Abstract
Purpose of Review
β cells represent one of many cell types in heterogeneous pancreatic islets and play the central role in maintaining glucose homeostasis, such that disrupting β cell function leads to diabetes. This review summarizes methods for isolating and characterizing β cells, and describes integrated “omics” approaches used to define the β cell by its transcriptome and proteome.
Recent Findings
RNA Sequencing and mass spectrometry-based protein identification have now identified RNA and protein profiles for mouse and human pancreatic islets and β cells, and for β cell lines. Recent publications have outlined these profiles and, more importantly, have begun to assign the presence or absence of specific genes and regulatory molecules to β cell function and dysfunction. Overall, researchers have focused on understanding the pathophysiology of diabetes by connecting genome, transcriptome, proteome, and regulatory RNA profiles with findings from genome wide association studies (GWAS).
Summary
Studies employing these relatively new techniques promise to identify specific genes or regulatory RNAs with altered expression as β cell function begins to deteriorate in the spiral toward the development of diabetes. The ultimate goal is to identify potential therapeutic targets to prevent β cell dysfunction and thereby better treat the individual with diabetes.
Keywords: β-cell transcriptome, β-cell function, β-cell dysfunction, gene expression, diabetes
Introduction
The human pancreas performs both endocrine and exocrine functions. Pancreatic endocrine functions are served by the Islets of Langerhans, a heterogeneous arrangement of endothelial cells and several different hormone secreting cells including α, β, δ, PP, and epsilon cells (1,2). The pancreatic islets play the major role of maintaining glucose homeostasis through the regulated release of insulin and glucagon from β and α cells, respectively. Type 1 diabetes (T1D) is caused by an autoimmune attack that results in β cell destruction and a consequent loss of endogenous insulin production (3). Type 2 diabetes (T2D) occurs as a result of resistance to insulin activity in muscle and adipose tissue, coupled with β cell dysfunction (4,5). Thus, the pancreatic β cell is a prime target for gene expression analysis in healthy, deteriorating, and diseased states. Until recently, however, β cell specific studies have been difficult due to the heterogeneous arrangement of pancreatic islet cellular composition.
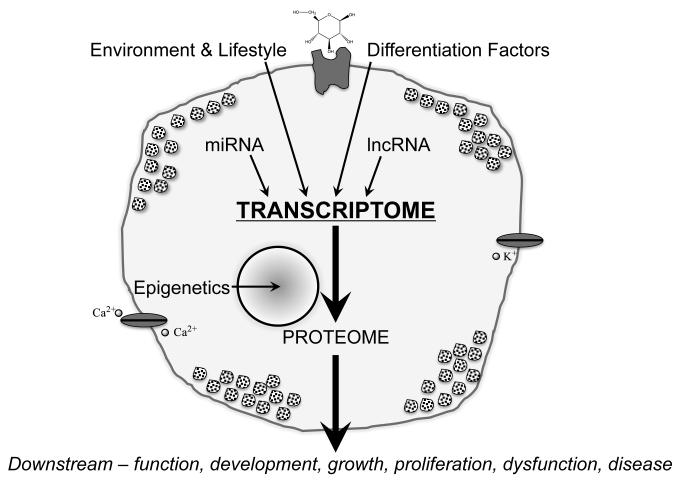
In order to determine how gene expression controls β cell growth, development, response to environmental stimuli, and function (as well as dysfunction), studies should ideally focus on purified β cells. Previous efforts to obtain such purified β cells from heterogeneous islet cellular composition have relied upon flow cytometry based sorting methodologies and unique β cell characteristics such as side scatter and granularity (6,7), zinc-content (8-11), mice genetically engineered such that their β cells express marker genes (12-14), combinations of surface markers (15-17), and intracellular hormone staining (18). More recent technological advances in islet cell separation techniques, next generation sequencing, and mass spectrometry have improved our understanding of the pancreatic β cell at the DNA, RNA, and protein levels. This review will focus on how new results using these techniques have suggested previously unsuspected effects from the environment, lifestyle, differentiation factors, and regulatory RNA molecules. The control of gene expression at the transcriptome level and the resultant proteome together show how these factors control β cell function, proliferation development, and dysfunction resulting in diabetes (Figure 1).
Figure 1. Factors controlling the β cell transcriptome.
External and internal factors control β cell gene expression as represented by the mRNA transcriptome. The connection between the transcriptome and the expressed proteins are manifested by the β cell phenotype.
SOURCE: ORIGINAL
Text of Review
β Cell Transcriptome
Transcriptome analyses from whole pancreatic islets or cultured β cell-lines using real-time PCR and microarrays (19-22) have long served as a surrogate for the β cell gene expression pattern. Advanced flow cytometry based methods for β cell sorting have improved the ability of researchers to measure cell-specific gene expression patterns (17,18). Most recently, the advent of next generation RNA Sequencing (RNASeq) (23) has provided an unbiased and high throughput method for examining gene expression profiles from enriched β cells (both rodent and human).
