Abstract
BACKGROUND
Deep dermatophytosis is a severe and sometimes life-threatening fungal infection caused by dermatophytes. It is characterized by extensive dermal and subcutaneous tissue invasion and by frequent dissemination to the lymph nodes and, occasionally, the central nervous system. The condition is different from common superficial dermatophyte infection and has been reported in patients with no known immunodeficiency. Patients are mostly from North African, consanguineous, multiplex families, which strongly suggests a mendelian genetic cause.
METHODS
We studied the clinical features of deep dermatophytosis in 17 patients with no known immunodeficiency from eight unrelated Tunisian, Algerian, and Moroccan families. Because CARD9 (caspase recruitment domain–containing protein 9) deficiency has been reported in an Iranian family with invasive fungal infections, we also sequenced CARD9 in the patients.
RESULTS
Four patients died, at 28, 29, 37, and 39 years of age, with clinically active deep dermatophytosis. No other severe infections, fungal or otherwise, were reported in the surviving patients, who ranged in age from 37 to 75 years. The 15 Algerian and Tunisian patients, from seven unrelated families, had a homozygous Q289X CARD9 allele, due to a founder effect. The 2 Moroccan siblings were homozygous for the R101C CARD9 allele. Both alleles are rare deleterious variants. The familial segregation of these alleles was consistent with autosomal recessive inheritance and complete clinical penetrance.
CONCLUSIONS
All the patients with deep dermatophytosis had autosomal recessive CARD9 deficiency. Deep dermatophytosis appears to be an important clinical manifestation of CARD9 deficiency. (Funded by Agence Nationale pour la Recherche and others.)
Deep dermatophytosis is a rare, invasive, sometimes life-threatening, fungal infection caused by dermatophytes.1 These filamentous fungi are ubiquitous and usually cause benign infections that are limited to keratinized tissues and lead to onychomycosis, tinea corporis, tinea cruris, tinea pedis, or tinea capitis.2 In deep dermatophytosis, dermatophytes invade the dermis and hypodermis and disseminate to the skin, hair, nails, lymph nodes, and brain.3 Deep dermatophytosis has been reported in patients with the human immunodeficiency virus and patients who are receiving immunosuppressive therapy.3 It was first described in 1959 in otherwise apparently healthy persons as “dermatophytic disease.”1 Forty-five cases have been reported to date in persons from North Africa.1,4-11 Twenty-four of these patients were from consanguineous families; 5 patients had sporadic disease, and 19 patients from eight multiplex families had familial disease. The remaining 21 patients were from families not reported to be consanguineous; 14 patients had sporadic disease and 7 had familial disease. This strongly suggests that predisposition to idiopathic deep dermatophytosis is inherited as an autosomal recessive trait. In addition, 18 cases of sporadic disease in patients from nonconsanguineous families have been reported in England, Russia, Denmark, Mexico, Brazil, the United States, and Japan.12-18
Genetic susceptibility to fungal diseases in otherwise healthy patients has gained interest in recent years.19 In particular, various inborn errors of interleukin-17 immunity (e.g., autosomal recessive interleukin-17RA deficiency, autosomal dominant interleukin-17F deficiency, and monoallelic gain-of-function mutations in STAT1) have been reported to underlie chronic mucocutaneous candidiasis disease.20-25 Autosomal recessive CARD9 (caspase recruitment domain– containing protein 9) deficiency has been reported in two unrelated kindreds with candida species meningitis,26,27 chronic mucocutaneous candidiasis, and cutaneous dermatophytosis.26 However, deep dermatophytosis has not been reported in association with any of these disorders. We used a candidate-gene approach, including the sequencing of CARD9, to investigate 17 patients, from eight unrelated kindreds, who had deep dermatophytosis and no known immunodeficiency.
METHODS
Details regarding recruitment of patients, ethical considerations, selection of controls, analysis of founder effects, Western blotting, and methods of histologic analysis are described in the Supplementary Appendix, available with the full text of this article at NEJM.org. This work was approved by the institutional review board, and all patients (or their family members) provided written informed consent for participation in the study (see the Supplementary Appendix).
MOLECULAR GENETICS
CARD9 was amplified with specific primers. Polymerase-chain-reaction (PCR) amplification conditions and primer sequences are described in the Supplementary Appendix.
STIMULATION OF WHOLE-BLOOD CELLS
Whole blood was diluted at a 1:2 ratio, and samples were incubated for 24 hours and 48 hours with medium alone, zymosan (5 μg per milliliter), heat-killed Candida albicans (cell density, 106 per milliliter), heat-killed Saccharomyces cerevisiae (106 per milliliter), lipopolysaccharide (1 ng per milliliter), and — as a positive control of activation — phorbol 12-myristate 13-acetate plus ionomycin (0.2 μg per milliliter and 2.10−4 μg per milliliter, respectively). Interleukin-6 production was measured in the supernatants by enzyme-linked immunosorbent assay.
MONOCYTE-DERIVED DENDRITIC CELLS
Monocyte-derived dendritic cells were prepared as previously described.28 On day 8, flow cytometry and analysis of total protein extracts were performed.
