Abstract
The centromere is the chromosomal region that directs kinetochore assembly during mitosis in order to facilitate the faithful segregation of sister chromatids. The location of the human centromere is epigenetically specified. The presence of nucleosomes that contain the histone H3 variant, CENP-A, are thought to be the epigenetic mark that indicates active centromeres. Maintenance of centromeric identity requires the deposition of new CENP-A nucleosomes with each cell cycle. During S-phase, existing CENP-A nucleosomes are divided among the daughter chromosomes, while new CENP-A nucleosomes are deposited during early G1. The specific assembly of CENP-A nucleosomes at centromeres requires the Mis18 complex, which recruits the CENP-A assembly factor, HJURP. We will review the unique features of centromeric chromatin as well as the mechanism of CENP-A nucleosome deposition. We will also highlight a few recent discoveries that begin to elucidate the factors that temporally and spatially control CENP-A deposition.
Keywords: CENP-A, Centromere, Nucleosome, Chromatin, Epigenetics, Chromosome
Accurate chromosome segregation is controlled by the centromere. The centromere is the chromosomal domain that directs kinetochore assembly thereby coupling microtubule-pulling forces to each chromosome. In most eukaryotes, centromeres exist as a single locus on each chromosome, and a chromosome lacking a centromere will fail to segregate properly. Segregation errors lead to aneuploidy, which in turn causes cellular stress and greater genomic instability [1]. On the other hand, a chromosome with too many centromeres leads to chromosome breakage when one chromatid is attached to opposite spindle poles during mitosis and is torn apart. Such breakages can lead to breakage-fusion-bridge (BFB) cycles described originally nearly 71 years ago by Barbara McClintock [2]. BFB cycles may play a critical role in the creation of complex chromosome rearrangements often observed in cancers [3]. Therefore, a cell must dedicate and maintain a single contiguous locus as the centromere among the millions of possible base pairs present on each chromosome. As we will describe in the following review, this process requires the involvement of a large, multi-protein centromere complex, which is directed by the cell cycle-controlled assembly of centromere-specific nucleosomes, as well as chromatin remodeling and modifying activities, and the destabilization of centromeric nucleosomes at non-centromeric loci.
The evolutionarily conserved mark of centromeres is the presence of a unique nucleosome in which canonical histone H3 is replaced by CENP-A (Cse4 in budding yeast, Cnp1 in fission yeast, and CID/CenH3 in fruit flies). CENP-A is absolutely essential for viability in all organisms tested [4, 5]. While the presence of a centromere-specific nucleosome is conserved throughout eukaryotic evolution, several features of the centromere including its organization, underlying DNA sequence, and mechanism of assembly are quite divergent. The budding yeast, S. cerevisiae, and a number of its relatives, determine the location of their centromeres through a specific DNA sequence, a so-called “point centromere” that consists of 125 base pairs that position the centromere-specific nucleosome [6, 7]. Fission yeast, S. pombe, and higher eukaryotes have more expansive genomic regions assigned as centromeres and employ epigenetic mechanisms to specify centromeric location at a single site on each chromosome. In contrast to both of these cases, the nematode, C. elegans, assembles centromeres along the entire length of the chromosome instead of at a single site [8]. Despite these differences, all known centromeres depend on the presence of the centromere-specific nucleosome that contains the H3 variant, CENP-A. Only recently have we begun to appreciate that as dissimilar as point and regional centromeres are, many of the proteins and mechanisms involved in CENP-A deposition are highly conserved [9].
The epigenetic specification of the centromere
The heritability of experimentally induced and naturally occurring neocentromeres and pseudodicentric chromosomes is the best evidence that the site of centromere formation and maintenance is epigenetically determined. There are numerous examples in humans where an initially non-centromeric locus, outside of the endogenous alpha-satellite region, becomes an active centromere. These regions are called “neocentromeres” and they arise stochastically, at very low frequency, and without rearrangements of the underlying DNA [10]. Pseudodicentric chromosomes can occur through DNA translocations or inverted duplication and result in a single chromosome with two alpha-satellite centromere-containing regions. In psuedodicentrics, only one of the two alpha-satellite regions remains active. Neocentromeres and active centromeres of pseudodicentric chromosomes recruit CENP-A and all other centromere proteins that have been tested, except the sequence-specific binding protein, CENP-B [11–14]. The epigenetic inheritance of regional centromeres has been experimentally demonstrated in fission yeast and flies through the generation of stable neocentromeres on chromosomal fragments that lack an original centromeric locus [15, 16]. These data suggest that DNA is neither necessary nor sufficient for centromere specification, but that the proteins associated with the centromere are the determinants of centromere identity.
