Abstract
As part of a continuous, standardized programme of monitoring the Leishmania vectors in German military camps in northern Afghanistan between 2007 and 2009, a detailed taxonomic analysis of the endemic sandfly fauna, as sampled using light and odour-baited traps, was conducted. Of the 10 sandfly species that were recorded, six may serve as enzootic and/or zooanthroponotic vectors of parasites causing human leishmaniasis.
The use of a simple DNA-‘barcoding’ technique based on the mitochondrial cyt b gene, to identify the collected sandflies to species level, revealed (1) a clear discrimination between the potential vector species, (2) clustering of species within most subgenera, and (3) particularly high heterogeneity within the subgenus Paraphlebotomus (Phlebotomus alexandri being grouped with Ph. papatasi rather than with other Paraphlebotomus species). The data also indicate a high level of genetic heterogeneity within the subgenus Sergentomyia but close similarity between Sergentomyia sintoni and Sergentomyia murgabiensis. The morphological similarity of many medically important sandflies can make species identification difficult, if not impossible. The new DNA-barcoding techniques may provide powerful discriminatory tools in the future.
Sandflies of the genus Phlebotomus (Diptera: Psychodidae) can serve as obligate vectors of protozoan Leishmania parasites (Kinetoplastida: Trypanosomatidae), some of which are the causative agents of human leishmaniasis. In Central Asia, Phlebotomus (Phlebotomus) papatasi is widely distributed and confirmed to be the primary vector of Leishmania major, the causative agent of zoonotic cutaneous leishmaniasis (zCL). Although the major ‘reservoir’ host of Le. major in this region is the great gerbil, Rhombomys opimus (Rodentia: Muridae), other rodents can be infected with the parasite (Nadim et al., 1979). Since the deployment of international military forces to Iraq and, subsequently, to Afghanistan, hundreds of the soldiers involved have developed zCL (Coleman et al., 2006; Faulde et al., 2006). Appropriate preventive schemes based on monitoring and control need to include sandfly trapping, assessments of the prevalences of infection in local sandflies, reduction in the numbers of potential breeding sites for the vector species, and personal protection against sandfly bites (Faulde et al., 2009).
The identification of vector species is essential for a good understanding of the epidemiology of any vector-borne disease. Until the last few years, the most recent information on the sandfly fauna of Afghanistan and its taxonomy dated back to the 1970s, when 44 species — half assigned to the genus Sergentomyia and half to the genus Phlebotomus — were recorded (Artemiev, 1981; Seccombe et al., 1993). The genus Sergentomyia comprises predominantly non-anthropophilic species, some of which are known to feed on reptiles or birds. All of the primary or secondary vectors of parasites causing human leishmaniasis in the Old World [i.e. zCL, anthroponotic CL (aCL; caused by Le. tropica) and visceral leishmaniasis (VL; caused by Le. donovani s.l.) — all of which occur, at varying levels of endemicity, in different parts of Afghanistan] are Phlebotomus species. Apart from Ph. papatasi, the only other proven vector of parasites causing human leishmaniasis in Afghanistan is Ph. sergenti, which transmits Le. tropica (Artemiev, 1978). Several other Phlebotomus species have, however, been reported as vectors in neighbouring countries or other Old-World regions (e.g. some Phlebotomus species in the subgenus Laroussius are the vectors of Le. infantum in the Mediterranean basin; Seccombe et al., 1993). Unfortunately, several sandfly taxa (such as ‘Ph. caucasicus/andrejevi/mongolensis’) have so far only been identified as vectors of Le. major, Le. tropica and/or Le. donovani s.l. on the basis of untyped promastigotes found in wild-caught adult females.
In a recent analysis of 8442 sandflies caught in Balkh province in northern Afghanistan, at least 2·5% of the female Ph. papatasi were found infected with Le. major (Faulde et al., 2008b) whereas the other sandfly species (Ph. caucasicus and Se. sintoni) were never found infected. Balkh not only has very high densities of the great gerbil (the highest known) but also high densities of Ph. papatasi, the most important vector of Le. major (Faulde et al., 2008a). In contrast, the mountainous surroundings of Feyzabad, in the north–eastern province of Badakhshan province, do not favour the same vector–reservoir combination, with great gerbils not detected in this area until 2009 (M. Faulde, unpubl. obs.).
The study of the bionomics and vector potential of sandflies is hampered by the nocturnal activity, cryptic life-cycles (with the sub-adult stages often in animal burrows), small size and general morphological similarity of the insects. In the initial study of wild-caught Old-World sandflies, Phlebotomus species are usually separated from Sergentomyia (Coleman et al., 2009), and the female sandflies from the males (the males being easily recognised by their terminalia). If based, as usual, on morphology, any further identification of the sandflies requires the insects to be mounted on slides, with the time-consuming micro-dissection of the female spermathecae and head capsule (cibarium and pharynx) and the male terminalia, genital pump and head capsule (cibarium) often necessary to make an accurate identification to species level. In many cases, the antennae and palpi (which are easily lost or damaged during capture) also have to be examined.
Over the last couple of decades, molecular methods have been increasingly employed to explore the taxonomy and phylogeny of many insects, including sandflies. A prerequisite for the development of molecular taxonomy is a reference collection of specimens that have been correctly identified to species (usually morphologically) and from which DNA can be extracted. Even then, the development of molecular identification tools can be hindered by intraspecific and geographical genetic variation. So far, large numbers of sandflies from the Mediterranean region (Aransay et al., 1999; Di Muccio et al., 2000; Hamarsheh et al., 2007), the Middle East (Moin-Vaziri et al., 2007a, b; Parvizi et al., 2010), Africa (Depaquit et al., 2008), Madagascar (Depaquit et al., 2002, 2007) and Latin America (Ready et al., 1997; Uribe Soto et al., 2001) have been studied using DNA-based tools. In Afghanistan, however, only two specimens of Ph. papatasi have been investigated using such tools (Esseghir et al., 1997).
One of the most recently employed tools in molecular systematics is DNA ‘barcoding’, which uses standardized, well-conserved, short sequences of mitochondrial DNA for the identification of unknown specimens. Although cytochrome c oxidase I (COI) is the gene most frequently investigated (Hebert et al., 2003; Boudabous et al., 2009), the cytochrome b (cyt b) gene has also been used (Hajibabaei et al., 2007), and the cyt b gene of several sandfly species has already been characterised (Esseghir et al., 1997; Ready et al., 1997; Parvizi et al., 2003; Hamarsheh et al., 2007; Moin-Vaziri et al., 2007a, b).
