Abstract
BACKGROUND
CCR5 is the major coreceptor for human immunodeficiency virus (HIV). We investigated whether site-specific modification of the gene (“gene editing”) — in this case, the infusion of autologous CD4 T cells in which the CCR5 gene was rendered permanently dysfunctional by a zinc-finger nuclease (ZFN) — is safe.
METHODS
We enrolled 12 patients in an open-label, nonrandomized, uncontrolled study of a single dose of ZFN-modified autologous CD4 T cells. The patients had chronic aviremic HIV infection while they were receiving highly active antiretroviral therapy. Six of them underwent an interruption in antiretroviral treatment 4 weeks after the infusion of 10 billion autologous CD4 T cells, 11 to 28% of which were genetically modified with the ZFN. The primary outcome was safety as assessed by treatment-related adverse events. Secondary outcomes included measures of immune reconstitution and HIV resistance.
RESULTS
One serious adverse event was associated with infusion of the ZFN-modified autologous CD4 T cells and was attributed to a transfusion reaction. The median CD4 T-cell count was 1517 per cubic millimeter at week 1, a significant increase from the preinfusion count of 448 per cubic millimeter (P<0.001). The median concentration of CCR5-modified CD4 T cells at 1 week was 250 cells per cubic millimeter. This constituted 8.8% of circulating peripheral-blood mononuclear cells and 13.9% of circulating CD4 T cells. Modified cells had an estimated mean half-life of 48 weeks. During treatment interruption and the resultant viremia, the decline in circulating CCR5-modified cells (−1.81 cells per day) was significantly less than the decline in unmodified cells (−7.25 cells per day) (P = 0.02). HIV RNA became undetectable in one of four patients who could be evaluated. The blood level of HIV DNA decreased in most patients.
CONCLUSIONS
CCR5-modified autologous CD4 T-cell infusions are safe within the limits of this study. (Funded by the National Institute of Allergy and Infectious Diseases and others; ClinicalTrials.gov number, NCT00842634.)
The ability to make site-specific modifications to (or “edit”) the human genome has been an objective in medicine since the recognition of the gene as the basic unit of heredity.1,2 The challenge of genome editing is the ability to generate a single double-strand break at a specific point in the DNA molecule. Numerous agents, including meganucleases, oligonucleotides that form DNA triplexes, and peptide nucleic acids, have been tested and shown to be limited by inefficiency.3–5 Another class of reagents, the zinc-finger nucleases (ZFNs), have proved versatile for genome editing, and the use of ZFNs is now well established in a number of model organisms and in human cells.6,7
ZFNs are well suited for genome engineering because they combine the DNA recognition specificity of zinc-finger proteins (ZFPs) with the robust but restrained enzymatic activity of the cleavage domain of the restriction enzyme FokI (a nuclease).6,7 ZFPs, which provide DNA-binding specificity, contain a tandem array of Cys2His2 zinc fingers, each recognizing approximately 3 base pairs of DNA.8 By comparison, the bacterial type IIS restriction endonuclease, FokI, has no sequence specificity and must dimerize to cut the DNA.9 After the ZFN-mediated double-strand cut, the DNA can be repaired by either homologous recombination or nonhomologous end joining. Homologous recombination repairs the break while preserving the original DNA sequence. However, these cells are susceptible to recutting by ZFNs. In contrast, nonhomologous end joining can result in random insertion or deletion of nucleotides at the break site, resulting in permanent disruption of the primary DNA sequence. Therefore, nonhomologous end joining can be exploited to mutate a specific gene, leading to its functional knockout.6,7
The design of a ZFN pair consisting of two 4-finger proteins that bind to a target site within the human chemokine (C-C motif) receptor 5 gene (CCR5) was reported previously.10 In preclinical tests, CCR5-modified CD4 T cells expanded and functioned normally in response to mitogens, were protected from human immunodeficiency virus (HIV) infection, and reduced HIV RNA levels in a humanized mouse model (involving xenotransplantation) of HIV infection.10
We selected CCR5, which encodes a coreceptor for HIV entry,11,12 for several reasons. First, its disruption seemed likely to increase the survival of CD4 T cells; persons homozygous for a 32-bp deletion (delta32/delta32) in CCR5 are resistant to HIV infection.13 In vitro, CD4 T cells from such persons are highly resistant to infection with CCR5-using strains of HIV, which are the dominant strains in vivo.14 Moreover, persons who are heterozygous for CCR5 delta32 and who have HIV infection have a slower progression to the acquired immunodeficiency syndrome.15,16 Furthermore, the effectiveness of blocking or inhibiting CCR5 with the use of small-molecule inhibitors has been shown in humans.17 Finally, one person who underwent allogeneic transplantation with progenitor cells homozygous for the CCR5-delta32 deletion has remained off antiviral therapy for more than 4 years, with undetectable HIV RNA and proviral DNA in the blood, bone marrow, and rectal mucosa.18,19 Although the mechanism responsible for the apparent cure associated with this procedure remains to be established, acquired CCR5 deficiency is one possibility.20 Here we report the partial induction of acquired genetic resistance to HIV infection after targeted gene disruption (i.e., the infusion of autologous CD4 T cells modified at CCR5 by a ZFN).