FLOW CYTOMETRY
Antibodies against human CARD9 (Epitomics 5281) or its control isotype and an Alexa Fluor 488–conjugated secondary goat antirabbit antibody (Epitomics 3064-1) were used according to the manufacturers’ protocols. T cells producing interleukin-17A and interleukin-22 were evaluated by intracellular staining, as previously described.29
RESULTS
CASE SUMMARY
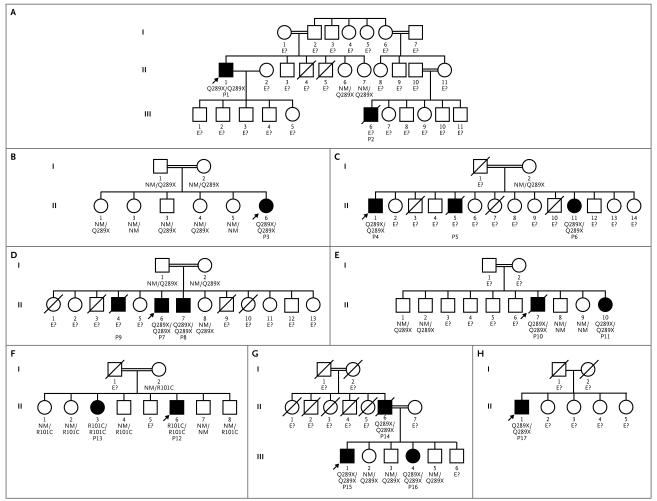
We report on 17 patients with deep dermatophytosis and no known immunodeficiency. The patients were from eight unrelated kindreds, seven of which were known to be consanguineous. The eight families (Fig. 1) originated from Morocco (1 family), Tunisia (2 families), and Algeria (5 families) (Table 1), and the main characteristics of the patients are reported in Table 2. Case reports are detailed in the Supplementary Appendix. In all patients, first symptoms appeared in childhood or early adulthood (age range, 2 to 21 years). Four patients (Patients 1, 2, 3, and 17) had adenitis caused by dermatophyte infection, which was diagnosed on the basis of histologic analysis or positive dermatophyte culture, and 13 patients had documented cutaneous deep dermatophytosis (Patients 1 through 5, Patients 7 through 10, and Patients 12, 15, 16, and 17). Patient 14 had extensive skin, scalp, and nail lesions, but no histologic studies or fungal cultures could be performed. However, this patient had two children (Patients 15 and 16) with proven deep dermatophytosis. Finally, 3 index patients (Patients 4, 10, and 12) had three sisters (Patients 6, 11, and 13, respectively) who had had chronic onychomycosis and tinea since childhood but did not have deep dermatophytosis per se; the sisters were nevertheless considered to be affected.
Figure 1. Pedigrees of the 17 Patients from Eight Kindreds with Deep Dermatophytosis and CARD9 Mutations.
Panels A through H represent the eight kindreds. Each generation is designated by a Roman numeral, and each family member by an Arabic numeral. Circles denote female family members, squares male family members, solid squares and circles patients with deep dermatophytosis, double horizontal lines consanguinity in a married couple, and slashes deceased family members. The probands are indicated by arrows. The CARD9 genotype is indicated below each family member. E? denotes no DNA available, NM nonmutated, and P patient.
Table 1.
Description of the 17 Patients with Deep Dermatophytosis.*
| Patient No. |
Case No.† |
Yr of Age at Symptom Onset |
Yr of Age at Last Follow-up |
Sex | Country of Origin |
Organ Involvement | Fungus | Status At Time of Study |
CARD9 Mutation |
Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A-II-1 | 6 | 75 | M | Algeria | Skin, scalp, nails, lymph nodes | Trichophyton violaceum | Alive | Q289X/Q289X | Boudghène- Stambouli and Mérad-Boudia6 |
| 2 | A-III-6 | 2 | 29 | M | Algeria | Skin, scalp, nails, lymph nodes, brain |
T. violaceum | Dead | E? | Boudghène- Stambouli and Mérad-Boudia,7 Boudghène- Stambouli et al.8 |
| 3 | B-II-6 | 9 | 40 | F | Algeria | Skin, scalp, nails, lymph nodes | T. rubrum | Alive | Q289X/Q289X | Boudghène- Stambouli and Mérad-Boudia9 |
| 4 | C-II-1 | 8 | 56 | M | Algeria | Skin, scalp, nails | T. violaceum | Alive | Q289X/Q289X | |
| 5 | C-II-5 | 8 | 34 | M | Algeria | Skin, scalp, nails, lymph nodes | T. violaceum | Dead | E? | Pruszkowski et al.10 |
| 6 | C-II-11 | 8 | 41 | F | Algeria | Nails | T. violaceum | Alive | Q289X/Q289X | |
| 7 | D-II-6 | 19 | 43 | M | Algeria | Skin, scalp, nails, lymph nodes | Fungal hyphae on biopsies | Alive | Q289X/Q289X | |
| 8 | D-II-7 | 21 | 40 | M | Algeria | Skin, perineum, scalp, lymph nodes |
Fungal hyphae on biopsies | Alive | Q289X/Q289X | |
| 9 | D-II-4 | NA | 28 | M | Algeria | Skin, scalp | Fungal hyphae on biopsies | Dead | E? | |
| 10 | E-II-7 | NA | 39 | M | Algeria | Skin, scalp, lymph nodes | T. violaceum | Dead | Q289X/Q289X | Boudghène- Stambouli and Mérad-Boudia11 |
| 11 | E-II-10 | NA | 37 | F | Algeria | Nails | Dermatophyte | Alive | Q289X/Q289X | |
| 12 | F-II-6 | NA | 40 | M | Morocco | Skin, bone, lymph nodes, nails | T. rubrum | Alive | R101C/R101C | |
| 13 | F-II-3 | NA | 49 | F | Morocco | Scalp, nails | Dermatophyte | Alive | R101C/R101C | |
| 14 | G-II-6 | 6 | 91 | M | Tunisia | Skin, scalp, nails | Dermatophyte | Dead | Q289X/Q289X | |
| 15 | G-III-1 | 12 | 44 | M | Tunisia | Scalp, nails | T. rubrum | Alive | Q289X/Q289X | |
| 16 | G-III-4 | 5 | 52 | F | Tunisia | Skin, scalp, nails, lymph nodes | T. rubrum and T. violaceum | Alive | Q289X/Q289X | |
| 17 | H-II-1 | 6 | 62 | M | Tunisia | Skin, scalp, nails, lymph nodes | T. rubrum and T. violaceum | Alive | Q289X/Q289X |
E? denotes no DNA available, and NA not available.