The most recent β cell transcriptome studies have focused on: 1.) Establishing the gene expression profile for each islet cell subtype (24-26), 2.) Determining the cellular location of each genome wide association screen (GWAS) gene identified from populations with T1D and T2D (25-27), and 3.) Assessing the effect of immune pressure on β cell health and gene expression patterns (27).
On average, β cells comprise approximately 30-75% of the endocrine pancreatic islet in humans, whereas in mice, β cells constitute about 60-80% of the islet cells (28). Transgenic mice expressing green fluorescent protein controlled by the mouse insulin I gene promoter (so called MIP-GFP mice) can be used to isolate a 95% pure β cell population based upon GFP expression (12). A limitation of such approaches is, of course, that the expressed transgene could influence the expression of other transcripts. With that caveat in mind, Ku et al. performed RNASeq on both whole islets and sorted GFP-positive β cells to identify genes specifically expressed in β cells (*24). Of the over 12,000 genes detected in the β cell, 43 demonstrated greater than 4-fold more gene expression in the β cell versus whole islets, whereas approximately 1,400 whole islet-specific genes displayed a 4-fold decreased expression in the β cell enriched population. Interestingly, they identified 16 genes that were exclusively expressed in the enriched β cells and not in any other organ tissue.
Transitioning from mouse to human islets, Bramswig et al. used surface antibodies to greatly enrich α, β, and exocrine cell populations for transcriptome analysis. These results provide a census of genes expressed in each cell population along with their relative, and in some cases apparently cell-specific gene expression (*25). In another study, Nica et al. used Newport Green staining to compare gene expression profiles of enriched human β cell populations to both the non-β cell and whole islet fractions. They applied a mathematical modeling strategy based upon these populations in an effort to account for α and δ cells contaminating the β cells, and specifically identified 614 islet genes not expressed in the β cell and 526 genes unique to the β cell (**26). As previously speculated based upon the innervated nature of pancreatic islets and other β cell characteristics, the functional annotation analysis found that the β cell specific gene expression pattern demonstrated numerous similarities to neuronal cells (29,30).
GWAS studies examining single nucleotide polymorphisms have identified over 70 candidate genes potentially linked to T1D or to T2D (31-33). RNASeq has permitted tissue specific, high throughput detection of mRNA from those genetic loci. Using RNASeq followed by real time PCR confirmation, over 60% of the identified T1D genes (such as the insulin gene), have been localized to the pancreatic β cell (26,**27). In T2D, greater than 90% of the associated genes, including VEGFA and SLC30A8, are specifically expressed in the β cell (26).
In other studies, overexpression of inflammatory, innate, and autoimmune response genes have been found in affected islets from individuals with T1D (34). Upon exposure to cytokines, the islet responded by increasing expression of apoptosis and inflammation related genes and T1D candidate gene expression was altered specifically in the β cell (27). These findings further establish the link between the endocrine β cell and the body’s immune system.
Regulatory RNAs in the β Cell
A network of transiently expressed transcription factors control pancreatic organogenesis and subsequent differentiation, including the development and proliferation of the endocrine pancreas (including the β cell subpopulation). Non-coding RNAs have been implicated as major regulators of gene expression and include both small microRNA (miRNA) and long noncoding RNA (lncRNA) (35,36).
MicroRNAs have been intricately studied in mouse pancreatic development and maintenance. miR-375 has been emphasized as a key miRNA regulating α and β cell mass, insulin secretion, and therefore total body glucose homeostasis (37). Furthermore, miR-375 expression has recently been reported in an enriched human β cell population, highlighting its likely importance in β cell health (**38,**39). When looking at human miRNA profiles, 366 miRNAs were found to be expressed in the human islet, and 85% were expressed in both whole islets and β cells. As compared with α cells, β cells expressed 19-fold more of these miRNAs (39), and that expression was inhibited to a greater degree by α-cell specific transcription factors. Klein et al. overexpressed β cell specific miRNAs in α-TC cells and inhibited those same putative β cell miRNAs in MIN6 cells (a mouse β cell line). They reported that overexpressing β cell miRNAs decreased glucagon mRNA expression by silencing α cell specific transcription factors, whereas suppressing the β cell specific miRNAs increased the levels of α-cell transcription factors (39). Similarly, Barbagallo et al. showed that suppressing miRNA expression in α-TC cells in response to environmental stress resulted in overexpression of targets (such as IGF1R, IRS-1 and ERK-1) and prevented apoptosis (40). Overall, these recent studies show that miRNAs play a key role in promoting the β cell phenotype by preventing expression of many genes that impair proper β cell function, previously termed “β cell disallowed genes” by Schuit et al (41). A study by van de Bunt et al. [**38] showing how islet specific miRNA target sequences matched GWAS-identified genes and variations associated with T2D demonstrates that repressing specific genes is required for proper β-cell function. Given the sequence specific regulatory function of miRNAs, variation in the target sequence appears to alter miRNA regulatory mechanisms and may well contribute to T2D pathogenesis.