METHODS
We enrolled 12 patients in two case series (cohort 1 and cohort 2), each with 6 patients (Table 1). The patients had chronic aviremic HIV infection while they were receiving highly active antiretroviral therapy (HAART). Patients were infused with SB-728-T (Sangamo BioSciences), consisting of autologous CD4-enriched T cells that have been modified at the CCR5 gene locus by ZFNs. The investigational ZFN was donated by Sangamo BioSciences, which had no role in any aspect of the study design, the writing of the manuscript, or the decision to submit the manuscript for publication; the ZFN-modified cells were manufactured at the University of Pennsylvania. The primary objective of the study was to assess the safety and side-effect profile of a single dose of autologous CD4-enriched T cells modified at CCR5 by ZFNs. Secondary objectives included the assessment of increases in the CD4 T-cell count, persistence of the modified cells, homing to gut mucosa, and effects on viral load. Details of a concurrent control cohort are outlined in Table S3 in the Supplementary Appendix, available with the full text of this article at NEJM.org. Details of the methods and the statistical analysis are provided in the Supplementary Appendix. All patients provided written informed consent. All the authors vouch for the accuracy and completeness of the data and the fidelity of the study to the protocol.
Table 1.
Patient Demographics and Cell Manufacturing.*
| Cohort and Patient No.† | Age | Race or Ethnic Group |
Sex | Duration of HIV Infection |
Baseline CD4 T-Cell Count |
Baseline CD4:CD8 T-Cell Ratio |
SB-728-T Dose‡ |
SB-728-T CD3§ |
SB-728-T Cell Modification |
SB-728-T Pentamer Duplication |
|---|---|---|---|---|---|---|---|---|---|---|
| yr | yr | per mm3 | % | % | per 106 cells | |||||
| Cohort 1 | ||||||||||
| 201 | 54 | White | M | 20.2 | 665 | 1.38 | l.00×l010 | 97.0 | 14.6 | 57,000 |
| 203 | 50 | Black | M | 21.1 | 659 | 0.59 | 1.08×l010 | 97.7 | 24.5 | 81,000 |
| 204 | 31 | White | M | 4.3 | 621 | 1.43 | l.00×l010 | 97.9 | 10.9 | 47,000 |
| 205 | 50 | White | M | 2.4 | 955 | 1.99 | l.00×l010 | 99.1 | 19.1 | 64,000 |
| 251 | 56 | White | M | 23.1 | 554 | 1.67 | l.00×l010 | 98.6 | 14.4 | 57,000 |
| 253 | 38 | Asian | M | 2.8 | 997 | 0.98 | l.00×l010 | 94.3 | 18.4 | 61,000 |
| Median | 50 | 12.3 | 662 | 1.41 | l.00×l010 | 97.8 | 16.5 | 59,000 | ||
| Cohort 2 | ||||||||||
| 306 | 50 | White | M | 19.1 | 271 | 0.99 | 0.77×l010 | 93.1 | 20.9 | 60,000 |
| 308 | 37 | Black | M | 13.1 | 328 | 0.78 | 0.50×l010 | 97.5 | 25.3 | 67,000 |
| 309 | 48 | Asian Indian | F | 2.9 | 341 | 0.54 | 0.80×l010 | 92.3 | 27.7 | 67,000 |
| 351 | 60 | Black | M | 14.1 | 193 | 1.25 | l.00×l010 | 97.7 | 25.4 | 73,000 |
| 354 | 41 | Hispanic | M | 15.6 | 220 | 0.50 | 0.90×l010 | 99.3 | 26.7 | 69,000 |
| 355 | 44 | Black | F | 16.2 | 272 | 0.65 | l.00×l010 | 94.1 | 26.9 | 72,000 |
| Median | 46 | 14.9 | 272 | 0.72 | 0.85×l010 | 95.8 | 26.0 | 68,000 | ||
SB-728-T consists of autologous CD4 T cells in which the CCR5 gene was rendered permanently dysfunctional by zinc-finger nucleases.
Cohort 1 comprised patients with adequate CD4 T-cell recovery after highly active antiretroviral therapy (HAART), defined as those with CD4 T-cell counts above 450 per cubic millimeter at screening, with a documented nadir of not lower than 300 per cubic millimeter. Cohort 2 comprised patients with inadequate CD4 T-cell recovery after HAART, defined as those with CD4 T-cell counts persistently between 200 per cubic millimeter and 500 per cubic millimeter at screening, despite 2 or more years of HAART.
A single dose of autologous CD4 T cells modified at CCR5 by SB-728-T consisted of an infusion of a median of l.00×l010 total cells in cohort 1, and a median of 0.85xl010 in cohort 2.
The percentages of total cells expressing the T-cell marker CD3 are listed.
RESULTS
ADVERSE EVENTS
One serious adverse event occurred in a single patient from cohort 2. Fever, chills, joint pain, and back pain developed in the patient and precipitated a visit to the emergency department within 24 hours after infusion of the study drug. We attributed the symptoms to a transfusion reaction related to the study drug (see the Supplementary Appendix for further details).