The aim of the present study was to assess the usefulness of cyt-b-based DNA barcoding for the identification of the vector and non-vector sandflies that live in and around German military camps in northern Afghanistan and in nearby southern Uzbekistan.
MATERIALS AND METHODS
Study Area
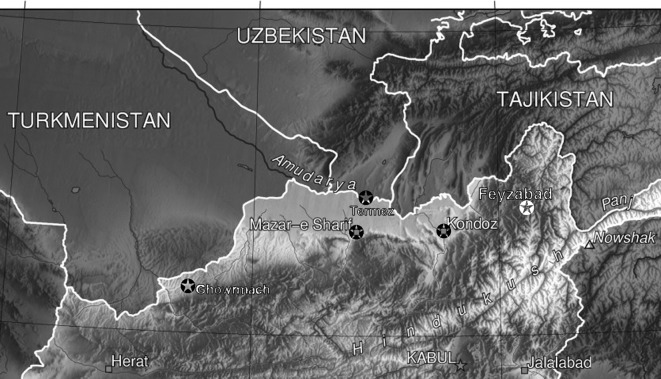
Sandflies were collected in and around German military camps in northern Afghanistan and nearby southern Uzbekistan between 2007 and 2009 (see Figure 1 and Table 1). These camps were located, at altitudes of 400–1800 m, north of the Hindukush Mountains (except the site near Kabul) and south of the Turanian Basin. They included the semi-arid grasslands of the Turkistan plains in the east, the ephemeral desert of the northern foothills of the Marmal Mountains, and the mountains of Badakhshan in the west (Breckle, 2007). The Uzbek city of Termez lies on the banks of the Amu-Darya River, which marks the international border between Afghanistan and Uzbekistan. Sandfly collections were made between May and September, when the activity of adult sandflies peaks. At this time of the year, maximum daily temperatures normally reach 30–40°C and do not drop below 20°C at night, and there are occasional sandstorms but virtually no rainfall.
Figure 1.
Study area in northern Afghanistan/southern Uzbekistan. Collection sites are marked by asterisks.
Table 1. The morphologically identified specimens and their origins.
| No. of female/male specimens morphologically identified as: | |||||||||||||||
| Province | Site | Altitude (m) | Co-ordinates | Date | P.p. | P.a. | P.s. | P.c.* | A.t. | L.k. | S.m. | S.g. | S.c. | G.d. | Total no. of sandflies |
| Badakhshan | Feyzabad | 1160 | 37°7′N, 70°31′E | Aug 2007 | 1/0 | 45/10 | 5/1 | 0/1 | 1/0 | 4/0 | 16/4 | 88 | |||
| Aug 2008 | 4/0 | 4 | |||||||||||||
| Jul–Sep 2009 | 0/8 | 0/3 | 0/5 | 0/7 | 23 | ||||||||||
| Kondoz | Kondoz | 430 | 36°42′N, 68°53′E | May 2007 | 10/10 | 1/0 | 0/1 | 3/0 | 3/0 | 28 | |||||
| Sep 2008 | 6/0 | 5/0 | 3/0 | 14 | |||||||||||
| May–Jun 2009 | 10/18 | 0/1 | 5/4 | 0/1 | 1/0 | 40 | |||||||||
| Aug–Sep 2009 | 0/127 | 0/5 | 0/15 | 0/1 | 148 | ||||||||||
| Balkh | Mazar-e Sharif | 375 | 36°43′N, 67°14′E | Aug 2007 | 2/0 | 2 | |||||||||
| May–Jun 2009 | 32/13 | 2/5 | 4/12 | 6/2 | 0/1 | 77 | |||||||||
| Sep–Nov 2009 | 0/2 | 0/1 | 0/1 | 4 | |||||||||||
| Badghis | Ghowrmach | 640–690 | 35°44′N, 63°47′E | May 2009 | 3/4 | 24/4 | 6/6 | 47 | |||||||
| Kabul | Kabul | 1780 | 34°32′N, 69°18′E | 2009 | 1/1 | 1/0 | 1/0 | 1/0 | 5 | ||||||
| Surxondarya Uzbekistan | Termez | 310 | 37°17′N, 67°20′E | Jun 2009 | 13/10 | 1/0 | 4/2 | 1/0 | 31 | ||||||
| All investigated | All investigated | 78/193 | 46/19 | 9/22 | 27/11 | 11/27 | 1/0 | 21/6 | 8/3 | 7/1 | 17/4 | 511 | |||
A.t., Phlebotomus (Adlerius) turanicus; G.d., Sergentomyia (Grassomyia) dreyfussi turkestanica; L.k., Ph. (Laroussius) keshishiani; P.a., Ph. (Paraphlebotomus) alexandri; P.c., Ph. (Para.) caucasicus; P.p., Ph. papatasi; P.s., Ph. (Para.) sergenti; S.c., Se. (Sintonius) clydei; S.g., Se. (Parrotomyia) grekovi; S.m., Sergentomyia murgabiensis; F, females; M, males.
*Female Ph. caucasicus are indistinguishable, morphologically, from female Ph. andrejevi and Ph. mongolensis (Artemiev, 1978).
Field Collections of Sandflies
Adult sandflies were usually collected using CDC miniature light traps (Model 512; John W. Hock, Gainesville, FL) but some additional collections were made using the BG-Sentinel mosquito traps (BioGents, Regensburg, Germany) baited with BG-lure (BioGents), which produces an odour resembling that of human skin. The traps were run overnight from 17·00 to 07·00 hours, mostly on the periphery of each camp. The CDC traps were hung at a height of about 1 m, attached to a tripod. After each collection night, any sandflies caught were put in a freezer, to kill them, and then either preserved in absolute ethanol (about 50% of each collection) or dried.
Morphotaxonomy
Single specimens were left in 70% ethanol for about 5 min, and then the distal parts of the abdomen (for the examination of the genitalia) and the entire head (for the examination of the cibarium and pharynx) were removed with fine forceps. The heads and distal parts of the abdomens were cleared by incubation in 1 m KOH until the dark-reddish eye pigment had mostly faded (about 30 min). The remaining body parts were kept in absolute ethanol, for subsequent DNA extraction. After being cleared, the genitalia and heads were transferred onto a slide, slightly squashed under a coverslip, and then immediately examined and photographed under a light microscope with an attached digital camera. Each specimen was thus documented photographically. Each was identified morphologically, usually to species, using the keys provided by Artemiev (1978).