The case numbers correspond to the pedigrees.
Table 2.
Characteristics of the 17 Patients.
| Variable | No. of Patients |
|---|---|
| Male sex | 12 |
| Female sex | 5 |
| Country of origin | |
| Morocco | 2 |
| Algeria | 11 |
| Tunisia | 4 |
| First symptoms* | |
| Severe or recurrent tinea capitis | 14 |
| Severe or recurrent tinea corporis | 10 |
| Onychomycosis | 6 |
| Presentations in adulthood | |
| Lymph node enlargement | 10 |
| Central nervous system invasion | 1 |
| Local organ invasion (bone, diges- tive tract) |
2 |
| Associated infection: thrush | 6 |
| Deaths† | 5 |
| Dermatophyte identified | |
| T. rubrum | 5 |
| T. violaceum | 8 |
| Histologic features | |
| Granuloma | 10 |
| Necrosis | 6 |
| Hyphae on biopsy | 12 |
| Biologic exploration‡ | |
| Hypereosinophilia (>500/mm3) | 9/10 |
| High IgE levels (>500,000 IU/ml) | 4/4 |
| Lymphocyte subset‡ | |
| Normal CD4+ T-lymphocyte subset | 4/4 |
| Normal CD8+ T-lymphocyte subset | 4/4 |
| Normal B-lymphocyte subset | 3/3 |
| Normal NK-lymphocyte subset | 3/3 |
The median age at first symptoms was 8 years (range, 2 to 21).
The median age at death was 34 years (range, 28 to 91).
For biologic exploration and lymphocyte subset, the number and total number of patients tested are shown for each category.
The median age of the patients at recruitment was 41 years (range, 28 to 91). Skin lesions subsequently included extensive erythematosquamous lesions and nodular subcutaneous or ulcerative fistulized infiltrations (Fig. 2B; and Fig. S4.1A, S4.1J through S4.1M, S4.1P, S4.1Q, S4.1R, and S4.1T in the Supplementary Appendix). Two patients had contiguous locoregional extension to the bone or digestive tract (Fig. 2B and 2C; and Fig. S4.1O, S4.1U, S4.1V, and S4.1W in the Supplementary Appendix). Fifteen patients had severe onychomycosis (Fig. S4.1B, S4.1C, and S4.1S in the Supplementary Appendix). Manifestations of the disease in other extradermatologic locations were also observed — lymphadenopathies in 10 patients and probable brain involvement in 1 patient (Fig. S4.1F in the Supplementary Appendix).
Figure 2. Clinical and Histologic Features of Patients with CARD9 Deficiency.
A skin-biopsy specimen from Patient 13 (Panel A, periodic acid–Schiff) shows irregularly branched septate hyphae (arrowhead) in the center of a granuloma containing multinucleated giant cells (asterisk). Clinical features of Patient 12 (Panel B) and Patient 8 (Panel C) are shown.
Histologic examination of the skin revealed a multifocal-to-coalescing granulomatous dermatitis. The dermatitis extended throughout the dermis, was characterized by infiltrates of activated macrophages and epithelioid cells that can fuse to form multinucleated giant cells, and was associated with lymphocytes, plasma cells, neutrophils, and eosinophils. In the center of such granulomas and sometimes even in the cytoplasm of multinucleated giant cells, pseudohyphae and irregularly branched hyaline septate hyphae can be seen (Fig. 2A, and Fig. S4.2A and S4.2B in the Supplementary Appendix). Immunohistochemical analyses were positive when a primary antidermatophyte monoclonal antibody was used (Fig. S4.2C in the Supplementary Appendix), and PCR assay of a skin-biopsy specimen was positive for Trichophyton rubrum. Histologic examination of the lymph nodes revealed granulomas containing hyphae and necrosis in four patients (Patients 1, 2, 3, and 17). Dermatophytes also grew from the lymph nodes of Patients 2 and 3 (T. violaceum and T. rubrum, respectively). The only other associated infectious condition was oral candidiasis in six patients that was confirmed by mycologic evaluation. Four patients with clinically active deep dermatophytosis died at the ages of 28, 29, 37, and 39 years. One patient died in a bedridden state at the age of 91 years. None of the patients had any detectable T-cell immunodeficiency known to confer a predisposition to severe dermatophyte infection (Table 2).