CHANGES IN CIRCULATING LYMPHOCYTES
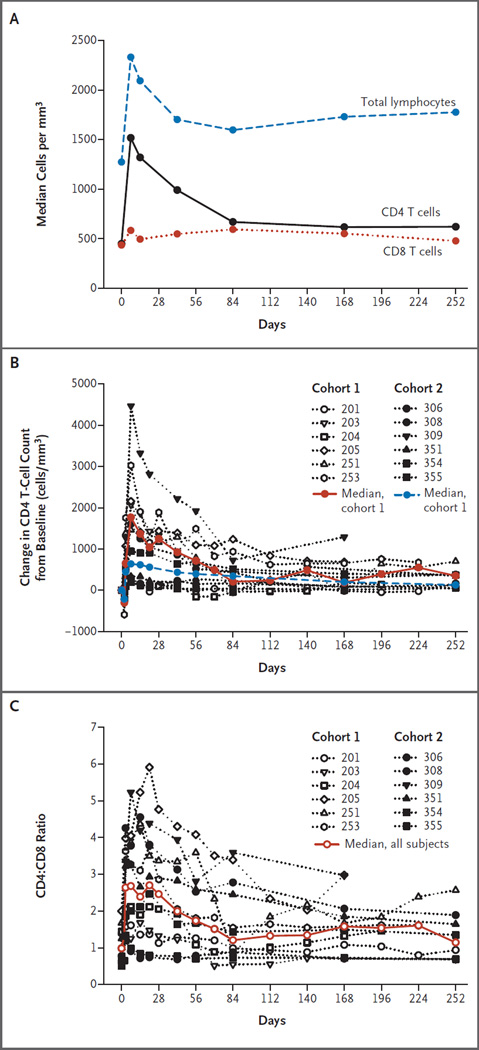
The median total lymphocyte counts within the vascular compartment significantly increased in the 12 patients, from 1.27×103 per cubic millimeter at baseline to 2.33×103 per cubic millimeter 1 week after the infusion of SB-728-T (P = 0.002 with the use of a sign test) (Fig. 1A). Subsequently, the median circulating lymphocyte count progressively declined to 1.70×103 per cubic millimeter by 6 weeks and was stable thereafter (1.60×103, 1.73×103, and 1.78×103 per cubic millimeter at 12, 24, and 36 weeks, respectively). The increase in CD8 T-cell counts was moderate, with a median of 435 per cubic millimeter at baseline as compared with 582 per cubic millimeter at week 1. By comparison, the CD4 T-cell counts in these patients significantly increased, from a median of 448 per cubic millimeter at baseline to 1517 per cubic millimeter at week 1 (P<0.001 with the use of a sign test) (Fig. 1A, and Fig. S1 in the Supplementary Appendix). All patients had increased CD4 T-cell counts after infusion (Fig. 1B, and Tables S1 and S2 in the Supplementary Appendix), but we observed heterogeneity between participants in both cohorts, with most of the increase in CD4 T-cell counts derived from 7 participants who had large increases in CD4 T-cell counts. Median changes in CD4 T-cell count from baseline, according to cohort, are shown in Figure 1B. We observed a median (±SD) increase of 1201±1350 cells per cubic millimeter at week 1 that progressively declined to a median of 615 cells per cubic millimeter at the end of 36 weeks but remained above the baseline levels by 256 cells per cubic millimeter (Fig. 1B, and Fig. S1 in the Supplementary Appendix). The median increase in CD4 T-cell count at week 1 was 1765±1138 per cubic millimeter in participants with adequate CD4 T-cell recovery after HAART (sometimes referred to as immune responders, defined as those with CD4 T-cell counts >450 per cubic millimeter at screening, with a documented nadir of not lower than 300 per cubic millimeter) and 637±1638 per cubic millimeter in participants with inadequate CD4 T-cell recovery after HAART (sometimes referred to as immune nonresponders, defined as those with CD4 T-cell counts persistently between 200 per cubic millimeter and 500 per cubic millimeter at screening, despite ≥2 years of HAART), but the difference between the two groups was not significant (P = 0.75 by the Mann–Whitney test). A finding consistent with these observations was that the median ratio of CD4 T cells to CD8 T cells more than doubled, from 0.99 at baseline to 2.62 at week 1 (Fig. 1C); the median ratio declined to 1.14 by week 36.
Figure 1. Lymphocyte Values.
In Panel A, median total lymphocyte, CD4 T-cell, and CD8 T-cell values for all study participants are shown (see Fig. S1 in the Supplementary Appendix). The increase in total lymphocyte count is due to an increase in the number of CD4 T cells, since changes in CD8 counts are negligible. In Panel B, the change in CD4 T-cell count from baseline is plotted for all participants, and the median change is plotted for each cohort. In Panel C, the median ratios of CD4 T cells to CD8 T cells are plotted for all participants.