DNA Extraction and Analysis
Genomic DNA was extracted, from the unmounted parts (thorax and proximal parts of the abdomen) of each sandfly of interest, by homogenization, lysis and Chelex treatment, as described, for blackflies, by Dumas et al. (1998) and Krueger and Hennings (2006). This DNA was used as the template in a PCR that targeted a fragment, of approximately 550 bp, that included the mitochondrial cyt b gene, a transfer-RNA gene and part of the ND1 gene (Ready et al., 1997). The amplicons produced using DNA from representatives of each sandfly species collected in Afghanistan (and reference specimens of species from Kosovo; see Table 2) were purified with the NucleoSpin Extract II kit (Macherey-Nagel, Düren, Germany) and commercially sequenced by direct sequencing [either by Eurofins (Ebersberg, Germany) or Seqlab (Göttingen, Germany)]. The respective sequences were deposited in GenBank under accession numbers HM803186–217 and HQ204193–94 (Table 2). These sequences and several reference sequences that were already available from GenBank (see Figures 2 and 3) were aligned with ClustalW2 (Larkin et al., 2007). The alignment files were uploaded into the PAUP 4·0b10 software package (Swofford, 2002), which was then used to produce taxon-identity trees. As the goal was not phylogenetic estimation, a simple neighbour-joining algorithm was generally considered sufficient for tree construction. A maximum-parsimony analysis was, however, used for an intraspecific analysis of Ph. sergenti.
Table 2. Collection details of DNA specimens.
| Species | Specimen code(s) and (gender) | Locality | Date | Accession number(s) |
| test samples from Afghanistan | ||||
| Phlebotomus (Paraphlebotomus) alexandri | 01 and 04 (♀♀) | Feyzabad | Aug 2007 | HM803186, HM803188 |
| Ph. (Para.) caucasicus* | 31 and 32 (♀♀) | Ghowrmach | May 2009 | HM803204, HM803206 |
| 40 and 41 (♂♂) | Kondoz | 2009 | HM803213, HM803215 | |
| Ph. papatasi | 08d (♀) | Feyzabad | Aug 2007 | HM803194 |
| 44 (♂) | Ghowrmach | May 2009 | HM803211 | |
| 45 (♀) | Kondoz | Jun 2009 | HM803210 | |
| 48 (♀) | Mazar-e Sharif | Jun 2009 | HM803212 | |
| Ph. (Para.) sergenti | 03, 06, 08, 08a, 08c and 13 (♀♀) | Feyzabad | Aug 2007 | HM803187, HM803189, HM803191–3, HM803197 |
| 07, 42 and 43 (♂♂) | Kondoz | 2007, Aug 2009 | HM803190, HM803214, HM803216 | |
| Ph. (Adlerius) turanicus | 29 (♀) | MeS | May 2009 | HM803195 |
| Ph. (Adlerius) sp. | 46 (♀) | Kabul | 2009 | HM803196 |
| Ph. (Laroussius) keshishiani | 10 (♀) | Feyzabad | Aug 2007 | HQ204193 |
| Sergentomyia murgabiensis | 17 (♀) | Kondoz | 2007 | HM803201 |
| Se. (Parrotomyia) sp. | 24 (♂) | Kondoz | Aug 2007 | HM803203 |
| Se. (Grassomyia) dreyfussi turkestanica | 15 (♀) | Feyzabad | Aug 2007 | HM803200 |
| 50 (♀) | Kondoz | May 2009 | HM803199 | |
| Se. (Sintonius) clydei | 20 (♀) | Kondoz | 2007 | HQ204194 |
| Se. (Parrotomyia) grekovi | 19 (♀) | Feyzabad | Aug 2007 | HM803202 |
| reference samples from Kosovo | ||||
| Ph. (Laroussius) neglectus | 35 (♂) | Prizren | Aug 2009 | HM803208 |
| Ph. (Laroussius) perfiliewi | 38 (♀) | Prizren | Aug 2009 | HM803217 |
| Se. (Sergentomyia) minuta | 36 and 39 (♂♂) | Prizren | Aug 2009 | HM803209, HM803205 |
*Female Ph. caucasicus are indistinguishable from female Ph. andrejevi and Ph. mongolensis, both morphologically (Artemiev, 1978) and in terms of their cyt b sequences (Parvizi et al., 2010).
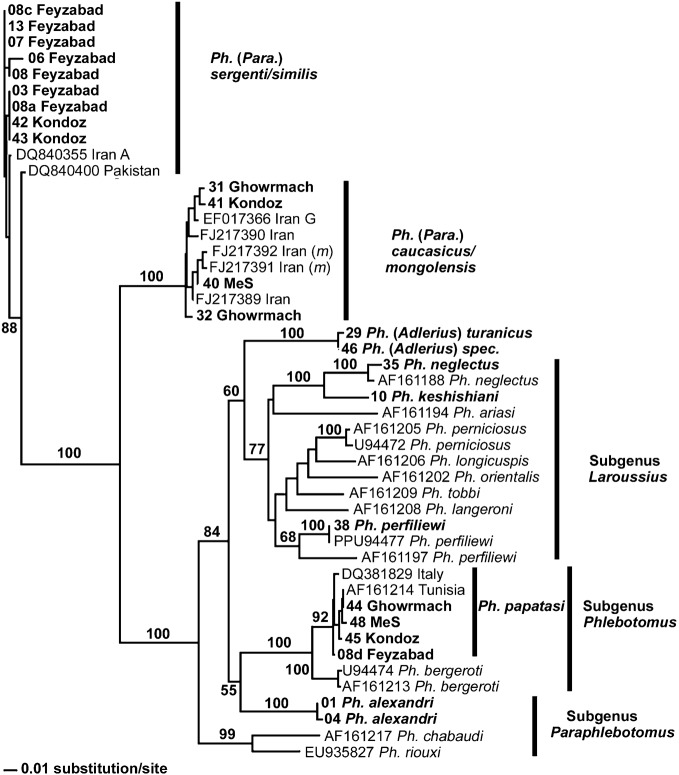
Figure 2.
An unrooted (neighbour-joining) tree of Afghan Phlebotomus taxa, based on comparison of 270-bp sequences of the mitochondrial cyt B gene. The specimens included in the present study are indicated by bold text (see Table 2 for details). Specimens of Ph. mongolensis are indicated (m). Bootstrap values of >50% are shown at the corresponding branches.