Much like the miRNA clusters that occur within small regions of genomic DNA and are transcribed together (42-44), lncRNAs with tissue-specific expression patterns are found adjacent to protein coding genes, especially transcription factors (45-47). In addition to mRNA profiles, transcriptome analysis by next generation sequencing has also discovered, identified, and cataloged lncRNA that are specifically expressed in whole islet or enriched β cell populations (24,25,**48). In whole islets and in enriched β cells, lncRNAs appear to be much more tissue- or cell-specific than protein coding genes (24,48). Two separate studies by Ku et al. and Morán et al. show that lncRNAs are co-located in the same genomic region as specific islet and β cell proteins and transcription factors and include intergenic and antisense RNA located near FOXA2, GATA1, HNF1A, INSM1, ISL1, MAFB, NEUROD1, NKX2-2, NKX6-1, PAX6, PCSK1, PDX1, and RFX6 (24,48). Further, lncRNA expression may be more prominent during endocrine differentiation and maturation versus early development, and lncRNAs may serve as potential biomarkers in embryonic or induced pluripotent stem (iPS) cell differentiation into β cells (48). Last, like miRNAs, many lncRNAs appear to be associated with T2D-gene variants, and the regulatory effect on gene expression may contribute to β cell dysfunction and diabetes (48).
β Cell Proteome
Similar to what RNASeq has allowed for RNA analysis, mass spectrometry serves as an unbiased method to detect nearly all proteins expressed in a particular cell type: at baseline, in response to environmental conditions, or in the disease state (49,50). The proteome serves as the ultimate functional result of miRNA and lncRNA regulation on β cell gene expression. As our understanding of the genomics of β cell biology advances, characterizing the β cell proteome has become even more critical to promote understanding β cell function and dysfunction.
Specific proteomic studies of whole islets, cultured cell lines, and enriched β cells have used two approaches: 2-dimensional gel electrophoresis (2DGE) combined with mass spectrometry (MS) (51) and liquid chromatography in line with tandem MS (LC-MS/MS) (52). Ahmed et al. generated the first proteomic gel maps of isolated human islets, which opened the door to studying whole-cell protein expression across multiple conditions and disease states (53). More recently, the same group used 2DGE/MS to demonstrate altered expression of 75 mitochondrial proteins in the INS1E rat β cell line subjected to chronic hyperglycemia (54). Similarly, Maris and colleagues identified 74 differentially expressed proteins in INS1E cells cultured in hyperglycemic conditions, suggesting potential targets responsible for β cell dysfunction from chronic high glucose exposure (55). Due to their frequent use as a model system for studying insulin secretion and β cell dysfunction, D’Hertog et al. developed a 2DGE/MS proteomic reference map for INS1E cells (56). In the MIN6 mouse β cell line, Diraison et al. used 2DGE/MS to demonstrate that translationally controlled tumor protein (TCTP) is glucose regulated, suppressed by cell stress, and that it therefore appeared to play a protective role for β cells during hypoglycemia (57). In studying islet cell differentiation from islet and bone marrow-derived mesenchymal stem cells (MSCs), Zanini et al. showed that different sources of MSCs did not yield identical proteomic profiles when the cells were differentiated into islet-like cells (58). Coppola and colleagues generated a cytokine-resistant mouse β cell line and profiled protein differences between the cytokine sensitive and resistant β cells in order to better understand the autoimmune destruction of β cells in T1D (59). Specifically relevant to T1D studies, proteomics has been used to identify potential antibody targets for therapeutics (60). For instance, Massa et al. attempted to identify new autoantigen targets by blotting human pancreatic islet cells against sera from patients with T1D (*61). Finally, Maris et al used 2DGE/MS to highlight the changes in the β cell proteome induced by the fatty acid palmitate, implicating it as a cause of β cell dysfunction (62).
Sample availability and quantity are often limiting factors for β cell biologists (and in particular for human β cell studies), but newer LC-MS/MS based methods have recently been employed to resolve the β cell proteome (52,63). The first analysis of the human islet proteome by LC-MS/MS by Metz and colleagues identified an unprecedented 3,365 proteins (64). Waanders et al. later identified 6,873 unique proteins using mouse pancreatic islets, requiring only 3-4,000 cells in total (65). Using these general proteomic profiles and whole human islets, Schrimpe-Rutledge et al. identified 256 proteins that are upregulated or downregulated after chronic hypoglycemia (*66). Similar profiling of rat β cells showed that both regulators of protein biosynthesis and glycolytic enzymes were upregulated during hypoglycemia (67). Consistent with observations about the neuronal nature of the β cell, Schvartz and colleagues used INS1E cells grown in hyperglycemic conditions and identified, as one of 11 proteins upregulated by glucose stress, neuronal pentraxin 1 (NP1), a protein previously thought to be expressed only in the brain (68).