ENGRAFTMENT OF CCR5-MODIFIED CD4 T CELLS
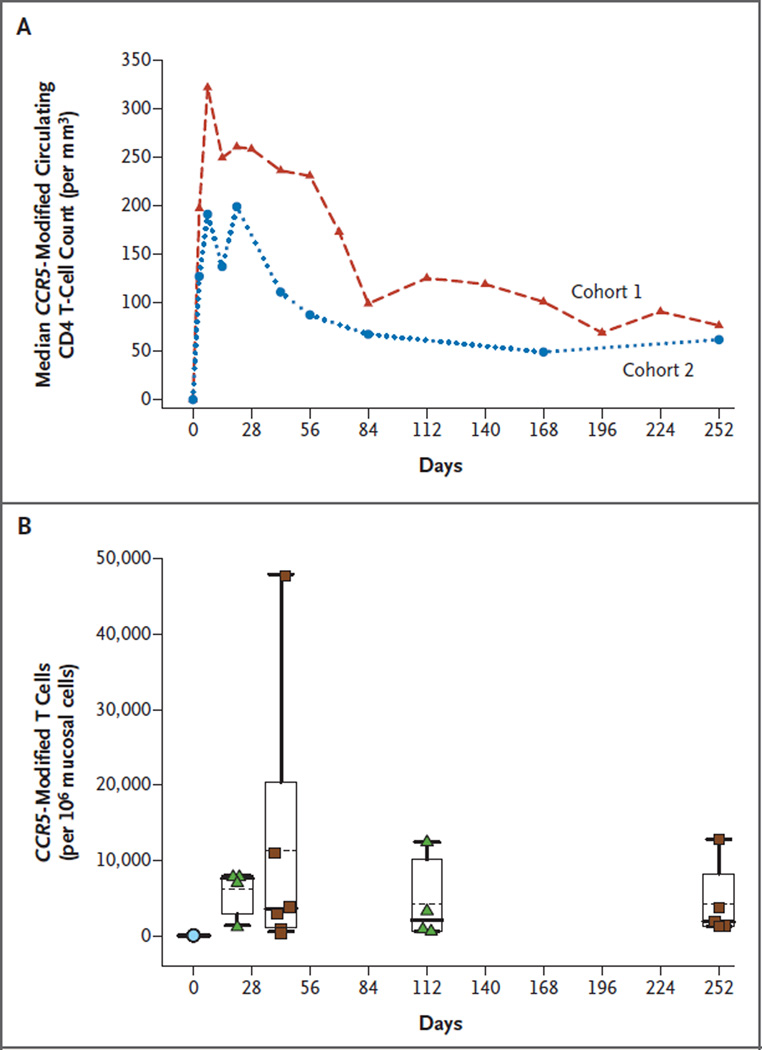
CCR5-modified CD4 T cells could be tracked after infusion because of the creation of a five-nucleotide (pentamer) duplication that occurred in approximately 25% of the modified cells.10 Therefore, the total number of gene-modified cells is calculated by multiplying the number of cells with the pentamer duplication by four (see the Supplementary Appendix). After infusion, we observed an increase in the number of CCR5-modified circulating CD4 T cells (Fig. 2A), with peak levels observed at week 1 (range among the 12 patients, 30 to 1106 cells per cubic millimeter). The median concentration of CCR5-modified CD4 T cells at 1 week was 250 cells per cubic millimeter. This constituted a median of 8.8% of peripheral-blood mononuclear cells (PBMCs) and 13.9% of the CD4 T cells in the vascular compartment (Fig. S2 in the Supplementary Appendix). The number of CCR5-modified CD4 T cells in the circulation constituted a similar percentage of the circulating CD4 T cells and PBMCs in the participants with and in those without adequate CD4 T-cell recovery after HAART (Fig. S2 in the Supplementary Appendix). The time to peak level (known as Tmax) of gene-modified cells ranged from 3 to 14 days (median, 7). The number of gene-modified cells in the vascular compartment decreased moderately, with an estimated mean half-life of 48 weeks at a median follow-up of 64 weeks (range, 24 to 142). The gene-modified T cells could be detected in all patients at all subsequent time points examined during the long-term follow-up study, the longest to date being 42 months in the first patient, at which time CCR5-modified CD4 T cells were present at a concentration of 13 cells per cubic millimeter, representing 0.6% of circulating PBMCs and 1.7% of circulating CD4 T cells, respectively.
Figure 2. CCR5-Modified CD4 T Cells in the Circulation and Mucosal Tissues.
Panel A shows that the median absolute number of CCR5-modified circulating CD4 T cells was similar in participants with adequate CD4 T-cell recovery after highly active antiretroviral therapy (HAART) (cohort 1) and in those with inadequate CD4 T-cell recovery after HAART (cohort 2). Panel B shows CCR5-modified cell traffic to rectal mucosal tissues. Patients in cohort 1 (green triangles) underwent a rectal biopsy at baseline (blue circle) and on days 21 and 112, and those in cohort 2 (brown squares) underwent biopsies at baseline (blue circle) and on days 42 and 252. Box plots show the 25th percentile (lower edge of the box), mean (dotted line in the box), median (solid line in the box), 75th percentile (upper edge of the box), and 90th percentile (whisker). CCR5-modified CD4 T-cells constituted a mean of 0.6%, 1.1%, 0.4%, and 0.4% (and a median of 0.8%, 0.4%, 0.2%, and 0.2%) of rectal mucosal mononuclear cells on days 21, 42, 112, and 252, respectively.