IDENTIFICATION OF HOMOZYGOUS NONSENSE OR MISSENSE CARD9 MUTATIONS
Using a candidate-gene approach, we investigated 14 of the 17 patients who had deep dermatophytosis and for whom genetic material was available. We first sequenced CARD9 and found homozygous mutations in all 14 patients. Eight Algerian patients and 4 Tunisian patients had a homozygous c.C865T mutation in exon 6, resulting in a premature termination codon in position 289, Q289X (Fig. S1 in the Supplementary Appendix), in the region encoding the coiled-coil domain of CARD9 (Fig. S2 in the Supplementary Appendix). The 2 patients from the Moroccan kindred had a homozygous CARD9 missense mutation, c.C301T, in exon 3, resulting in the replacement of the arginine residue in position 101 with a cysteine residue (R101C) (Fig. S1.A in the Supplementary Appendix). This amino acid substitution is located only a few amino acids after the end of the CARD domain (Fig. S2 in the Supplementary Appendix). Finally, all healthy members of the eight kindreds were found to be either homozygous for the nonmutated allele or heterozygous and had no unusual infections, fungal or otherwise.
The segregation of the two mutations in the eight kindreds was consistent with autosomal recessive CARD9 deficiency with complete clinical penetrance. These two mutations in patients with deep dermatophytosis were different from the Q295X mutation previously reported in an Iranian kindred with CARD9 deficiency and the compound heterozygous missense mutations G72S and R373P found in a Dutch girl originating from Asia.26,27 The missense and nonsense mutations reported here were not found in any of the various public databases searched (Human Gene Mutation Database, Ensembl, and 1000 Genomes Project) or in our in-house whole-exome-sequencing database (>1000 exomes). We also sequenced CARD9 in 1052 controls from the Human Genome Diversity Project–Centre d’Etude du Polymorphisme Humain panel, as well as 138 persons from Morocco, 100 from Tunisia, and 83 from Algeria; all were found to be homozygous for nonmutated CARD9, thus decreasing substantially the possibility that the Q289X and R101C variants were irrelevant polymorphisms. With the use of Polymorphism Phenotyping, version 2 (PolyPhen-2),30 and Sorting Intolerant from Tolerant (SIFT 2) software, the missense mutation was predicted in silico to be deleterious.
FOUNDER-EFFECT ANALYSIS OF THE Q289X MUTATION
The eight Algerian patients and four Tunisian patients harbored the same previously unknown homozygous premature termination codon (Q289X), a finding suggestive of a founder effect. An analysis of Affymetrix 250K Nsp Array data performed for seven Q289X/Q289X unrelated patients showed a common homozygous haplotype surrounding CARD9 (Fig. S3 in the Supplementary Appendix). The largest common haplotype upstream from the mutation identified in Patients 1 and 3 encompassed 1.2 megabases (corresponding to 33 single-nucleotide polymorphisms [SNPs]). The largest common haplotype downstream from the mutation identified in Patients 4 and 8 encompassed 1.6 megabases (29 SNPs). The ESTIAGE program was used to estimate the age of the most recent common ancestor to 39 generations (95% confidence interval [CI], 23 to 70). Assuming a generation time of 25 years, the most recent common ancestor of the patients therefore lived approximately 975 years ago (95% CI, 575 to 1750).
EFFECT OF CARD9 MUTATIONS ON PROTEIN LEVELS
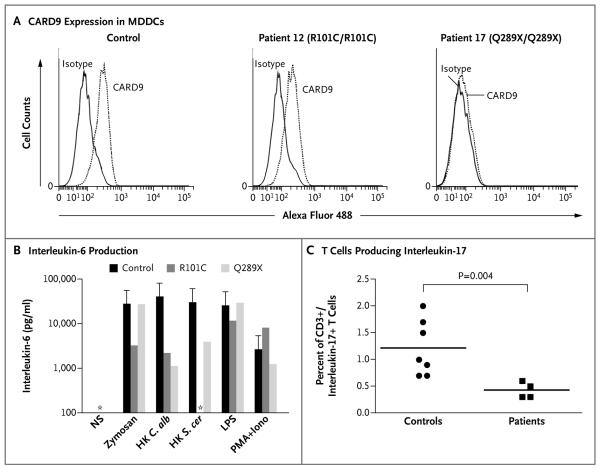
By transfection of the various CARD9 alleles in human embryonic kidney (HEK) 293T cells, we found that the R101C protein expression pattern, as assessed with the use of Western blotting, was similar to nonmutant protein, unlike truncated Q289X (Fig. S5 in the Supplementary Appendix). We also performed Western blotting on cultured monocyte-derived dendritic cells from two patients homozygous for the Q289X mutation (Patients 15 and 17). The two controls had a 62 kDa protein corresponding to the nonmutant CARD9 protein, whereas no protein was detected in monocyte-derived dendritic cells from Patients 15 and 17, not even at a lower molecular weight (Fig. S6.1 in the Supplementary Appendix). Flow-cytometric analysis of CARD9 levels in monocyte-derived dendritic cells from Patient 12 (R101C/R101C) showed this protein to be less abundant than in monocyte-derived dendritic cells from a healthy control that was tested in parallel (33% of monocyte-derived dendritic cells were CARD9-positive in Patient 12, whereas 71% were CARD9-positive in the control); Patient 17 (Q289X/Q289X) produced no CARD9 protein at all (Fig. 3A). Thus, in monocyte-derived dendritic cells, the Q289X allele leads to an absence of CARD9 protein, whereas the R101C allele results in much lower levels of CARD9 than normal.