In recent years, subcellular β cell proteomics has become a popular target for study. Danzer et al. used a cell surface protein isolation method combined with LC-MS/MS to characterize the proteome of membrane-resident glycoproteins in both MIN6 cells and human whole islets (69). Han et al. used INS1E cells to identify 683 novel β cell phosphorylation sites from over 1,419 identified phosphoproteins (70). Also using INS1E cells, Schvartz et al. were able to identify not one, but three separate proteomic profiles exclusive to mature insulin secretory vesicles (ISCs), immature ISCs, and proteins common to both ISCs and the rest of the cell (71). In all of these studies, the proteins identified have great potential as therapeutic targets to prevent or overcome β cell dysfunction.
Conclusion
Recent technological advances have made it possible to separate the heterogeneous cells comprising an islet into the α, β, and δ cell subpopulations thereby making it possible to independently investigate the gene expression profiles and regulatory mechanisms specific to β cells (and other cell types). Similarly, advanced technologies to discover and catalog mRNA, miRNA, lncRNA, and proteins specific to β cells in an unbiased fashion will identify potential therapeutic targets responsible for β cell function, differentiation, proliferation, or dysfunction. As we better characterize the β cell, the techniques promise to identify transcripts, regulatory RNAs, and/or proteins that control how environmental, genetics, and epigenetic factors contribute to diabetes pathogenesis. Scientists hope to apply these findings to determine what molecules are necessary to promote embryonic or iPS cells to differentiate into insulin producing β cells, to identify biomarkers of β cell dysfunction or autoimmune destruction, and to understand how β cells and the other endocrine cell types maintain glucose homeostasis.
Key points.
Pancreatic islets are very complex ‘miniorgans’ composed of many different cell types: endocrine and nonendocrine. Flow cytometry and antibody staining techniques enable the separation of highly enriched subpopulations (particularly β cells) for detailed analysis.
Next generation sequencing and mass spectrometry analysis have generated β cell specific mRNA, miRNA, lncRNA, and protein profiles.
β-cell transcriptome and proteome results show how associated DNA variants can alter gene expression and regulatory mechanisms.
β cell expression profiling efforts are beginning to identify genes, regulatory RNAs, and proteins that are differentially expressed in diseased versus healthy cells.
Expression profiles at multiple stages of development should help guide the directed differentiation of embryonic and induced pluripotent stem cells to glucose sensitive, insulin secreting β cells.
Acknowledgements
The authors thank Shaked Afik, Laura Alonso, Phil DiIorio, Manuel Garber, Dale Greiner, and Anetta Nowosielska for helpful discussions about the β cell transcriptome. The National Institutes of Health (grants U01 DK089572-02, DK032520, R24 DK-093437-01) provided the funding to support this work.
Disclosure of Funding: This effort was supported, in part, by NIH/NIDDK grants U01 DK089572-02 and 1 R24 DK093437-01
Abbreviations
- T1D
(Type 1 Diabetes)
- T2D
(Type 2 Diabetes)
- (RNASeq)
RNA Sequencing
- (GWAS)
Genome wide association study
- (MIP-GFP)
Transgenic mice expressing green fluorescent protein controlled by the mouse insulin I gene promoter
- (miRNA)
microRNA
- (lncRNA)
long noncoding RNA
- (iPS) cells
induced pluripotent stem
- (2DGE)
2-dimensional gel electrophoresis
- (LC-MS/MS)
liquid chromatography inline with tandem mass spectrometry
- (TCTP)
translationally controlled tumor protein
- (NP1)
neuronal pentraxin 1
- (ISCs)
insulin secretory vesicles
Footnotes
Conflict of Interest The authors declare no conflicts of interest.