TRAFFICKING OF CCR5-MODIFIED CD4 T CELLS TO RECTAL MUCOSA
In humans, the vascular compartment contains 1 to 2% of the T-cell mass, whereas the mucosal tissues are the largest lymphoid reservoir, containing at least 50% of the T-cell mass.21 In this study, CCR5-modified CD4 T cells were detected in all rectal-biopsy specimens. One patient in cohort 1 declined to undergo the scheduled biopsies; the remaining 11 participants underwent biopsies on two or more occasions. A total of 30 of 33 scheduled biopsies were performed. Gene-modified cells constituted a median of 0.8% of rectal mononuclear cells on day 21 and varied from 0.4% to 0.2% thereafter (Fig. 2B).
TREATMENT INTERRUPTION
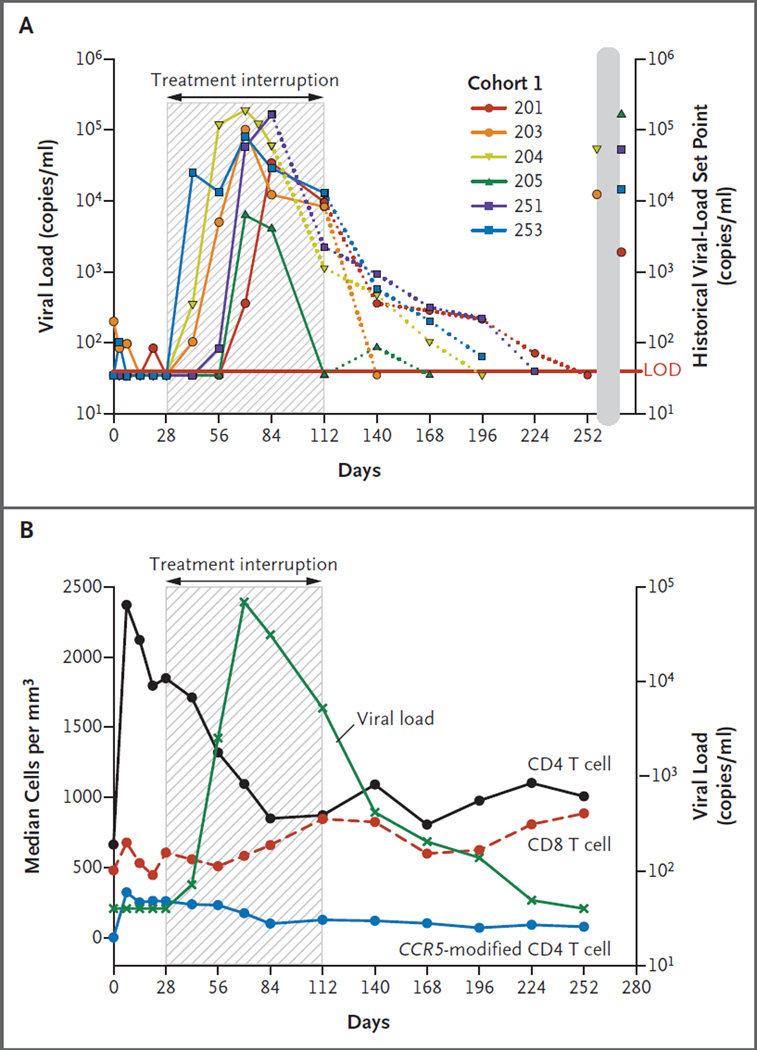
Participants in cohort 1 underwent a 12-week interruption in treatment that began 4 weeks after SB-728-T infusion. Viral load, as measured by HIV RNA, was below the limit of detection in all six patients at the start of the treatment interruption. Viral load became detectable in four of the six patients (66.7%) 2 to 4 weeks after the cessation of HAART and peaked at 6 to 8 weeks. The treatment interruption was terminated prematurely, at 8 weeks, in Patient 204 and Patient 251 (Fig. 3A). In Patient 204, HAART was reinitiated because of three consecutive HIV RNA values exceeding 100,000 copies per milliliter, which was a prospectively defined stopping rule. In Patient 251, HAART was reinitiated because the patient’s primary provider was concerned about the rapid increase in the viral load. In the four patients who completed the 12-week treatment interruption, the viral load decreased during the interruption by an average of 1.2 log10 (range, 0.5 to 2.1) from the peak level during the absence of HAART (P = 0.07). Patient 205, who had a pretreatment viral set point of 165,000 copies per milliliter and who we later discovered is heterozygous for CCR5 delta32, did not show an increase in viral load until week 6 of the treatment interruption, at which time the viral load peaked at 6247 copies per milliliter, a value below the set point. The viral load in this patient declined thereafter and was below the limit of detection before reinstitution of HAART.
Figure 3. Changes in Viremia during Treatment Interruption.
Panel A depicts HIV viral loads (HIV RNA) for the six patients in cohort 1. SB-728-T was infused on day 0. A 12-week (84-day) treatment interruption (shaded area) was initiated on day 28 and terminated on day 112. The treatment interruption was terminated prematurely, on day 84 (week 8 of the interruption period), in Patients 204 and 251. The dotted lines indicate reinstitu-tion of highly active antiretroviral therapy. The historical HIV-RNA set point for each patient is also shown. The limit of detection (LOD) for the viral-load assay is plotted at 50 copies. Patient 205 was heterozygous for CCR5 delta32. Panel B shows the median CD4 T-cell, CD8 T-cell, and CCR5-modi-fied T-cell counts in cohort 1 during the treatment interruption, as well as the viral load.