Figure 3.
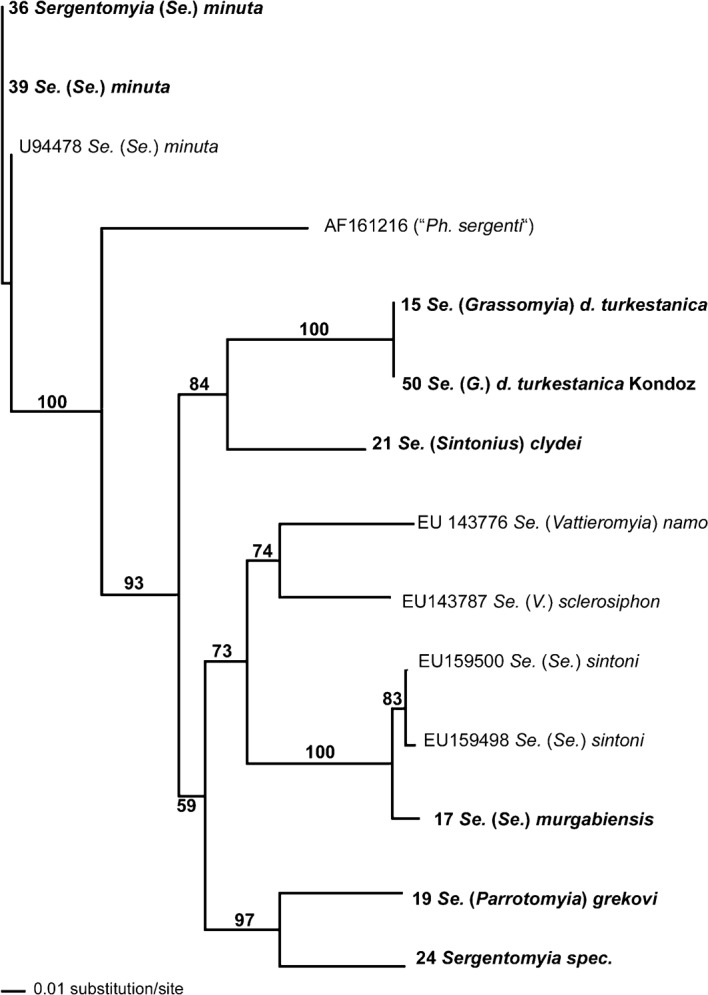
An unrooted (neighbour-joining) tree of Afghan Sergentomyia taxa, based on comparison of 270-bp sequences of the mitochondrial cyt B gene. The specimens included in the present study are indicated by bold text (see Table 2 for details). Bootstrap values of >50% are shown at the corresponding branches.
RESULTS
Overall, 286 male and 225 female sandflies were identified morphologically. In total, six species of the genus Phlebotomus (Ph. alexandri, Ph. caucasicus, Ph. keshishiani, Ph. papatasi, Ph. sergenti and Ph. turanicus) and four species of Sergentomyia (Se. clydei, Se. dreyfussi turkestanica, Se. grekovi and Se. murgabiensis) were identified (Table 1). Phlebotomus papatasi was the most widespread of the 10 species collected, and was also the most abundant species at Kondoz, Mazar-e Sharif (MeS) and Termez.
Phlebotomus alexandri, Ph. sergenti and, most notably, Se. dreyfussi turkestanica showed a relatively limited geographical distribution, being found only in one (Se. dreyfussi turkestanica) or two of the sampled camps. All three of these species were found in the eastern subalpine semidesert of Feyzabad, with Ph. alexandri and Ph. sergenti also being collected in Kondoz (desert–woodland mosaic). Phlebotomus caucasicus was most abundant in the eastern hills of Ghowrmach (semi-arid Pistacia vera woodland), rather scarce in MeS (ephemeral desert) and Kondoz, and not collected in Feyzabad or Termez (peri-urban lowland).
In terms of annual population fluctuations, Ph. turanicus was found to be quite numerous in 2009 (when it represented 23·5% of the Phlebotomus collected in MeS and 30·0% of those collected in Feyzabad) compared with 2007 (when it represented only 1·6% of the Phlebotomus collected in Feyzabad) [or compared with the collections made in 2006 by Faulde et al. (2008b), when it represented 0% of the Phlebotomus collected in MeS].
Twenty-seven specimens representing all 10 species collected in Afghanistan (plus one male of an unidentified Sergentomyia species, presumably of the subgenus Parrotomyia) were subjected to molecular analysis of their cyt b genes (Table 2). For comparison, four specimens of three sandfly species from Kosovo were also analysed (Table 2). For alignment and tree constructions, the full-length sequence of around 550 bp was reduced to an informative data-set of some 270 bp that spanned the cyt b gene. This data-set was aligned with 29 older GenBank entries representing 18 sandfly taxa but was subsequently split into a Phlebotomus data-set and a Sergentomyia data-set, for the construction of two trees. The resulting trees (Figures 2 and 3) show clear discrimination between the potential vector species collected in northern Afghanistan, most significantly between the known vector Ph. papatasi and potential or suspected vectors such as Ph. sergenti and ‘Ph. caucasicus/mongolensis’ [according to Parvizi et al. (2010), Ph. caucasicus and Ph. mongolensis are indistinguishable in terms of their cyt b sequences] and any other (non-vector) sympatric sandfly species.
The results also indicate that Ph. (Paraphlebotomus) alexandri is, in terms of its cyt b sequence, more similar to species in the subgenus Phlebotomus than to other species in the subgenus Paraphlebotomus (Fig. 2). Likewise, two species assigned to the subgenus Sergentomyia — Se. (Se.) murgabiensis/sintoni and Se. (Se.) minuta — showed less genetic similarity to each other than to other subgenera of Sergentomyia (Fig. 3).
The distance matrix for all the sequenced specimens (with data from other studies excluded) is shown in Table 3. Generally, the level of intrasubgeneric genetic differentiation lay between 0 and 15% while the level of intersubgeneric genetic differentiation exceeded 15% (reaching up to 27%). However, as well as the two exceptions mentioned above (Ph. alexandri and the subgenus Sergentomyia), sandflies in the subgenus Adlerius only differed by 13%–17% from the subgenera Phlebotomus and Laroussius, whereas subgeneric distances within the genus Sergentomyia lay between 12% and 20%.
Table 3. An uncorrected (‘p’) distance matrix for the cyt b sequences under study*.