References
Papers of particular interest, published within the annual period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Novak I. Purinergic receptors in the endocrine and exocrine pancreas. Purinergic Signal. 2008 Sep;4(3):237–53. doi: 10.1007/s11302-007-9087-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wierup N, Svensson H, Mulder H, Sundler F. The ghrelin cell: a novel developmentally regulated islet cell in the human pancreas. Regul. Pept. 2002 Jul 15;107(1-3):63–9. doi: 10.1016/s0167-0115(02)00067-8. [DOI] [PubMed] [Google Scholar]
- 3.Roep BO, Peakman M. Antigen targets of type 1 diabetes autoimmunity. Cold Spring Harb Perspect Med. 2012 Apr;2(4):a007781. doi: 10.1101/cshperspect.a007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kahn SE. The relative contributions of insulin resistance and beta-cell dysfunction to the pathophysiology of Type 2 diabetes. Diabetologia. 2003 Jan;46(1):3–19. doi: 10.1007/s00125-002-1009-0. [DOI] [PubMed] [Google Scholar]
- 5.Ferrannini E, Gastaldelli A, Miyazaki Y, Matsuda M, Mari A, DeFronzo RA. beta-Cell function in subjects spanning the range from normal glucose tolerance to overt diabetes: a new analysis. J. Clin. Endocrinol. Metab. 2005 Jan;90(1):493–500. doi: 10.1210/jc.2004-1133. [DOI] [PubMed] [Google Scholar]
- 6.Rabinovitch A, Russell T, Shienvold F, Noel J, Files N, Patel Y, et al. Preparation of rat islet B-cell-enriched fractions by light-scatter flow cytometry. Diabetes. 1982 Nov;31(11):939–43. doi: 10.2337/diacare.31.11.939. [DOI] [PubMed] [Google Scholar]
- 7.Nielsen DA, Lernmark A, Berelowitz M, Bloom GD, Steiner DF. Sorting of pancreatic islet cell subpopulations by light scattering using a fluorescence-activated cell sorter. Diabetes. 1982 Apr;31(4 Pt 1):299–306. doi: 10.2337/diab.31.4.299. [DOI] [PubMed] [Google Scholar]
- 8.Figlewicz DP, Forhan SE, Hodgson AT, Grodsky GM. 65Zinc and endogenous zinc content and distribution in islets in relationship to insulin content. Endocrinology. 1984 Sep;115(3):877–81. doi: 10.1210/endo-115-3-877. [DOI] [PubMed] [Google Scholar]
- 9.Lukowiak B, Vandewalle B, Riachy R, Kerr-Conte J, Gmyr V, Belaich S, et al. Identification and purification of functional human beta-cells by a new specific zinc-fluorescent probe. J. Histochem. Cytochem. 2001 Apr;49(4):519–28. doi: 10.1177/002215540104900412. [DOI] [PubMed] [Google Scholar]
- 10.Iglesias I, Bentsi-Barnes K, Umeadi C, Brown L, Kandeel F, Al-Abdullah IH. Comprehensive analysis of human pancreatic islets using flow and laser scanning cytometry. Transplant. Proc. 2008 Mar;40(2):351–4. doi: 10.1016/j.transproceed.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parnaud G, Bosco D, Berney T, Pattou F, Kerr-Conte J, Donath MY, et al. Proliferation of sorted human and rat beta cells. Diabetologia. 2008 Jan;51(1):91–100. doi: 10.1007/s00125-007-0855-1. [DOI] [PubMed] [Google Scholar]
- 12.Hara M, Wang X, Kawamura T, Bindokas VP, Dizon RF, Alcoser SY, et al. Transgenic mice with green fluorescent protein-labeled pancreatic beta -cells. Am. J. Physiol. Endocrinol. Metab. 2003 Jan;284(1):E177–83. doi: 10.1152/ajpendo.00321.2002. [DOI] [PubMed] [Google Scholar]
- 13.Katsuta H, Akashi T, Katsuta R, Nagaya M, Kim D, Arinobu Y, et al. Single pancreatic beta cells co-express multiple islet hormone genes in mice. Diabetologia. 2010 Jan;53(1):128–38. doi: 10.1007/s00125-009-1570-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quoix N, Cheng-Xue R, Guiot Y, Herrera PL, Henquin J-C, Gilon P. The GluCre-ROSA26EYFP mouse: a new model for easy identification of living pancreatic alpha-cells. FEBS Lett. 2007 Sep 4;581(22):4235–40. doi: 10.1016/j.febslet.2007.07.068. [DOI] [PubMed] [Google Scholar]
- 15.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat. Rev. Immunol. 2004 Aug;4(8):648–55. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 16.Dorrell C, Abraham SL, Lanxon-Cookson KM, Canaday PS, Streeter PR, Grompe M. Isolation of major pancreatic cell types and long-term culture-initiating cells using novel human surface markers. Stem Cell Res. 2008 Sep;1(3):183–94. doi: 10.1016/j.scr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Dorrell C, Grompe MT, Pan FC, Zhong Y, Canaday PS, Shultz LD, et al. Isolation of mouse pancreatic alpha, beta, duct and acinar populations with cell surface markers. Mol. Cell. Endocrinol. 2011 Jun 6;339(1-2):144–50. doi: 10.1016/j.mce.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pechhold S, Stouffer M, Walker G, Martel R, Seligmann B, Hang Y, et al. Transcriptional analysis of intracytoplasmically stained, FACS-purified cells by high-throughput, quantitative nuclease protection. Nat. Biotechnol. 2009 Nov;27(11):1038–42. doi: 10.1038/nbt.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quayum N, Kutchma A, Sarkar SA, Juhl K, Gradwohl G, Mellitzer G, et al. GeneSpeed Beta Cell: an online genomics data repository and analysis resource tailored for the islet cell biologist. Exp Diabetes Res. 2008;2008:312060. doi: 10.1155/2008/312060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kutlu B, Burdick D, Baxter D, Rasschaert J, Flamez D, Eizirik DL, et al. Detailed transcriptome atlas of the pancreatic beta cell. BMC Med Genomics. 2009;2:3. doi: 10.1186/1755-8794-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bensellam M, Van Lommel L, Overbergh L, Schuit FC, Jonas JC. Cluster analysis of rat pancreatic islet gene mRNA levels after culture in low-, intermediate- and high-glucose concentrations. Diabetologia. 2009 Mar;52(3):463–76. doi: 10.1007/s00125-008-1245-z. [DOI] [PubMed] [Google Scholar]
- 22.Dorrell C, Schug J, Lin CF, Canaday PS, Fox AJ, Smirnova O, et al. Transcriptomes of the major human pancreatic cell types. Diabetologia. 2011 Nov;54(11):2832–44. doi: 10.1007/s00125-011-2283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mutz K-O, Heilkenbrinker A, Lönne M, Walter J-G, Stahl F. Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotechnol. 2013 Feb;24(1):22–30. doi: 10.1016/j.copbio.2012.09.004. [DOI] [PubMed] [Google Scholar]
- *24.Ku GM, Kim H, Vaughn IW, Hangauer MJ, Myung Oh C, German MS, et al. Research resource: RNA-Seq reveals unique features of the pancreatic β-cell transcriptome. Mol. Endocrinol. 2012 Oct;26(10):1783–92. doi: 10.1210/me.2012-1176. This study uses RNASeq to reveal the islet and beta cell transcriptomes (both mRNA and lncRNA) in mice.