In cohort 1, the median circulating CD4 T-cell count changed from 1849 per cubic millimeter (range, 720 to 2881) at the onset of the treatment interruption to 1711 per cubic millimeter (range, 719 to 2341) at the onset of viremia (after 2 weeks of treatment interruption) (P = 0.38). Relative to the start of the interruption, at week 4, the median CD4 T-cell count continued to decline, to 1095 per cubic millimeter at the peak of viremia, at week 10 (P = 0.06) (Fig. 3B). At the end of the 12-week treatment interruption, the median CD4 T-cell count was 872 per cubic millimeter; the median decrease from the start of the interruption was 237±315 cells per cubic millimeter (P = 0.69). Similarly, the median number of circulating CCR5-modified CD4 T cells declined from 259 per cubic millimeter at the onset of treatment interruption to 126 per cubic millimeter at the end of the treatment interruption (P = 0.03), with a median decrease of 126±76 cells per cubic millimeter. Concomitant with HIV replication, the median CD8 T-cell count, which had been 605 per cubic millimeter at the start of the treatment interruption, peaked at 845 per cubic millimeter at the end of the treatment interruption, at week 16 (P = 0.69).
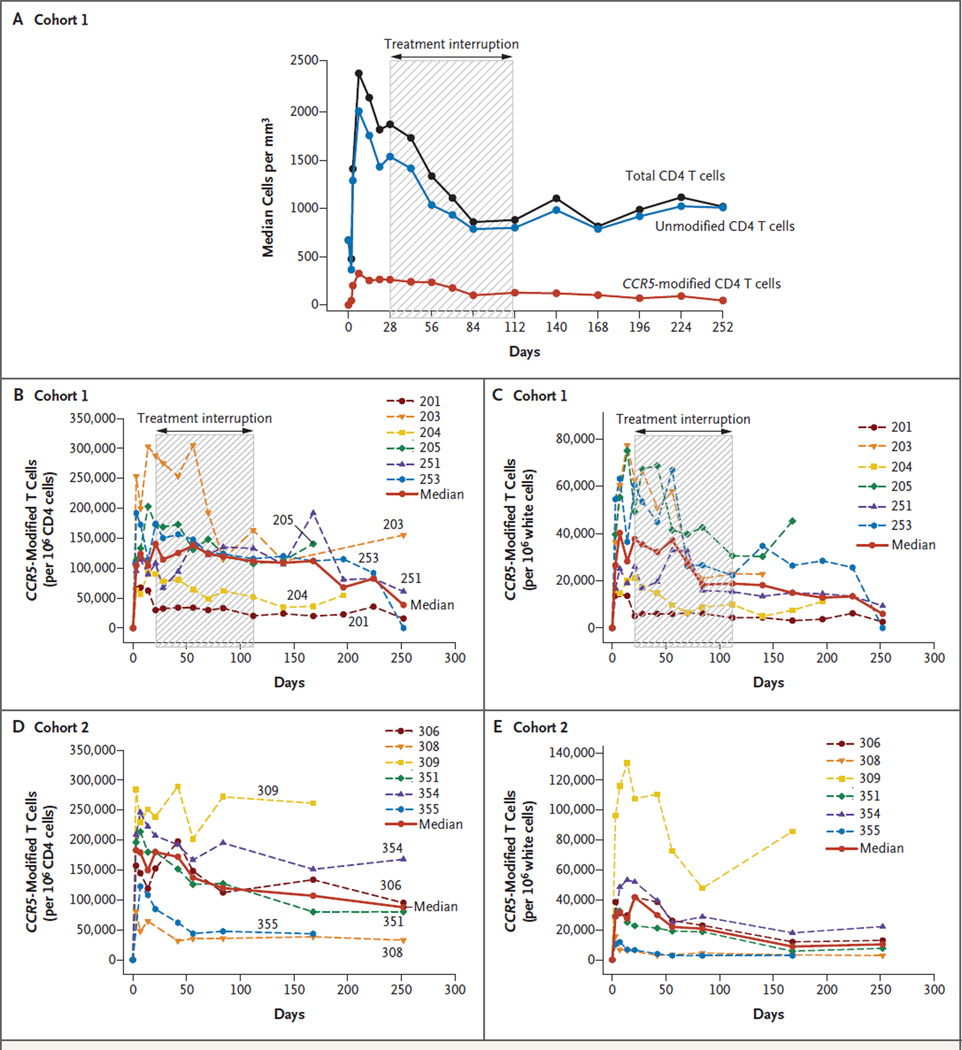
Thereafter, with the resumption of antiretroviral treatment, the median CD8 T-cell count decreased to pre–treatment-interruption levels at week 24, and the median CD4 T-cell count progressively increased to 1007 per cubic millimeter by the end of the 36-week study. The kinetics of the decline in CCR5-modified CD4 T cells and in unmodified CD4 T cells may differ (Fig. 4A). With the use of mixed quantile regression, the rate of decline during the treatment interruption was significantly greater for the median unmodified CD4 T cells (−7.25 cells per day; 95% confidence interval [CI], −12.14 to −2.94) than for the CCR5-modified CD4 T cells (−1.81 cells per day; 95% CI, −3.08 to −0.46) (P = 0.02). However, the difference in the decline for the mean cell counts over time did not reach significance when they were compared by means of a linear mixed-effects model (P = 0.08).