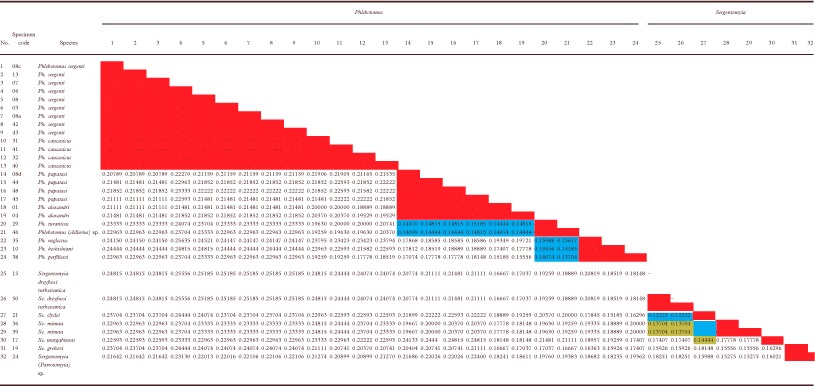
*The shaded cells in the matrix indicate intrasubgeneric clusters ( ) or intersubgeneric clusters (
) or intersubgeneric clusters ( ,
,  ) that are mentioned in the Results section.
) that are mentioned in the Results section.
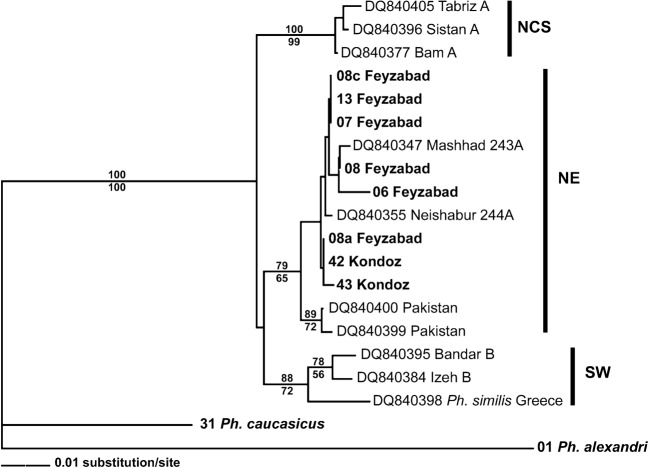
Because no specimens of Ph. sergenti from Afghanistan have been included in previous DNA studies, a more detailed comparison of about 430 bp of the cyt b genes of 19 specimens of this species was conducted. Ten published Ph. sergenti sequences (mainly from neighbouring Iran and Pakistan; Moin-Vaziri et al., 2007a), nine sequences of Ph. sergenti caught in the present study, one Ph. caucasicus sequence and one Ph. alexandri sequence (serving as an outgroup) were aligned and phylogenetically analysed (Fig. 4). The estimated neighbour-joining and maximum-parsimony trees resolved three clusters, one of which (tentatively named the NE lineage) contained all the Afghan specimens. The Afghan specimens resemble Iranian morphotype B in terms of the shape of the style (with one sub-terminal spine) and the width of the basal lobe of the coxite (data not shown).
Figure 4.
A neighbour-joining tree of Afghan Phlebotomus sergenti, based on comparison of 434-bp sequences of the mitochondrial cyt B gene. The specimens included in the present study are indicated by bold text (see Table 2 for details). The tree is rooted with the two Paraphlebotomus species: Ph. caucasicus and Ph. alexandri. The lineage abbreviations NCS, NE and SW refer to the nomenclature of Moin-Vaziri et al. (2007a). Bootstrap values of >50% are shown above the corresponding branches (the values below branches are from simultaneous maximum-parsimony analysis).
DISCUSSION
Accurate vector identification is an essential factor in the effective surveillance and control of any vector-borne disease. Sandfly-transmitted Le. major is a significant public-health threat in northern Afghanistan, causing rural zCL, and both anthroponotic CL and visceral leishmaniasis may also occur in the country, albeit at lower prevalences (Leslie et al., 2006; Faulde et al., 2008a). The present results indicate that Ph. papatasi (the primary vector of Le. major) is widely distributed in northern Afghanistan and particularly abundant in MeS and Kondoz, which are both foci of endemic zCL (Faulde et al., 2008a).
Phlebotomus sergenti is a typical element of urban or peri-urban highland environments and is the main vector of the Le. tropica that causes urban aCL in Kabul (Omar et al., 1969; Killick-Kendrick et al., 1995) and probably, in recent years, also in urban Feyzabad (Reithinger et al., 2004). In the present study, Ph. sergenti was also detected in Kondoz, where aCL has occurred only sporadically in recent years (Faulde et al., 2008a). The results of the detailed molecular analysis of Afghan, Iranian and Pakistani Ph. sergenti (Fig. 4) indicate that the distribution of this species extends from Iran (Moin-Vaziri et al., 2007a) into northern Afghanistan. The Afghan Ph. sergenti specimens that were investigated clearly cluster with the Iranian ‘NE’ lineage that probably originated in the Iranian province of Khorassan (which shares a border with Herat province, in north–western Afghanistan, and southern Turkmenistan). It remains unclear to what extent morphological and molecular characters truly define the subspecies of Ph. sergenti. Morphologically, the male specimens of Ph. sergenti caught in northern Afghanistan resembled morphotype B (Moin-Vaziri et al., 2007a), probably indicating that they were Ph. sergenti similis, whereas most specimens in the Iranian ‘NE’ lineage are of morphotype A and therefore Ph. sergenti sergenti.
Artemiev (1981) reported 44 sandfly species and subspecies from Afghanistan, with some members of the subgenus Adlerius endemic, but had very few data for remote areas such as Feyzabad, or Ghowrmach. Although Artemiev (1981) listed six species from the greater Feyzabad area, two of these (Ph. longiductus and Se. sodgiana) were not collected (from Feyzabad or anywhere else) in the present study, and the Ph. papatasi, Ph. turanicus and Se. dreyfussi turkestanica collected in Feyzabad during the present study are apparently the first records of these species for this location.
Although Ph. (Adlerius) turanicus (apparently the only Adlerius species collected in the present study) is normally considered xerophilic, in 2009 it was collected in the slightly cooler subalpine setting of Feyzabad as well as in the hot semi-desert of Ghowrmach and MeS. As all 27 male Adlerius collected during the present study were identified as Ph. (A.) turanicus, it was assumed that the single Adlerius female (collected from MeS) that was used for sequencing belonged to this species [although morphologically indistinguishable from female Ph. (A.) kabulensis].