Figure 3. Effect of CARD9 Mutations on CARD9 Expression and Function.
Panel A shows flow cytometric expression of CARD9 in monocyte-derived dendritic cells (MDDCs) in Patients 12 and 17 and controls. Panel B shows interleukin-6 production by whole-blood cells from patients with Q289X/Q289X and R101C/R101C CARD9 genotypes after 48 hours, as measured by enzyme-linked immunosorbent assay on stimulation with zymosan, heat-killed Candida albicans (HK C. alb), heat-killed Saccharomyces cerevisiae (HK S. cer), lipopolysaccharide (LPS), and phorbol 12-myristate 13-acetate plus ionomycin (PMA+Iono). NS denotes unstimulated. The asterisks indicate values below 100 pg per milliliter, and the T bars the standard deviation. Panel C shows impaired development of interleukin-17–producing T cells in patients homozygous for the R101C or Q289X CARD9 mutations.
EFFECT OF CARD9 MUTATIONS ON PROTEIN FUNCTION
We evaluated the functional consequence of CARD9 mutations by studying interleukin-6 production by whole-blood cells after 24 hours and after 48 hours of stimulation with zymosan (an agonist of dectin-1 and toll-like receptor 2 [TLR2]), heat-killed Candida albicans, heat-killed Saccharomyces cerevisiae, lipopolysaccharide (TLR4 agonist), and phorbol 12-myristate 13-acetate plus ionomycin. We tested three controls, two patients homozygous for the CARD9 Q289X allele, and one patient homozygous for the CARD9 R101C allele. All patients had markedly low levels of interleukin-6 production after 24 hours (Fig. S6.2 in the Supplementary Appendix) and after 48 hours (Fig. 3B) of whole-blood stimulation with heat-killed C. albicans and heat-killed S. cerevisiae. Finally, for three patients homozygous for the CARD9 Q289X allele and one patient homozygous for the CARD9 R101C allele, we used flow cytometry ex vivo to evaluate the proportion of T cells expressing interleukin-17A. These cells were significantly less common in these patients than in the seven healthy controls tested in parallel (P = 0.004) (Fig. 3C).
DISCUSSION
We have identified autosomal recessive CARD9 deficiency as a potential genetic cause of deep dermatophytosis. This broadens the spectrum of severe fungal infections that are associated with CARD9 deficiency. Subsequent to the Q295X nonsense mutation and the compound heterozygote missense mutations G72S and R373P previously reported,26,27 we identified two new CARD9 mutations. One was a missense mutation (R101C), and the other was a nonsense mutation (Q289X). Patients homozygous for these deleterious alleles were found in four countries (Iran,26 Morocco, Tunisia, and Algeria); the prevalence of parental consanguinity is high in these countries. Five Algerian families and two Tunisian families carried the same Q289X mutation because of a founder effect, with the most recent common ancestor living approximately 975 years ago. None of the heterozygous persons had any clinical signs, whereas all persons homozygous for the mutated allele had signs, findings that were consistent with an autosomal recessive mode of inheritance and complete clinical penetrance.