Figure 4. CCR5-Modified CD4 T Cells during Treatment Interruption.
The median count for total CD4 T cells, unmodified CD4 T cells, and CCR5-modified CD4 T cells for all patients in cohort 1 is plotted in Panel A. CCR5-modified CD4 T cells in blood are shown as a fraction of CD4 cells (Panels B and D) and as a fraction of white cells (Panels C and E).
To further assess the safety of the infusions and treatment interruption, we also measured HIV DNA in the patients’ PBMCs by droplet digital polymerase-chain-reaction (PCR) assay, which is more sensitive by a factor of approximately 100 than are conventional assays.22 Despite an approximate 3-log10 increase in the mean HIV-RNA copy number in the plasma, the HIV DNA level did not change in five patients during treatment interruption, was significantly increased in one patient during the treatment interruption, and returned to baseline in all six patients after the resumption of antiretroviral treatment (Fig. S3 in the Supplementary Appendix).
We also assayed the number of copies of gag, a gene encoded by HIV DNA, in the peripheral blood of participants in cohort 2 (Fig. S4 in the Supplementary Appendix). The gag levels decreased in four patients, remained stable in one patient, and were below the limit of detection in one patient. Using the same assay, we assessed the rates of HIV DNA decay in a series of aviremic patients (i.e., those with an undetectable level of HIV RNA [<75 copies per milliliter], according to ultrasensitive PCR assay) who did not receive SB-728 T-cell infusions (Table S3 in the Supplementary Appendix). The demographics of these patients are similar to those in cohorts 1 and 2. The rates of HIV DNA decline in the concurrent control cohort are shown in Figures S5 and S6 in the Supplementary Appendix. A comparison of the median and mean changes in gag copies indicates that the rate of HIV DNA decline is approximately 10 times as rapid in the cohort 2 patients as in the aviremic control cohort, although the difference in slopes was not significant (P = 0.65) (Table S4 in the Supplementary Appendix).
DISCUSSION
In this study, we tested the safety and feasibility of inducing acquired genetic resistance to HIV infection in an attempt to mimic the known inherited resistance displayed by persons with the CCR5-delta32 mutation.13,15,16 The infusion of autologous CD4 T cells in which the CCR5 receptor had been rendered dysfunctional by ZFNs targeting CCR5 was generally safe, although the small cohort sizes in the study render null the generalizability of this conclusion. The gene-modified cells readily engrafted and persisted after adoptive transfer. Potential beneficial effects associated with the infusion of SB-728-T included increased levels of CD4 T cells. The observed relative survival advantage of the gene-modified cells during treatment interruption suggests that genome editing at the CCR5 locus confers a selective advantage to CD4 T cells in patients infected with HIV.
The long-term persistence of the CCR5-modified CD4 T cells suggests that the cells were not rendered immunogenic as a result of CCR5 disruption. Immune deficiency is unlikely to explain the lack of rejection of the CCR5-modified cells, because previous studies have shown that gene-modified T cells engineered with immunogenic viral vectors can be rapidly eliminated by immune-mediated clearance in patients with late-stage HIV.23 Although the persistence of CCR5-modified cells as measured by the concentration in blood is similar in the two cohorts, the percentage of CCR5-modified CD4 T cells appears to be more stable than does the concentration of unmodified CD4 T cells in the blood, suggesting differential rates of cell death or trafficking to extravascular compartments. The decline in CCR5-modified CD4 T cells in the PBMC compartment may reflect, at least in part, egress from the vascular compartment and accumulation in the mucosal immune system. Since the vascular compartment contains only 1 to 2% of the T-cell mass, it is possible that the total body content of CCR5-modified cells is underestimated by the counts in the blood compartment, given the presence of CCR5-modified cells in the rectal mucosa and the large size of the mucosal immune component.21
The study participant with the longest delay in viral recrudescence was later discovered to be heterozygous for the CCR5 deletion. Preliminary analyses suggest that the degree of biallelic disruption of CCR5 may correlate with control of viral load (Fig. S7 in the Supplementary Appendix). Successful clinical application will most likely depend on biallelic knockout in persons who have two nonmutated CCR5 alleles.
We have applied the principles of synthetic biology24 to the goal of creating an immune system that is resistant to HIV infection. Our strategy is to repopulate the immune system with CCR5-deficient central memory T lymphocytes by infusion of SB-728-T. Alternative approaches that are promising include infusions of autologous CCR5-modified hematopoietic stem cells,25 stem-cell transplantation procedures, and the use of CCR5-specific ribozymes and short hairpin RNA.26–28 We anticipate that our strategy will elucidate the contribution of acquired CCR5 deficiency to the antiviral effect that led to the functional eradication of HIV after CCR5-delta32 stem-cell transplantation.18,19
Our study supports the feasibility of targeted genome editing to introduce a disease-resistance allele. Future studies directed toward increasing the engraftment of these gene-modified cells are warranted.