- *25.Bramswig NC, Everett LJ, Schug J, Dorrell C, Liu C, Luo Y, et al. Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J. Clin. Invest. 2013 Mar 1;123(3):1275–84. doi: 10.1172/JCI66514. This study uses surface staining of dissociated human islets to obtain a detailed characterization of the transcriptome and epigenome of alpha and beta cells. These findings show how the alpha cell appears to be particularly amenable to potential epigenome manipulation for transdifferentiation.
- **26.Nica AC, Ongen H, Irminger J-C, Bosco D, Berney T, Antonarakis SE, et al. Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Res. 2013 Sep;23(9):1554–62. doi: 10.1101/gr.150706.112. This study provided a human beta-cell specific gene expression profile versus whole islets. The details in this study allowed for further comparison of beta cell specific genes identified in diabetes genome wide association studies. Additionally, the results of this study showed that the beta cell gene expression profile is quite similar to the neuronal profile.
- 27.Eizirik DL, Sammeth M, Bouckenooghe T, Bottu G, Sisino G, Igoillo-Esteve M, et al. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genet. 2012 Mar;8(3):e1002552. doi: 10.1371/journal.pgen.1002552. This study focused on T1D and the effects of cytokine exposure on islet gene expression, especially the genes identified in T1D genome wide association studies.
- 28.Brissova M, Fowler MJ, Nicholson WE, Chu A, Hirshberg B, Harlan DM, et al. Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. J. Histochem. Cytochem. 2005 Sep;53(9):1087–97. doi: 10.1369/jhc.5C6684.2005. [DOI] [PubMed] [Google Scholar]
- 29.Schwartz MW, Woods SC, Porte D, Seeley RJ, Baskin DG. Central nervous system control of food intake. Nature. 2000 Apr 6;404(6778):661–71. doi: 10.1038/35007534. [DOI] [PubMed] [Google Scholar]
- 30.Ahrén B. Autonomic regulation of islet hormone secretion--implications for health and disease. Diabetologia. 2000 Apr;43(4):393–410. doi: 10.1007/s001250051322. [DOI] [PubMed] [Google Scholar]
- 31.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009 Jun;41(6):703–7. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, Steinthorsdottir V, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012 Sep;44(9):981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jain P, Vig S, Datta M, Jindel D, Mathur AK, Mathur SK, et al. Systems biology approach reveals genome to phenome correlation in type 2 diabetes. PLoS ONE. 2013;8(1):e53522. doi: 10.1371/journal.pone.0053522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Planas R, Pujol-Borrell R, Vives-Pi M. Global gene expression changes in type 1 diabetes: insights into autoimmune response in the target organ and in the periphery. Immunol. Lett. 2010 Oct 30;133(2):55–61. doi: 10.1016/j.imlet.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006 Apr 15;15(Spec No 1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 36.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009 Feb 20;136(4):629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Dumortier O, Van Obberghen E. MicroRNAs in pancreas development. Diabetes Obes Metab. 2012 Oct;14(Suppl 3):22–8. doi: 10.1111/j.1463-1326.2012.01656.x. [DOI] [PubMed] [Google Scholar]
- **38.van de Bunt M, Gaulton KJ, Parts L, Morán I, Johnson PR, Lindgren CM, et al. The miRNA profile of human pancreatic islets and beta-cells and relationship to type 2 diabetes pathogenesis. PLoS ONE. 2013;8(1):e55272. doi: 10.1371/journal.pone.0055272. This study identified microRNAs in both human islets and specific to human beta cells. This profile was compared to genome wide association studies identified SNPs to identify patterns of microRNAs associated with T2D.