Sergentomyia (Grassomyia) dreyfussi turkestanica was previously only known from a few places in northern Afghanistan (Kondoz) and central–eastern Afghanistan (Artemiev, 1976, 1981). The collection of this species in Feyzabad (present study) appears to be an unusual occurrence, since this species is described as thermo- and polyhygrophilic (Artemiev, 1983), two characters that also define Ph. papatasi (Artemiev, 1983), which was collected only very rarely in Feyzabad.
Factors such as micro-habitat differences (e.g. vicinity of the lizard-harbouring walls that favour Sergentomyia species), trap type (e.g. light v. sticky), geographical region and seasonality can greatly alter the species composition and sex ratios of collections of Phlebotomus and Sergentomyia (Yuval et al., 1988; Kamhawi et al., 1995; Wheeler et al., 1996; Burkett et al., 2007; Hanafi et al., 2007; Fahmy et al., 2009; Kasap et al., 2009; Kasili et al., 2010). It is important to remember, therefore, that the sex ratios and species compositions reported here only represent trap-related samples and do not necessarily reflect the natural abundances of each gender and species.
In the past, Afghan sandflies have been poorly studied using molecular techniques. The simple barcoding method employed in the present study appeared to be generally adequate for their identification to species level. The cyt b gene was chosen merely because it had been used by other researchers in the investigation of various sandfly taxa, and so there was already a useful data-set of cyt b sequences available in GenBank (Esseghir et al., 1997; Ready et al., 1997; Parvizi et al., 2003; Hamarsheh et al., 2007; Moin-Vaziri et al., 2007a, b). As well as permitting the successful and unambiguous typing of most of the sandfly specimens collected in the camps, the present results also allowed for further taxonomic hypotheses. Though not a phylogenetic analysis, the genetic-distance calculations indicated that the subgenus Paraphlebotomus is paraphyletic, confirming the observations made by Depaquit et al. (1998, 2000) in an investigation of internal-transcribed-spacer-2 (ITS2) ribosomal DNA. In a taxon-identity tree produced in a recent comparison of ITS2 sequences that included the subgenera Laroussius and Anaphlebotomus, Ph. alexandri was placed, with high bootstrap support, at the base of the subgenus Phlebotomus as a sister clade of Paraphlebotomus (unpubl. obs.), indicating a similar relationship to that seen with the cyt b sequences (present study).
Another example of possible paraphyly occurs in the subgenus Sergentomyia, where the two species Se. (Se.) sintoni/murgabiensis (see discussion below) and Se. (Se.) minuta do not cluster together. This result was clearly foreshadowed by Seccombe et al. (1993) who stated: ‘However, Sergentomyia is clearly a genus of practical convenience: its larger subgenera (… Sergentomyia …) are not well-defined …’.
There is a taxonomic inconsistency in the literature concerning the use of the names Se. (Se.) sintoni and Se. (Se.) murgabiensis (plus subspecies). According to Artemiev (1978) and Seccombe et al. (1993), Se. murgabiensis is a valid species, with Se. (Se.) murgabiensis sintoni as one of three valid subspecies. Recently, however, Parvizi and Amirkhani (2008), for unspecified reasons, re-used the name Se. sintoni for specimens from Iran. In the present study, an Afghan specimen of ‘Se. murgabiensis’ was found to share 97%–99% nucleotide identity with Iranian Se. sintoni in its cyt b gene, indicating possible conspecificity. If the synonymy of Se. sintoni with Se. murgabiensis is proven in the future, Se. sintoni will become a junior synonym. This taxonomic problem merits further attention, since both Se. murgabiensis (as ‘Se. arpaklensis’) and Se. sintoni seem to play a role in the enzootic cycles of Le. major and other Leishmania species (Seccombe et al., 1993; Parvizi and Amirkhani, 2008).
Acknowledgments
This study formed part of the M.Sc thesis of one of the authors (L.S.) at Hamburg University’s Department of Biology; Professor B. Misof is thanked for his supervision. The authors are also grateful to M. Fiedler, K. Hepke, L. Vergnes, S. Priesnitz and A. Crecelius for fieldwork and laboratory assistance. The laboratory support of the Bernhard Nocht Institute for Tropical Medicine is also gratefully acknowledged.
REFERENCES
- Aransay AM, Scoulica E, Chaniotis B, Tselentis Y.(1999)Typing of sandflies from Greece and Cyprus by DNA polymorphism of 18S rRNA gene. Insect Molecular Biology 8179–184. [DOI] [PubMed] [Google Scholar]
- Artemiev MM.(1976)Sandflies (Diptera, Psychodidae, Phlebotominae) of eastern Afghanistan. Communication III. Genus Sergentomyia, subgenera Sintonius, Rondanomyia and Grassomyia. Meditsinskaya Parazitologiya i Parazitarnye Bolezni 4535–41.(In Russian) [PubMed] [Google Scholar]
- Artemiev MM.(1978)Sandflies (Diptera, Psychodidae, Phlebotominae) of Afghanistan Kabul: Ministry of Public Health [Google Scholar]
- Artemiev MM.(1981)Sandflies (Diptera, Phlebotominae) of Afghanistan, Report 1. Fauna and Zoogeography Moscow: Martsinovski Institute for Medical Parasitology and Tropical Medicine; (In Russian) [Google Scholar]
- Artemiev MM.(1983)Sandflies (Diptera, Phlebotominae) of Afghanistan, Report 2. Distribution by topography. Meditsinskaya Parazitologiya I Parazitarnye Bolezni 5225–33. [PubMed] [Google Scholar]