Neocentromeres have been observed on almost every chromosome but appear to cluster around certain regions within a given chromosome [10]. The non-random distribution of neocentromeres across the human genome suggests not all sites have an equal potential to support centromere activity. This may be due either to unknown DNA sequences that are refractory to centromere formation or to chromatin states that may be more or less favorable for stable centromere formation. Such plasticity in centromere location means that not only can CENP-A nucleosomes be deposited in a variety of chromosomal domains, but the machinery that deposits CENP-A nucleosomes into chromatin must also be able to function at these various sites.
The CENP-A nucleosome
The CENP-A nucleosome is sufficient to specify the site of centromere formation and distinguish it as the location for kinetochore assembly during mitosis [17, 18]. The centromere is occupied throughout the cell cycle by a large multi-subunit complex of proteins termed the CCAN (constitutive centromere-associated network) comprised of 16 centromere proteins (CENPs C, H, I, K through U(50), W, and X) [19–21]. It is the CCAN which is thought to mediate the assembly of kinetochore structure in mitosis. The CCAN assembles only at centromeres and therefore distinguishes CENP-A nucleosomes from the H3-containing nucleosomes found in general chromatin. The overall structure of the CENP-A nucleosome as well as particular sequences within have been proposed to be the defining features that mediate CENP-A chromatin-specific CCAN recruitment.
Several provocative forms of the CENP-A nucleosome, other than the canonical octamer, have been proposed in an attempt to describe the uniqueness of the CENP-A nucleosome. The different proposed forms include a heterotetrameric form containing a single copy of each histone (CENP-A, histone H4, H2A and H2B) in flies and humans, as well as a hexameric form in yeast that excludes H2A and H2B but contains the chaperone Scm3 [22–24]. These and others have been extensively discussed in a previous review [25]. Recently, the crystal structure of the human CENP-A nucleosome was solved, revealing an octameric nucleosome that wraps DNA in a left-handed manner similar to the H3-containing nucleosome [26]. Additional evidence for an octameric nucleosome structure comes from in-depth mutational studies of CENP-A. Mutations that disrupt the CENP-A–CENP-A interface in humans and flies preclude the stable incorporation of CENP-A into chromatin (Fig. 1) [27, 28]. While these data do not rule out the possibility that CENP-A nucleosomes exist in multiple forms, it appears that the formation of an octameric structure is possible and important for initial stable CENP-A incorporation.
Fig. 1.

a Primary sequences of human CENP-A and H3.1 are compared at single amino acid resolution. Dashes have been added at the relative position for H3.1. Known posttranslational modifications are mapped onto the sequence of both histones. Serine 7 phosphorylation by Aurora B in human cells is the only known post-translational modification on CENP-A. However, the serine at position seven is not a well-conserved feature of CENP-A even within vertebrates. Binding sites of HJURP, CENP-N, and CENP-C are highlighted on the CENP-A sequence, as well as the CENP-A-CENP-A dimerization domain (labeled nucleosome self-association). b Space-filling models of the CENP-A nucleosome as well as a CENP-A–CENP-A dimer from different perspectives (PDB ID: 3AN2). Highlighted amino acids are shaded to match the colors of the binding sites mapped in a. Two additional, non-conserved residues (R80, G81) in CENP-A constitute a bulge relative to H3
The overall protein structure of the CENP-A nucleosome is very similar to the histone H3-containing nucleosome; however, there are several features of the CENP-A nucleosome that distinguish it from the canonical H3 nucleosome, which are highlighted in the schematic of human CENP-A in Fig. 1a. The CENP-A targeting domain (CATD) consists of the unique residues within the loop 1 and alpha-2 helix of the CENP-A histone fold. The CATD is sufficient for centromere localization and confers a unique rigidity to the CENP-A nucleosome that may be a defining characteristic [29, 30]. Structural studies have identified two other regions of the human CENP-A nucleosome that stand out relative to canonical nucleosomes. The most N-terminal helix of CENP-A, the alpha-N helix, contains three fewer residues and is therefore approximately one helical turn shorter than the comparable helix in histone H3 [26]. This region of the nucleosome is interesting because it is also the DNA entry/exit site. Consistent with this is the observation by several groups that CENP-A nucleosomes protect a smaller fragment of DNA in nuclease assays due to a partial unwrapping of the DNA at the entry/exit sites [26, 31, 32]. Correlative data from hydrogen–deuterium exchange mass spectrometry experiments measuring protein dynamics show that CENP-A exchanges protons tenfold faster than H3 at the α-N helix, even in extended nucleosome arrays [31].