This study and previous studies26,27 have identified a total of 25 patients from 10 families in five countries, with five different alleles. Four different clinical phenotypes have now been reported in patients with autosomal recessive CARD9 deficiency. The seven related Iranian patients and the Dutch patient had candida infection of the central nervous system,26,27 chronic mucocutaneous candidiasis, and superficial dermatophytosis.26 We now show that CARD9 deficiency is a potential genetic cause of deep dermatophytosis, since all patients with idiopathic deep dermatophytosis studied to date have been shown to carry biallelic, rare, deleterious mutations in CARD9. Therefore, it is possible that CARD9 mutations will be identified in other patients with deep dermatophytosis.1,4,12,31,32
Dermatophytes usually infect keratinized tissues. There are different types of so-called dermatophyte-related invasive infections. Majocchi’s granuloma is a limited perifollicular granuloma.33,34 Its histologic characteristics are perifollicular granulomatous inflammation with dermal abscesses and dermatophyte hyphae. In contrast, deep dermatophytosis refers to dermal invasions that are not localized and in which the granulomatous reaction extends beyond the perifollicular area.35-38
In all the patients we have described, idiopathic deep dermatophytosis was diagnosed on clinical, histologic, and mycologic grounds according to the 2008 European Organization for Research and Treatment of Cancer–Mycoses Study Group consensus group definition (see the Supplementary Appendix, section 1). However, kindreds with CARD9 deficiency had phenotypic variability in dermatophytic infection, as already described.26 The clinical signs of deep dermatophytosis in the patients described here began in childhood, with recurrent and severe tinea and onychomycosis, and worsened during adolescence, leading to invasive disease. Survival was poor in these patients — 4 of the 17 patients we studied died between the ages of 28 and 39 years with clinically active deep dermatophytosis, and a fifth patient died at 91 years of age. However, no other severe infections were reported; in particular, there were no infections due to mycobacterium species or listeria, in contrast to reports of infections in CARD9-deficient mice.39,40
Twenty-five patients bearing biallelic deleterious CARD9 alleles have now been reported, and these patients had various fungal infections, including superficial and invasive fungal diseases. These findings highlight the role of CARD9 in the human immune responses controlling fungal infection. The molecular and cellular bases of susceptibility to fungal disease, including deep dermatophytosis in particular, remain unclear. A defect in macrophages, dendritic cells, or keratinocytes might account for the invasion of the dermis by dermatophytes in CARD9-deficient patients, since the Q289X mutation leads to a loss of expression and the R101C mutation leads to lower levels of expression in the patients’ monocyte-derived dendritic cells.
CARD9 is an adaptor in the signaling pathway downstream from dectin-1, dectin-2, macrophage-inducible C-type lectin, and probably other as yet unknown receptors involved in antifungal immunity.40-47 However, it remains unclear which receptors are actually involved in immunity to dermatophytes. The diverse clinical presentations of CARD9 deficiency, ranging from dermatophytosis to candida meningitis, suggest that multiple molecular pathways in multiple cell types are controlled by CARD9. The patients tested had impaired interleukin-6 production in response to whole-blood stimulation with fungal ligands (heat-killed C. albicans and S. cerevisiae), which indicates that the CARD9 alleles are deleterious for at least some cellular responses that may underlie deep dermatophytosis. Patients with CARD9 deficiency have also been reported to have low proportions of interleukin-17 T cells.26,27 We found that patients had significantly lower proportions of interleukin-17 T cells than normal. However, this may not necessarily be a consequence of CARD9 deficiency, since deep dermatophytosis may result in the trapping of interleukin-17–producing T cells in skin lesions.
Additional studies are required to characterize the CARD9-dependent pathways in both myeloid and lymphoid cells in humans, as well as the other genes responsible for controlling host defense against candida species and dermatophytes. In any case, deep dermatophytosis has been shown to be associated with biallelic, rare, deleterious CARD9 mutations in all kindreds tested to date, providing yet another example of a life-threatening infectious disease associated with single-gene inborn errors of immunity.48,49
Supplementary Material
Acknowledgments
Supported by grants from l’Agence Nationale pour la Recherche (ANR GENCMCD 11-BSV3-005-01, to Dr. Puel); the European Commission Sixth and Seventh Framework Programs (EURO-GENE-SCAN 223293 and EURO-PADnet HEALTH-F2-2008-201549); the Marie Curie Excellence program (MEXT-CT-2006-042316); the German Federal Ministry of Education and Research (BMBF 01 EO 0308); the National Center for Research Resources and the National Center for Advancing Translational Sciences (8UL1TR00004), National Institutes of Health; Rockefeller University; INSERM, Paris Descartes University; the St. Giles Foundation; the French Government Investissement d’Avenir program, Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases (ANR-10-LABX-62-IBEID); and the CMIT (French Faculties College of Infectious Diseases) and INSERM (both to Dr. Lanternier). Janssen-Cilag France provided itraconazole for compassionate use in the treatment of some patients, thanks to the intervention of Dr. Bessis.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
We thank the patients and their families; Aziz Belkadi for assistance with exome data analysis; Drs. Antignac, Helal, Ben Chibani, and Ben Rayana for providing control DNA; Drs. Glocker and Birmelin for technical advice; Dr. Lahfa for assistance with the collection of patients’ samples; and Dr. Roth (Xceltis) for the anti-dermatophyte monoclonal antibody.
Footnotes
The authors’ affiliations are listed in the Appendix.