- **39.Klein D, Misawa R, Bravo-Egana V, Vargas N, Rosero S, Piroso J, et al. MicroRNA expression in alpha and beta cells of human pancreatic islets. PLoS ONE. 2013;8(1):e55064. doi: 10.1371/journal.pone.0055064. This study compared the miRNA profiles for α and β cells isolated from human islets and identified a much higher number of β-cell specific miRNAs. They showed how changes in miRNA expression in cell lines affected the gene expression pattern of known ‘β cell disallowed genes’.
- 40.Barbagallo D, Piro S, Condorelli AG, Mascali LG, Urbano F, Parrinello N, et al. miR-296-3p, miR-298-5p and their downstream networks are causally involved in the higher resistance of mammalian pancreatic α cells to cytokine-induced apoptosis as compared to β cells. BMC Genomics. 2013;14:62. doi: 10.1186/1471-2164-14-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuit F, Van Lommel L, Granvik M, Goyvaerts L, de Faudeur G, Schraenen A, et al. β-cell-specific gene repression: a mechanism to protect against inappropriate or maladjusted insulin secretion? Diabetes. 2012 May;61(5):969–75. doi: 10.2337/db11-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, et al. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33(8):2697–706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc. Natl. Acad. Sci. U.S.A. 2007 Nov 6;104(45):17719–24. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, Haubrock M, Cao K-M, Hua X, Zhang C-Y, Wingender E, et al. Regulatory coordination of clustered microRNAs based on microRNA-transcription factor regulatory network. BMC Syst Biol. 2011;5:199. doi: 10.1186/1752-0509-5-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009 Aug;5(8):e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009 Jul 1;23(13):1494–504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009 Mar 12;458(7235):223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **48.Morán I, Akerman I, van de Bunt M, Xie R, Benazra M, Nammo T, et al. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012 Oct 3;16(4):435–48. doi: 10.1016/j.cmet.2012.08.010. This study provided a detailed list of human islet and pancreatic beta cell specific lncRNAs. The authors identified glucose regulated expression of lncRNA, identified conserved mouse orthologs, and showed the closed proximity of lncRNAs to genome areas associated with T2D.
- 49.Gstaiger M, Aebersold R. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat. Rev. Genet. 2009 Sep;10(9):617–27. doi: 10.1038/nrg2633. [DOI] [PubMed] [Google Scholar]
- 50.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003 Mar 13;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 51.Rabilloud T, Chevallet M, Luche S, Lelong C. Two-dimensional gel electrophoresis in proteomics: Past, present and future. J Proteomics. 2010 Oct 10;73(11):2064–77. doi: 10.1016/j.jprot.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 52.Patel VJ, Thalassinos K, Slade SE, Connolly JB, Crombie A, Murrell JC, et al. A comparison of labeling and label-free mass spectrometry-based proteomics approaches. J. Proteome Res. 2009 Jul;8(7):3752–9. doi: 10.1021/pr900080y. [DOI] [PubMed] [Google Scholar]
- 53.Ahmed M, Forsberg J, Bergsten P. Protein profiling of human pancreatic islets by two-dimensional gel electrophoresis and mass spectrometry. J. Proteome Res. 2005 May;4(3):931–40. doi: 10.1021/pr050024a. [DOI] [PubMed] [Google Scholar]
- 54.Nyblom HK, Thorn K, Ahmed M, Bergsten P. Mitochondrial protein patterns correlating with impaired insulin secretion from INS-1E cells exposed to elevated glucose concentrations. Proteomics. 2006 Oct;6(19):5193–8. doi: 10.1002/pmic.200600137. [DOI] [PubMed] [Google Scholar]
- 55.Maris M, Ferreira GB, D’Hertog W, Cnop M, Waelkens E, Overbergh L, et al. High glucose induces dysfunction in insulin secretory cells by different pathways: a proteomic approach. J. Proteome Res. 2010 Dec 3;9(12):6274–87. doi: 10.1021/pr100557w. [DOI] [PubMed] [Google Scholar]
- 56.D’Hertog W, Maris M, Thorrez L, Waelkens E, Overbergh L, Mathieu C. Two-dimensional gel proteome reference map of INS-1E cells. Proteomics. 2011 Apr;11(7):1365–9. doi: 10.1002/pmic.201000006. [DOI] [PubMed] [Google Scholar]
- 57.Diraison F, Hayward K, Sanders KL, Brozzi F, Lajus S, Hancock J, et al. Translationally controlled tumour protein (TCTP) is a novel glucose-regulated protein that is important for survival of pancreatic beta cells. Diabetologia. 2011 Feb;54(2):368–79. doi: 10.1007/s00125-010-1958-7. [DOI] [PubMed] [Google Scholar]
- 58.Zanini C, Bruno S, Mandili G, Baci D, Cerutti F, Cenacchi G, et al. Differentiation of mesenchymal stem cells derived from pancreatic islets and bone marrow into islet-like cell phenotype. PLoS ONE. 2011;6(12):e28175. doi: 10.1371/journal.pone.0028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coppola A, Tomasello L, Pizzolanti G, Pucci-Minafra I, Albanese N, Di Cara G, et al. In vitro phenotypic, genomic and proteomic characterization of a cytokine-resistant murine β-TC3 cell line. PLoS ONE. 2012;7(2):e32109. doi: 10.1371/journal.pone.0032109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lindskog C, Korsgren O, Pontén F, Eriksson JW, Johansson L, Danielsson A. Novel pancreatic beta cell-specific proteins: antibody-based proteomics for identification of new biomarker candidates. J Proteomics. 2012 May 17;75(9):2611–20. doi: 10.1016/j.jprot.2012.03.008. [DOI] [PubMed] [Google Scholar]
- *61.Massa O, Alessio M, Russo L, Nardo G, Bonetto V, Bertuzzi F, et al. Serological Proteome Analysis (SERPA) as a tool for the identification of new candidate autoantigens in type 1 diabetes. J Proteomics. 2013 Apr 26;82:263–73. doi: 10.1016/j.jprot.2013.02.030. This is the first known study using an antibody-based proteomics screen to probe for new autoantigens in T1D using sera from living T1D patients. It opens up the possibility for the development of future, non-invasive proteomics studies in living patients.