Supplementary Material
Acknowledgments
Supported in part by a grant from the National Institute of Allergy and Infectious Diseases (U19 AI066290), the Penn Center for AIDS Research (P30 AIO45008), the Penn Clinical Trials Unit (AIO69534), and Sangamo BioSciences.
We thank Don Siegel, Division of Transfusion Medicine and Therapeutic Pathology, Department of Pathology and Laboratory Medicine, and the nurses of the Apheresis Unit; Larisa Zifchak, Joe Quinn, Amber Nelson, Jenna Lewis, and Angelo Seda, who served as the research nurses on the study; Farida Shaheen of the Center for AIDS Research; Mark Sudell and Adonna Mackley for data collection; Reddy Malapati for graphics; Elizabeth Veloso, Lester Lledo, and Anne Chew for assistance in clinical research support; Zhaohui Zheng, Andrea Brennan, Julio Cotte, and members of the Clinical Cell and Vaccine Production Facility; Ro Kappes and Colleen Stewart for regulatory support and guidance; James Hoxie, Michael Lederman, Stephen Goff, Paul Johnson, Luis Montaner, Guido Silvestri, and David Weiner for helpful discussions as advisors; and program officers for constructive input from the National Institute of Allergy and Infectious Diseases.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Scherer S, Davis RW. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Igoucheva O, Alexeev V, Yoon K. Oligo-nucleotide-directed mutagenesis and targeted gene correction: a mechanistic point of view. Curr Mol Med. 2004;4:445–463. doi: 10.2174/1566524043360465. [DOI] [PubMed] [Google Scholar]
- 3.Ashworth J, Havranek JJ, Duarte CM, et al. Computational redesign of endonu- clease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin JY, Glazer PM. Repair of DNA lesions associated with triplex-forming oligonucleotides. Mol Carcinog. 2009;48:389–399. doi: 10.1002/mc.20501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim KH, Nielsen PE, Glazer PM. Site-specific gene modification by PNAs conjugated to psoralen. Biochemistry. 2006;45:314–323. doi: 10.1021/bi051379a. [DOI] [PubMed] [Google Scholar]
- 6.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 8.Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci U S A. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez EE, Wang J, Miller JC, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alkhatib G, Combadiere C, Broder CC, et al. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 12.Deng H, Liu R, Ellmeier W, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 13.Liu R, Paxton WA, Choe S, et al. Homo-zygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 14.Samson M, Libert F, Doranz BJ, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 15.Eugen-Olsen J, Iversen AK, Garred P, et al. Heterozygosity for a deletion in the CKR-5 gene leads to prolonged AIDS-free survival and slower CD4 T-cell decline in a cohort of HIV-seropositive individuals. AIDS. 1997;11:305–310. doi: 10.1097/00002030-199703110-00007. [DOI] [PubMed] [Google Scholar]
- 16.Cohen OJ, Vaccarezza M, Lam GK, et al. Heterozygosity for a defective gene for CC chemokine receptor 5 is not the sole determinant for the immunologic and vi-rologic phenotype of HIV-infected long-term nonprogressors. J Clin Invest. 1997;100(1):581–589. doi: 10.1172/JCI119682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gulick RM, Lalezari J, Goodrich J, et al. Maraviroc for previously treated patients with R5 HIV-1 infection. N Engl J Med. 2008;359:1429–1441. doi: 10.1056/NEJMoa0803152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hütter G, Nowak D, Mossner M, et al. Long-term control of HIV by CCR5 delta32/ delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 19.Allers K, Hütter G, Hofmann J, et al. Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood. 2011;117:2791–2799. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- 20.Deeks SG, McCune JM. Can HIV be cured with stem cell therapy? Nat Biotech-nol. 2010;28:807–810. doi: 10.1038/nbt0810-807. [DOI] [PubMed] [Google Scholar]
- 21.Douek DC, Roederer M, Koup RA. Emerging concepts in the immunopatho-genesis of AIDS. Annu Rev Med. 2009;60:471–484. doi: 10.1146/annurev.med.60.041807.123549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riddell SR, Elliott M, Lewinsohn DA, et al. T-cell mediated rejection of gene-modified HIV-specific cytotoxic T lymphocytes in HIV-infected patients. Nat Med. 1996;2:216–223. doi: 10.1038/nm0296-216. [DOI] [PubMed] [Google Scholar]
- 24.Benner SA, Sismour AM. Synthetic biology. Nat Rev Genet. 2005;6:533–543. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holt N, Wang J, Kim K, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitsuyasu RT, Merigan TC, Carr A, et al. Phase 2 gene therapy trial of an anti-HIV ribozyme in autologous CD34+ cells. Nat Med. 2009;15:285–292. doi: 10.1038/nm.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimizu S, Hong P, Arumugam B, et al. A highly efficient short hairpin RNA potently down-regulates CCR5 expression in systemic lymphoid organs in the hu-BLT mouse model. Blood. 2010;115:1534–1544. doi: 10.1182/blood-2009-04-215855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DiGiusto DL, Krishnan A, Li L, et al. RNA-based gene therapy for HIV with lenti-viral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med. 2010;2:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.