References
- 1.Hadida E, Schousboe A. Dermatophytic disease aspects. Alger Med. 1959;63:303–36. [Google Scholar]
- 2.Seebacher C, Bouchara JP, Mignon B. Updates on the epidemiology of dermatophyte infections. Mycopathologia. 2008;166:335–52. doi: 10.1007/s11046-008-9100-9. [DOI] [PubMed] [Google Scholar]
- 3.Marconi VC, Kradin R, Marty FM, Hospenthal DR, Kotton CN. Disseminated dermatophytosis in a patient with hereditary hemochromatosis and hepatic cirrhosis: case report and review of the literature. Med Mycol. 2010;48:518–27. doi: 10.3109/13693780903213512. [DOI] [PubMed] [Google Scholar]
- 4.Cheikhrouhou F, Makni F, Masmoudi A, Sellami A, Turki H, Ayadi A. A fatal case of dermatomycoses with retropharyngeal abscess. Ann Dermatol Venereol. 2010;137:208–11. doi: 10.1016/j.annder.2010.01.006. (In French.) [DOI] [PubMed] [Google Scholar]
- 5.Liautaud B, Marill FG. Dermatophytic disease: recent Algerian observations. Bull Soc Pathol Exot Filiales. 1984;77:637–48. (In French.) [PubMed] [Google Scholar]
- 6.Boudghène-Stambouli O, Mérad-Boudia A. Dermatophytic disease in Algeria: a new case and review of the literature. Ann Dermatol Venereol. 1991;118:17–21. (In French.) [PubMed] [Google Scholar]
- 7.Antifungal agents in dermatophytic disease: failure of griseofulvin, ketoconazole and itraconazole. Bull Soc Pathol Exot. 1990;83:170–6. Idem. (In French.) [PubMed] [Google Scholar]
- 8.Boudghène-Stambouli O, Mérad-Boudia A, Allal M. Cerebral injury in der matophytic disease. J Mycol Med. 1992;2:106–8. [Google Scholar]
- 9.Boudghène-Stambouli O, Mérad-Boudia A. Trichophyton rubrum dermatophytic disease: a new case. Ann Dermatol Venereol. 1989;116:725–7. (In French.) [PubMed] [Google Scholar]
- 10.Pruszkowski A, Bourgault Villada I, Cremer G, Ammar-Khodja A, Emilie D, Revuz J. Dermatophytic disease: role of type TC2 CD8 lymphocytes. Ann Dermatol Venereol. 1995;122(Suppl 1):55. [Google Scholar]
- 11.Boudghène-Stambouli O, Mérad-Boudia A. Dermatophytic disease: exuberant hyperkeratosis with cutaneous horns. Ann Dermatol Venereol. 1998;125:705–7. (In French.) [PubMed] [Google Scholar]
- 12.Beirana L, Novales J. Tinea universal y granulomatosa por T. tonsurans. Dermatol Rev Mex. 1959;3:4–16. [Google Scholar]
- 13.Oliveira H, Trincao R, Leitao A. Tricofitose cutanea generalizada com infeccao sistemica por T. violaceum. J Med (Porto) 1960;41:626–42. [Google Scholar]
- 14.Pelevine A, Tchernogouboff N. Mucosal, lymph node and bone dermatophytosis. Ann Derm Syph. 1927;8:403–24. [Google Scholar]
- 15.Sequeira JH. A case of granuloma trichophyticum. Br J Dermatol. 1912;207 [Google Scholar]
- 16.Hironaga M, Okazaki N, Saito K, Watanabe S. Trichophyton mentagrophytes granulomas: unique systemic dissemination to lymph nodes, testes, vertebrae, and brain. Arch Dermatol. 1983;119:482–90. doi: 10.1001/archderm.119.6.482. [DOI] [PubMed] [Google Scholar]
- 17.Araviysky AN, Araviysky RA, Eschkov GA. Deep generalized trichophytosis (endothrix in tissues of different origin) Mycopathologia. 1975;56:47–65. doi: 10.1007/BF00493584. [DOI] [PubMed] [Google Scholar]
- 18.Tejasvi T, Sharma VK, Sethuraman G, Singh MK, Xess I. Invasive dermatophytosis with lymph node involvement in an immunocompetent patient. Clin Exp Dermatol. 2005;30:506–8. doi: 10.1111/j.1365-2230.2005.01839.x. [DOI] [PubMed] [Google Scholar]
- 19.Vinh DC. Insights into human anti-fungal immunity from primary immunodeficiencies. Lancet Infect Dis. 2011;11:780–92. doi: 10.1016/S1473-3099(11)70217-1. [DOI] [PubMed] [Google Scholar]
- 20.Puel A, Cypowyj S, Bustamante J, et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 2011;332:65–8. doi: 10.1126/science.1200439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu L, Okada S, Kong XF, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med. 2011;208:1635–48. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van de Veerdonk FL, Plantinga TS, Hoischen A, et al. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med. 2011;365:54–61. doi: 10.1056/NEJMoa1100102. [DOI] [PubMed] [Google Scholar]
- 23.Puel A, Picard C, Cypowyj S, Lilic D, Abel L, Casanova JL. Inborn errors of mucocutaneous immunity to Candida albicans in humans: a role for IL-17 cytokines? Curr Opin Immunol. 2010;22:467–74. doi: 10.1016/j.coi.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puel A, Döffinger R, Natividad A, et al. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. J Exp Med. 2010;207:291–7. doi: 10.1084/jem.20091983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kisand K, Bøe Wolff AS, Podkrajsek KT, et al. Chronic mucocutaneous candidiasis in APECED or thymoma patients correlates with autoimmunity to Th17-associated cytokines. J Exp Med. 2010;207:299–308. doi: 10.1084/jem.20091669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glocker EO, Hennigs A, Nabavi M, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 2009;361:1727–35. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drewniak A, Gazendam RP, Tool AT, et al. Invasive fungal infection and impaired neutrophil killing in human CARD9 deficiency. Blood. 2013;121:2385–92. doi: 10.1182/blood-2012-08-450551. [DOI] [PubMed] [Google Scholar]