- 62.Maris M, Robert S, Waelkens E, Derua R, Hernangomez MH, D’Hertog W, et al. Role of the saturated nonesterified fatty acid palmitate in beta cell dysfunction. J. Proteome Res. 2013 Jan 4;12(1):347–62. doi: 10.1021/pr300596g. [DOI] [PubMed] [Google Scholar]
- 63.Zhu W, Smith JW, Huang C-M. Mass spectrometry-based label-free quantitative proteomics. J. Biomed. Biotechnol. 2010;2010:840518. doi: 10.1155/2010/840518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Metz TO, Jacobs JM, Gritsenko MA, Fontès G, Qian W-J, Camp DG, et al. Characterization of the human pancreatic islet proteome by two-dimensional LC/MS/MS. J. Proteome Res. 2006 Dec;5(12):3345–54. doi: 10.1021/pr060322n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waanders LF, Chwalek K, Monetti M, Kumar C, Lammert E, Mann M. Quantitative proteomic analysis of single pancreatic islets. Proc. Natl. Acad. Sci. U.S.A. 2009 Nov 10;106(45):18902–7. doi: 10.1073/pnas.0908351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **66.Schrimpe-Rutledge AC, Fontès G, Gritsenko MA, Norbeck AD, Anderson DJ, Waters KM, et al. Discovery of novel glucose-regulated proteins in isolated human pancreatic islets using LC-MS/MS-based proteomics. J. Proteome Res. 2012 Jul 6;11(7):3520–32. doi: 10.1021/pr3002996. A thorough comparative analysis of human islets cultured under low and high glucose conditions, this study highlights the power and resolution of LC-MS/MS technology in identifying protein targets under normal and pathological conditions.
- 67.Martens GA, Jiang L, Verhaeghen K, Connolly JB, Geromanos SG, Stangé G, et al. Protein markers for insulin-producing beta cells with higher glucose sensitivity. PLoS ONE. 2010;5(12):e14214. doi: 10.1371/journal.pone.0014214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schvartz D, Couté Y, Brunner Y, Wollheim CB, Sanchez J-C. Modulation of neuronal pentraxin 1 expression in rat pancreatic β-cells submitted to chronic glucotoxic stress. Mol. Cell Proteomics. 2012 Aug;11(8):244–54. doi: 10.1074/mcp.M112.018051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Danzer C, Eckhardt K, Schmidt A, Fankhauser N, Ribrioux S, Wollscheid B, et al. Comprehensive description of the N-glycoproteome of mouse pancreatic β-cells and human islets. J. Proteome Res. 2012 Mar 2;11(3):1598–608. doi: 10.1021/pr2007895. [DOI] [PubMed] [Google Scholar]
- 70.Han D, Moon S, Kim Y, Ho W-K, Kim K, Kang Y, et al. Comprehensive phosphoproteome analysis of INS-1 pancreatic β-cells using various digestion strategies coupled with liquid chromatography-tandem mass spectrometry. J. Proteome Res. 2012 Apr 6;11(4):2206–23. doi: 10.1021/pr200990b. [DOI] [PubMed] [Google Scholar]
- 71.Schvartz D, Brunner Y, Couté Y, Foti M, Wollheim CB, Sanchez J-C. Improved characterization of the insulin secretory granule proteomes. J Proteomics. 2012 Aug 3;75(15):4620–31. doi: 10.1016/j.jprot.2012.04.023. [DOI] [PubMed] [Google Scholar]