The second important region of distinction is loop 1 of the human CENP-A histone fold, which was observed to form a surface-accessible bulge using Arg80/Gly81 in the CENP-A heterotetramer and in the CENP-A nucleosome [26, 33]. The extra two amino acids that form the bulge are a conserved feature of CENP-A homologs across all species (although S. pombe contains an even larger expansion); however, conservation of the positively charged arginine is restricted to mammals and birds. This bulge is accessible on the surface of the CENP-A nucleosome and could therefore serve as a recognition motif (Fig. 1b). The surface bulge is not essential for centromeric targeting but it is required for stable incorporation of the CENP-A nucleosome. Coexpression of wild-type and CENP-AΔR80G81 confirmed that the mutant protein initially localized to centromeres. After 3 days, the number of cells with the mutant protein localized to centromeres decreased; however, those cells were still able to recruit wild-type CENP-A [26].
Two distinct components of the CCAN have been shown to “read” the unique structure of the CENP-A nucleosome. CENP-C and CENP-N both interact with the CENP-A nucleosome, but in different ways. CENP-C is recruited to centromeres via an interaction with the extreme carboxyl terminus of CENP-A (Fig. 1) [34, 35]. Replacement of the extreme C-terminus of histone H3 with the last six amino acids of CENP-A is sufficient to recruit CENP-C in vitro and in Xenopus extracts [34, 35]. However, the primary sequence of the CENP-A carboxyl terminus is not conserved between yeast, flies, zebrafish and humans. In addition, a chimeric histone H3 containing the CATD (H3CATD), which lacks the C-terminus of CENP-A, was sufficient to recruit CENP-C to centromeres at endogenous levels in human cells [36]. Therefore, it remains to be demonstrated whether recruitment of CENP-C through the C-terminus of CENP-A is a conserved method of CENP-A recognition. The interaction of CENP-N with CENP-A is through the CATD of CENP-A [37]. CENP-N selectively interacts with CENP-A in its nucleosomal form, suggesting that it recognizes a structural aspect of CENP-A that is only found in the intact nucleosome [34, 35, 37]. Both CENP-C and CENP-N appear to prefer nucleosomal CENP-A in vivo as well, as neither of these two proteins is found in prenucleosomal CENP-A fractions [20, 38].
Centro-chromatin: epigenetic context of CENP-A
Centromeres contain both CENP-A and histone H3 nucleosomes arranged in interspersed blocks [39–41]. Recently, another centromeric, chromatin-associated complex has been proposed that includes members of the CCAN, the CENP-T/W/S and X complex [42]. Each of these proteins contains a histone fold domain. Histone folds are not only found in histones, but are also found in several transcription factor complexes [43]. The members of the CENP-T/W/S/X complex use their histone folds to form a heterotetramer which has similarities to the structures of transcription factor complexes as well as the histone H3-H4 heterotetramer. Mutations in any of the tetramerization domains in this complex results in failed mitoses in vivo, suggesting that this complex is absolutely required for kinetochore formation in chickens and humans [42]. The CENP-T/W/S/X complex binds and protects 100 base pairs of DNA from nuclease digestion in vitro. While the data suggests that CENP-T/W/S/X may form a nucleosome-like structure at centromeres; CENP-T/W/S/X may simply bind to centromeric DNA, albeit in a discrete complex.