- 28.Filipe-Santos O, Bustamante J, Haverkamp MH, et al. X-linked susceptibility to mycobacteria is caused by mutations in NEMO impairing CD40-dependent IL-12 production. J Exp Med. 2006;203:1745–59. doi: 10.1084/jem.20060085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Beaucoudrey L, Puel A, Filipe-Santos O, et al. Mutations in STAT3 and IL12RB1 impair the development of human IL-17-producing T cells. J Exp Med. 2008;205:1543–50. doi: 10.1084/jem.20080321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blank F, Schopflocher P, Poirier P, Riopelle JL. Extensive Trichophyton infections of about fifty years’ duration in two sisters. Dermatologica. 1957;115:40–51. doi: 10.1159/000255985. [DOI] [PubMed] [Google Scholar]
- 32.Destombes P, Liautaud B, Marill FG. Histopathological study on the course of a dermatophytic disease. Bull Soc Pathol Exot Filiales. 1975;68:443–9. (In French.) [PubMed] [Google Scholar]
- 33.Majocchi D. Sepra una nuova tricofizia (granuloma tricofitio) studi clinici micologici. Bull R Acad Med Roma. 1883;9:220–3. [Google Scholar]
- 34.Ilkit M, Durdu M, Karakaş M. Majocchi’s granuloma: a symptom complex caused by fungal pathogens. Med Mycol. 2012;50:449–57. doi: 10.3109/13693786.2012.669503. [DOI] [PubMed] [Google Scholar]
- 35.Chastain MA, Reed RJ, Pankey GA. Deep dermatophytosis: report of 2 cases and review of the literature. Cutis. 2001;67:457–62. [PubMed] [Google Scholar]
- 36.Nir-Paz R, Elinav H, Pierard GE, et al. Deep infection by Trichophyton rubrum in an immunocompromised patient. J Clin Microbiol. 2003;41:5298–301. doi: 10.1128/JCM.41.11.5298-5301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demidovich CW, Kornfeld BW, Gentry RH, Fitzpatrick JE. Deep dermatophyte infection with chronic draining nodules in an immunocompromised patient. Cutis. 1995;55:237–40. [PubMed] [Google Scholar]
- 38.Smith KJ, Welsh M, Skelton H. Trichophyton rubrum showing deep dermal invasion directly from the epidermis in immunosuppressed patients. Br J Dermatol. 2001;145:344–8. doi: 10.1046/j.1365-2133.2001.04331.x. [DOI] [PubMed] [Google Scholar]
- 39.Dorhoi A, Desel C, Yeremeev V, et al. The adaptor molecule CARD9 is essential for tuberculosis control. J Exp Med. 2010;207:777–92. doi: 10.1084/jem.20090067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu YM, Zhang Y, You Y, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol. 2007;8:198–205. doi: 10.1038/ni1426. [DOI] [PubMed] [Google Scholar]
- 41.Drummond RA, Saijo S, Iwakura Y, Brown GD. The role of Syk/CARD9 coupled C-type lectins in antifungal immunity. Eur J Immunol. 2011;41:276–81. doi: 10.1002/eji.201041252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertin J, Guo Y, Wang L, et al. CARD9 is a novel caspase recruitment domain-containing protein that interacts with BCL10/CLAP and activates NF-kappa B. J Biol Chem. 2000;275:41082–6. doi: 10.1074/jbc.C000726200. [DOI] [PubMed] [Google Scholar]
- 43.Gross O, Gewies A, Finger K, et al. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature. 2006;442:651–6. doi: 10.1038/nature04926. [DOI] [PubMed] [Google Scholar]
- 44.Hara H, Ishihara C, Takeuchi A, et al. The adaptor protein CARD9 is essential for the activation of myeloid cells through ITAM-associated and Toll-like receptors. Nat Immunol. 2007;8:619–29. doi: 10.1038/ni1466. [DOI] [PubMed] [Google Scholar]
- 45.Hara H, Ishihara C, Takeuchi A, et al. Cell type-specific regulation of ITAM-mediated NF-kappaB activation by the adaptors, CARMA1 and CARD9. J Immunol. 2008;181:918–30. doi: 10.4049/jimmunol.181.2.918. [DOI] [PubMed] [Google Scholar]
- 46.Hara H, Saito T. CARD9 versus CARMA1 in innate and adaptive immunity. Trends Immunol. 2009;30:234–42. doi: 10.1016/j.it.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 47.LeibundGut-Landmann S, Gross O, Robinson MJ, et al. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007;8:630–8. doi: 10.1038/ni1460. [DOI] [PubMed] [Google Scholar]
- 48.Casanova JL, Abel L. The genetic theory of infectious diseases: a brief history and selected illustrations. Annu Rev Genomics Hum Genet. 2013;14:215–43. doi: 10.1146/annurev-genom-091212-153448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alcaïs A, Quintana-Murci L, Thaler DS, Schurr E, Abel L, Casanova JL. Life-threatening infectious diseases of childhood: single-gene inborn errors of immunity? Ann N Y Acad Sci. 2010;1214:18–33. doi: 10.1111/j.1749-6632.2010.05834.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.