP-1.RAG Tg mice were immunized i.p. with 50μg TRP1 peptide (CGTCRPGWRGAACNQKILTVR, 92% purity, Biomatik) in DPBS and 10μg LPS (Sigma-Aldrich) on Day 1 and 7. Control animals were immunized only with 10μg LPS in DPBS.
Lamina propria cell isolation and adoptive transfers
Lamina propria lymphocytes, intracellular cytokine staining, and Foxp3 staining were performed as previously described (Ivanov et al., 2008). For adoptive transfers, 5–10 × 106 MACS-purified CD4 T cells (Miltenyi Biotec; 95–98% purity) or 5 × 106 FACS sorted TCRβ+CD4+CD62LhiCD44lo naïve T cells (BD Influx cell sorter) were transferred intravenously into Ly5.1 WT recipients before or 10–14 days after SFB colonization. Th17 cell induction in transferred cells in different tissues was assessed 2 weeks after transfer unless otherwise noted. In some experiments cells were labeled with CellTrace Violet proliferation dye (Life Technologies).
In vitro co-culture experiments
LP TCRβ+CD4+ T cell subsets were purified by cell sorting and labeled with CellTrace Violet proliferation dye. 5 × 104 CD4 T cells were co-cultured in 96-well U-bottom plates with either 5 × 104 MACS purified splenic CD11c+ cells or 2 × 105 total TCRα-KO splenocytes as APCs in the presence or absence of autoclaved bacterial lysates. T cell proliferation was assessed 72 hours later by dye dilution. For SFB antigens, SFB filaments were purified from feces of SFB-monocolonized mice. Briefly, individual fecal pellets from SFB-monocolonized mice were homogenized in PBS. The supernatant was cleared from debris by several low-speed centrifugations and bacteria were pelleted by centrifugation at 4,000g and washed twice with PBS. After the final wash the pellet was resuspended in PBS and layered onto 60% w/v Nycodenz density gradient. SFB filaments were collected at the interphase and the procedure repeated. Finally, the SFB filaments were washed twice in PBS. SFB and other bacterial antigens were prepared by autoclaving bacterial suspensions and used at 1:200 dilution.
Hybridoma generation and screening
FACS purified SI LP TCRβ+CD4+GFP+ and TCRβ+CD4+GFP− T cells were stimulated in vitro for 3 days in tissue culture plates coated with 5ug/ml each of anti-CD3 and anti-CD28 mAbs, fused with BW5147 thymoma (White et al., 1989) and plated in limited dilutions in selective media. Individual clones were picked 10 days later and expanded in 24-well plates. The response of cloned hybridomas towards autoclaved bacterial lysates was measured using the HT-2 assay (Pacholczyk et al., 2007). In brief, 105 hybridoma cells were stimulated with plate-bound aCD3/aCD28 (positive control) or incubated with 105 bone-marrow-derived dendritic cells (or splenocytes from TCRα-KO mice) alone (no antigen control), or with SFB, E. coli, or Clostridium perfringens lysates. After 24 h, the amount of secreted IL-2 was measured with the detector HT-2 cell line. The proliferation of HT-2 cells in response to IL-2 was measured with the MTT (Sigma) assay (Pacholczyk et al., 2007), and the response for each hybridoma was plotted as percentage from the IL-2 response in the aCD3/aCD28-stimulated positive control.
Statistics
Significance was scored by using unpaired two-tailed t test unless otherwise noted. P values were represented on figures as follows: ns, p ≥ 0.05, * p < 0.05, ** p < 0.01, *** p < 0.005, **** p < 0.001, ***** p < 0.0005, ****** p < 0.0001. Error bars on all figures represent standard deviation of the mean.
Supplementary Material
HIGHLIGHTS.