- Boudabous R, Bounamous A, Jouet D, Depaquit J, Augot D, Ferté H, Berchi S, Couloux A, Veuille M, Babba H.(2009)Mitochondrial DNA differentiation between two closely related species, Phlebotomus (Paraphlebotomus) chabaudi and Phlebotomus (Paraphlebotomus) riouxi (Diptera: Psychodidae), based on direct sequencing and polymerase chain reaction-restriction fragment length polymorphism. Annals of the Entomological Society of America 102347–353. [Google Scholar]
- Breckle S.-W.(2007)Flora and vegetation of Afghanistan. Basic and Applied Dryland Research 1155–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkett DA, Knight R, Dennett JA, Sherwood V, Rowton E, Coleman RE.(2007)Impact of phlebotomine sand flies on U.S. military operations at Tallil air base, Iraq: 3. Evaluation of surveillance devices for the collection of adult sand flies. Jounral of Medical Entomology 44381–384. [DOI] [PubMed] [Google Scholar]
- Coleman RE, Burkett DA, Putnam JL, Sherwood V, Caci JB, Jennings BT, Hochberg LP, Spradling SL, Rowton ED, Blount K, Ploch J, Hopkins G, Raymond JL, O'Guinn ML, Lee JS, Weina PJ.(2006)Impact of phlebotomine sand flies on U.S. Military operations at Tallil Air Base, Iraq: 1. background, military situation, and development of a ‘Leishmaniasis Control Program’. Journal of Medical Entomology 43647–662. [DOI] [PubMed] [Google Scholar]
- Coleman RE, Hochberg LP, Putnam JL, Swanson KI, Lee JS, McAvin JC, Chan AS, O’Guinn ML, Ryan JR, Wirtz RA, Moulton JK, Dave K, Faulde MK.(2009)Use of vector diagnostics during military deployments: recent experience in Iraq and Afghanistan. Military Medicine 174904–920. [DOI] [PubMed] [Google Scholar]
- Depaquit J, Perrotey S, Lecointre G, Tillier A, Tillier S, Ferté H, Kaltenbach M, Léger N.(1998)Molecular systematics of Phlebotominae: a pilot study. Paraphyly of the genus Phlebotomus. Comptes Rendus de l’Académie des Sciences. Series III. Sciences de la Vie 321849–855. [DOI] [PubMed] [Google Scholar]
- Depaquit J, Ferté H, Léger N, Killick-Kendrick R, Rioux J.-A, Killick-Kendrick M, Hanafi HA, Gobert S.(2000)Molecular systematics of the phlebotomine sandflies of the subgenus Paraphlebotomus (Diptera, Psychodidae, Phlebotomus) based on ITS2 rDNA sequences. Hypotheses of dispersion and speciation. Insect Molecular Biology 9293–300. [DOI] [PubMed] [Google Scholar]
- Depaquit J, Léger N, Robert V.(2002)Première mention de Phlebotomus à Madagascar (Diptera: Psychodidae). Description de Phlebotomus (Anaphlebotomus) fertei n.sp. et de Phlebotomus (Anaphlebotomus) huberti n.sp. Parasite 9325–331. [DOI] [PubMed] [Google Scholar]
- Depaquit J, Léger N, Robert V.(2007)Les phlébotomes de Madagascar (Diptera: Psychodidae). V. Description de Sergentomyia majungaensis n.sp. Parasite 14219–223. [DOI] [PubMed] [Google Scholar]
- Depaquit J, Lienard E, Verzeaux-Griffon A, Ferté H, Bounamous A, Gantier JC, Hanafi HA, Jacobson RL, Maroli M, Moin-Vaziri V, Müller F, Özbel Y, Svobodova M, Volf P, Léger N.(2008)Molecular homogeneity in diverse geographical populations of Phlebotomus papatasi (Diptera, Psychodidae) inferred from ND4 mtDNA and ITS2 rDNA. Epidemiological consequences. Infection, Genetics and Evolution 8159–170. [DOI] [PubMed] [Google Scholar]
- Di Muccio T, Marinucci M, Frusteri L, Maroli M, Pesson B, Gramiccia M.(2000)Phylogenetic analysis of Phlebotomus species belonging to the subgenus Laroussius (Diptera, Psychodidae) by ITS2 rDNA sequences. Insect Biochemistry and Molecular Biology 30387–393. [DOI] [PubMed] [Google Scholar]
- Dumas V, Herder S, Bebba A, Cadoux-Barbabé C, Bellec C, Cuny G.(1998)Polymorphic microsatellites in Simulium damnosum s.l. and their use for differentiating two savannah populations: implications for epidemiological studies. Genome 41154–161. [DOI] [PubMed] [Google Scholar]
- Esseghir S, Ready PD, Killick-Kendrick R, Ben-Ismail R.(1997)Mitochondrial haplotypes and phylogeography of Phlebotomus vectors of Leishmania major. Insect Molecular Biology 6211–225. [DOI] [PubMed] [Google Scholar]
- Fahmy AR, Samy AM, Doha SA, Shehata MG.(2009)Preliminary field investigations on phlebotomine sandflies (Diptera: Psychodidae) from a recent cutaneous leishmaniasis focus in northern Sinai, Egypt. Egyptian Academic Journal of Biological Sciences 29–15. [Google Scholar]
- Faulde MK, Heyl G, Amirih ML.(2006)Zoonotic cutaneous leishmaniasis, Afghanistan. Emerging Infectious Diseases 121623–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulde M, Schrader J, Heyl G, Amirih M.(2008a)Differences in transmission seasons as an epidemiological tool for characterization of anthropopnotic and zoonotic cutaneous leishmaniasis in northern Afghanistan. Acta Tropica 105131–138. [DOI] [PubMed] [Google Scholar]
- Faulde M, Schrader J, Heyl G, Amirih M, Hoerauf A.(2008b)Zoonotic cutaneous leishmaniasis outbreak in Mazar-e Sharif, northern Afghanistan: an epidemiological evaluation. International Journal of Medical Microbiology 298543–550. [DOI] [PubMed] [Google Scholar]
- Faulde M, Schrader J, Heyl G, Hoerauf A.(2009)High efficacy of integrated preventive measures against zoonotic cutaneous leishmaniasis in northern Afghanistan, as revealed by quantified infection rates. Acta Tropica 11028–34. [DOI] [PubMed] [Google Scholar]
- Hajibabaei M, Singer GAC, Hebert PDN, Hickey DA.(2007)DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends in Genetics 23167–172. [DOI] [PubMed] [Google Scholar]
- Hamarsheh O, Presber W, Abdeen Z, Sawalha S, Al-Lahem A, Schönian G.(2007)Genetic structure of Mediterranean populations of the sandfly Phlebotomus papatasi by mitochondrial cytochrome b haplotype analysis. Medical and Veterinary Entomology 21270–277. [DOI] [PubMed] [Google Scholar]