Presentation of SFB antigens by iDC is required for Th17 cell induction by SFB
SFB-induced Th17 cells recognize SFB antigens
SFB prime CD4 T cells and induce Th17 cell differentiation in the gut mucosa
Loss of MHCII on RORγt ILC leads to Th17 cell differentiation in the absence of SFB
Acknowledgments
We thank members of the I.I. laboratory for their help in all aspects of the project. We thank Dr. Yoshinori Umesaki at the Yakult Central Institute for providing feces from SFB-monocolonized mice. We thank Kenya Honda and Seiko Narushima for developing and sharing the SFB sorting protocol. We thank Steve Reiner and Boris Reizis for reading the manuscript and for invaluable scientific discussions. We thank Siu-Hong Ho in the Columbia Center for Translational Immunology Flow Cytometry Core and Amir Figueroa at the Department of Microbiology and Immunology Flow Cytometry Core for help with sorting. This work was supported by the National Institute of Health 1R01DK098378 to I.I. and by the Crohn’s and Colitis Foundation of America SRA#259540 to I.I.. G.N. was supported by a long-term research grant from Toyobo Biofoundation. I.I. is a Pew Scholar in the Biomedical Sciences, supported by the Pew Charitable Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, Takeda K. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunology and cell biology. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, Wekerle H, Krishnamoorthy G. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- Bogunovic M, Mortha A, Muller PA, Merad M. Mononuclear phagocyte diversity in the intestine. Immunol Res. 2012 doi: 10.1007/s12026-012-8323-5. [DOI] [PubMed] [Google Scholar]
- Cardell S, Tangri S, Chan S, Kronenberg M, Benoist C, Mathis D. CD1-restricted CD4+ T cells in major histocompatibility complex class II-deficient mice. The Journal of experimental medicine. 1995;182:993–1004. doi: 10.1084/jem.182.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. The Journal of experimental medicine. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D, Gray D, Dierich A, Kaufman J, Lemeur M, Benoist C, Mathis D. Mice lacking MHC class II molecules. Cell. 1991;66:1051–1066. doi: 10.1016/0092-8674(91)90448-8. [DOI] [PubMed] [Google Scholar]
- Darrasse-Jeze G, Deroubaix S, Mouquet H, Victora GD, Eisenreich T, Yao KH, Masilamani RF, Dustin ML, Rudensky A, Liu K, Nussenzweig MC. Feedback control of regulatory T cell homeostasis by dendritic cells in vivo. The Journal of experimental medicine. 2009;206:1853–1862. doi: 10.1084/jem.20090746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G, Littman DR. Thymic origin of intestinal alphabeta T cells revealed by fate mapping of RORgammat+ cells. Science. 2004;305:248–251. doi: 10.1126/science.1096472. [DOI] [PubMed] [Google Scholar]
- Fagarasan S, Kinoshita K, Muramatsu M, Ikuta K, Honjo T. In situ class switching and differentiation to IgA-producing cells in the gut lamina propria. Nature. 2001;413:639–643. doi: 10.1038/35098100. [DOI] [PubMed] [Google Scholar]
- Farache J, Zigmond E, Shakhar G, Jung S. Contributions of dendritic cells and macrophages to intestinal homeostasis and immune defense. Immunology and cell biology. 2013;91:232–239. doi: 10.1038/icb.2012.79. [DOI] [PubMed] [Google Scholar]
- Fritz JH, Rojas OL, Simard N, McCarthy DD, Hapfelmeier S, Rubino S, Robertson SJ, Larijani M, Gosselin J, Ivanov II, et al. Acquisition of a multifunctional IgA+ plasma cell phenotype in the gut. Nature. 2012;481:199–203. doi: 10.1038/nature10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Grusby MJ, Johnson RS, Papaioannou VE, Glimcher LH. Depletion of CD4+ T cells in major histocompatibility complex class II-deficient mice. Science. 1991;253:1417–1420. doi: 10.1126/science.1910207. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Joshi SK, Koni PA. A conditional null allele of the major histocompatibility IA-beta chain gene. Genesis. 2002;32:152–153. doi: 10.1002/gene.10056. [DOI] [PubMed] [Google Scholar]
- Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, Mantegazza AR, Ma HL, Crawford A, Angelosanto JM, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg RM, Mayer LF. Antigen processing and presentation by intestinal epithelial cells - polarity and complexity. Immunol Today. 2000;21:123–128. doi: 10.1016/s0167-5699(99)01575-3. [DOI] [PubMed] [Google Scholar]
- Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal N, Oliver JR, Wagner FH, Lazenby AS, Elson CO, Weaver CT. T helper 1 and T helper 2 cells are pathogenic in an antigen-specific model of colitis. The Journal of experimental medicine. 2002;195:71–84. doi: 10.1084/jem.2001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, de Frutos RL, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Honda K. Intestinal commensal microbes as immune modulators. Cell host & microbe. 2012;12:496–508. doi: 10.1016/j.chom.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annual review of immunology. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annual review of immunology. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos MP, Fan L, Lo D, Laufer TM. CD8alpha+ and CD11b+ dendritic cell-restricted MHC class II controls Th1 CD4+ T cell immunity. Journal of immunology. 2003;171:5077–5084. doi: 10.4049/jimmunol.171.10.5077. [DOI] [PubMed] [Google Scholar]
- Lochner M, Berard M, Sawa S, Hauer S, Gaboriau-Routhiau V, Fernandez TD, Snel J, Bousso P, Cerf-Bensussan N, Eberl G. Restricted microbiota and absence of cognate TCR antigen leads to an unbalanced generation of Th17 cells. Journal of immunology. 2011;186:1531–1537. doi: 10.4049/jimmunol.1001723. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. The Journal of biological chemistry. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- Muranski P, Boni A, Antony PA, Cassard L, Irvine KR, Kaiser A, Paulos CM, Palmer DC, Touloukian CE, Ptak K, et al. Tumor-specific Th17-polarized cells eradicate large established melanoma. Blood. 2008;112:362–373. doi: 10.1182/blood-2007-11-120998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberry RD, McDonough JS, McDonald KG, Lorenz RG. Postgestational lymphotoxin/lymphotoxin beta receptor interactions are essential for the presence of intestinal B lymphocytes. Journal of immunology. 2002;168:4988–4997. doi: 10.4049/jimmunol.168.10.4988. [DOI] [PubMed] [Google Scholar]
- Nutsch KM, Hsieh CS. T cell tolerance and immunity to commensal bacteria. Current opinion in immunology. 2012;24:385–391. doi: 10.1016/j.coi.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacholczyk R, Kern J, Singh N, Iwashima M, Kraj P, Ignatowicz L. Nonself-antigens are the cognate specificities of Foxp3+ regulatory T cells. Immunity. 2007;27:493–504. doi: 10.1016/j.immuni.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Guo X, Chen ZM, He L, Sonnenberg GF, Artis D, Fu YX, Zhou L. Group 3 Innate Lymphoid Cells Inhibit T-Cell-Mediated Intestinal Inflammation through Aryl Hydrocarbon Receptor Signaling and Regulation of Microflora. Immunity. 2013;39:386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nature immunology. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, Littman DR, Ivanov II. The genome of Th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host & Microbe. 2011;10:1–13. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima T, Fukushima K, Setoyama H, Imaoka A, Matsumoto S, Hara T, Suda K, Umesaki Y. Differential effects of two probiotic strains with different bacteriological properties on intestinal gene expression, with special reference to indigenous bacteria. FEMS immunology and medical microbiology. 2008;52:69–77. doi: 10.1111/j.1574-695X.2007.00344.x. [DOI] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchinsky MB, Garaude J, Martin AP, Blander JM. Innate immune recognition of infected apoptotic cells directs T(H)17 cell differentiation. Nature. 2009;458:78–82. doi: 10.1038/nature07781. [DOI] [PubMed] [Google Scholar]
- Umesaki Y, Okada Y, Matsumoto S, Imaoka A, Setoyama H. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol Immunol. 1995;39:555–562. doi: 10.1111/j.1348-0421.1995.tb02242.x. [DOI] [PubMed] [Google Scholar]
- White J, Blackman M, Bill J, Kappler J, Marrack P, Gold DP, Born W. Two better cell lines for making hybridomas expressing specific T cell receptors. Journal of immunology. 1989;143:1822–1825. [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.