- Hanafi HA, Fryauff DJ, Modi GB, Ibrahim MO, Main AJ.(2007)Bionomics of phlebotomine sandflies at a peacekeeping duty site in the north of Sinae, Egypt. Acta Tropica 101106–114. [DOI] [PubMed] [Google Scholar]
- Hebert PDN, Ratnasingham S, deWaard JR.(2003)Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society London. Series B, Biological Sciences 270(Suppl. 1)S96–S99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamhawi S, Abdel-Hafez SK, Molyneux DH.(1995)A comprehensive account of species composition, distribution and ecology of phlebotomine sandflies in Jordan. Parasite 2163–172. [Google Scholar]
- Kasap OE, Belen A, Kaynas S, Simsek FM, Biler L, Ata N, Alten B.(2009)Activity patterns of sand fly (Diptera: Psychodidae) species and comparative performance of different traps in an endemic cutaneous leishmaniasis focus in Cukurova plain, southern Anatolia, Turkey. Acta Veterinaria Brno 78327–335. [Google Scholar]
- Kasili S, Ngumbi PM, Koka H, Ngere FG, Kioko E, Odemba N, Kutima HL.(2010)Comparative performance of light trap types, lunar influence and sandfly abundance in Baringo district, Kenya. Journal of Vector Borne Diseases 47108–112. [PubMed] [Google Scholar]
- Killick-Kendrick R, Killick-Kendrick M, Tang Y.(1995)Anthroponotic cutaneous leishmaniasis in Kabul, Afghanistan: the high susceptibility of Phlebotomus sergenti to Leishmania tropica. Transactions of the Royal Society of Tropical Medicine and Hygiene 89477. [DOI] [PubMed] [Google Scholar]
- Krueger A, Hennings IC.(2006)Molecular phylogenetics of blackflies of the Simulium damnosum complex and cytophylogenetic implications. Molecular Phylogenetics and Evolution 3983–90. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG.(2007)ClustalW and ClustalX version 2. Bioinformatics 232947–2948. [DOI] [PubMed] [Google Scholar]
- Leslie T, Saleheen S, Sami M, Mayan I, Mahboob N, Fiekert K, Lenglet A, Ord R, Reithinger R.(2006)Visceral leishmaniasis in Afghanistan. Canadian Medical Association Journal 175245–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moin-Vaziri V, Depaquit J, Yaghoobi-Ershadi MR, Oshaghi MA, Derakhshandeh-Peykar P, Ferté H, Kaltenbach M, Bargues MD, Léger N, Nadim A.(2007a)Intraspecific variation within Phlebotomus sergenti Parrot (1917) (Diptera: Psychodidae) based on mtDNA sequences in Islamic Republic of Iran. Acta Tropica 10229–37. [DOI] [PubMed] [Google Scholar]
- Moin-Vaziri V, Depaquit J, Yaghoobi-Ershadi MR, Oshaghi MA, Derakhshandeh-Peykar P, Ferté H, Kaltenbach M, Bargues MD, Nadim A, Javadian E, Rassi Y, Jafari R.(2007b)Geographical variation in populations of Phlebotomus (Paraphlebotomus) caucasicus (Diptera: Psychodidae) in Iran. Bulletin de la Société de Pathologie Exotique 100291–295. [PubMed] [Google Scholar]
- Nadim A, Javadian E, Noushin MK, Nayil AK.(1979)Epidemiology of cutaneous leishmaniasis in Afghanistan. Part I: Zoonotic cutaneous leishmaniasis. Bulletin de la Société de Pathologie Exotique et de ses Filiales 7231–35. [PubMed] [Google Scholar]
- Omar A, Saboor A, Amin FM, Šerý V.(1969)Preliminary study on the foci of cutaneous leishmaniasis in Kabul city. Zeitschrift für Tropenmedizin und Parasitologie 20293–302. [PubMed] [Google Scholar]
- Parvizi P, Amirkhani A.(2008)Mitochondrial DNA characterization of populations of Sergentomyia sintoni and finding mammalian Leishmania infections in this sandfly by using ITS-rDNA gene. Majallahi Tahqiqati Dampizishkii Iran 99–18. [Google Scholar]
- Parvizi P, Benlarbi M, Ready PD.(2003)Mitochondrial and Wolbachia markers for the sandfly Phlebotomus papatasi: little population differentiation between peridomestic sites and gerbil burrows in Isfahan province, Iran. Medical and Veterinary Entomology 17351–362. [DOI] [PubMed] [Google Scholar]
- Parvizi P, Taherkhani H, Ready PD.(2010)Phlebotomus caucasicus and Phlebotomus mongolensis (Diptera: Psychodidae): indistinguishable by the mitochondrial cytochrome b gene in Iran. Bulletin of Entomological Research 100415–420. [DOI] [PubMed] [Google Scholar]
- Ready PD, Day JC, de Souza AA, Rangel EF, Davies CR.(1997)Mitochondrial DNA characterization of populations of Lutzomyia whitmanni (Diptera: Psychodidae) incriminated in the peri-domestic and silvatic transmission of Leishmania species in Brazil. Bulletin of Entomological Research 87187–195. [Google Scholar]
- Reithinger R, Aadil K, Hami S, Kolaczinski J.(2004)Cutaneous leishmaniasis, northern Afghanistan. Emerging Infectious Diseases 10966–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seccombe AK, Ready PD, Huddleston LM.(1993)A Catalogue of Old World Phlebotomine Sandflies (Diptera: Psychodidae, Phlebotominae) Occasional Papers on Systematic Entomology No. 8London: Natural History Museum [Google Scholar]
- Swofford DL.(2002)PAUP*, Phylogenetic Analysis Using Parsimony (*and Other Methods): Version 4.0b10 Sunderland, MA: Sinauer Associates [Google Scholar]
- Uribe Soto SI, Lehmann T, Rowton ED, Vélez BID, Porter CH.(2001)Speciation and population structure in the morphospecies Lutzomyia longipalpis (Lutz & Neiva) as derived from the mitochondrial ND4 gene. Molecular Phylogenetics and Evolution 1884–93. [DOI] [PubMed] [Google Scholar]
- Wheeler AS, Feliciangeli MD, Ward RD, Maingon RDC.(1996)Comparison of sticky-traps and CDC light-traps for sampling phlebotomine sandflies entering houses in Venezuela. Medical and Veterinary Entomology 10295–298. [DOI] [PubMed] [Google Scholar]
- Yuval B, Warburg A, Schlein Y.(1988)Leishmaniasis in the Jordan Valley. V. Dispersal characteristics of the sandfly Phlebotomus papatasi. Medical and Veterinary Entomology 2391–395. [DOI] [PubMed] [Google Scholar]