The stability of the CENP-T/W/S/X complex is very different from that of CENP-A, which is stable throughout the cell cycle and is completely retained through S-phase. Localization of CENP-T and -W occurs during late S-phase or G2 [44, 45]. The CENP-T/W dimer does not remain stably bound to centromeres, but is instead completely replenished upon each new cell cycle [45]. The CENP-T/W/S/X complex appears to be interspersed between CENP-A domains in stretched chromatin fibers, and immunoprecipitations of the complex from MNase treated extracts pull down histone H3 [44, 46]. This suggests that the CENP-T/W/S/X complex may couple the kinetochore to the H3-containing domains of centromeric chromatin [44, 46].
A consensus of immunofluorescence microscopy data on stretched interphase chromatin shows that centromeres containing alternating stretches of H3 and CENP-A-containing nucleosomes is a quality conserved from flies to humans [47]. The amino-terminal tails of the interspersed histone H3 stretches are enriched for dimethylation on Lys4, Lys9 and Lys36 (H3K4me2, H3K9me2 and H3K36me2) (Fig. 2) [39, 48]. The pattern of centromeric histone post-translational modifications is different from that of general chromatin as well as pericentric heterochromatin and does not adhere to the characteristic “activating” or “silencing” patterns. The perturbation of histone marks within the centromeres of human artificial chromosomes (HAC) results in a loss of HAC stability, loss of centromere-specific proteins and an inhibition of the CENP-A deposition pathway [48–51]. This suggests that the unique combination of histone modifications present in centromeric chromatin may be important for centromere function and propagation.
Fig. 2.
Centromere and pericentromere chromatin organization depicted in interphase (1D model: left) and in mitosis (2D model: right). Histone H3 and CENP-A post-translational modifications are notated per cell cycle position, with black for constitutive modifications and red for mitosis-specific posttranslational modifications. In the 2D model, background shading distinguishes the organization of the centromere: pericentromere (dark grey), inner centromere (light grey), and outer centromere (pink, green/red)
During mitosis the higher-order organization of the centromere is speculated to resemble a cylinder or a multi-layered boustrophedon [40, 46]. CENP-A occupies 10 % of the DNA at the primary constriction, in a condensed space at the distal, kinetochore-facing aspect of centromeres [52]. A self-organization model has been proposed to generate this three-dimensional centromeric chromatin structure. In such a model centromeric chromatin folds into a specific three-dimensional structure that facilitates kinetochore formation above the exterior CENP-A clusters while the interspersed H3 regions are excluded as a looped or coiled structure to form the inner centromere below. Consistent with this idea, the N-terminal tail of H3 nucleosomes has been shown to dictate the three-dimensional folding of polynucleosome arrays [53]. CENP-A and H3 N-terminal tails are vastly divergent (Fig. 1a). In vitro folding experiments show that arrays of CENP-A nucleosomes were found to fold into more condensed, higher-ordered structures than H3 nucleosome arrays [31]. The differences in histone posttranslational modifications (PTMs) found in the inner and outer centromere and pericentric regions may influence this property. Therefore, histone variant-specific protein–protein interactions may confer a way for three-dimensional folding instructions to be laid out in the two-dimensional organization of the centromere.
The CCAN: coupling chromosomes to the kinetochore
The CCAN forms a bridge between the centromeric chromatin and the mitotic kinetochore. As discussed above, CENP-C contacts the CENP-A nucleosomes and CENP-T is embedded into the centromeric chromatin. Both CENP-C and -T also interact with proteins of the kinetochore. A major microtubule binding complex of the kinetochore is the KMN network which consists of KNL-1, the Mis12 complex, and the Ndc80 complex. CENP-C recruits the Mis12 complex through its N-terminus [54–56]. The amino terminus of CENP-T extends beyond the centromeric chromatin to interact with the Ndc80 complex and is subject to stretching forces during mitosis [57–59]. A third CCAN/KMN network interaction point involves a complex of CENP-H, -I, and -K which appears to become recruited distally to CENP-C and -N and participate in kinetochore formation through direct binding to KNL-1 [60]. When CENP-T and CENP-C are targeted to a non-centromeric locus using the LacO/LacI system, they are sufficient to assemble a functional kinetochore, including mitotic checkpoint signaling; however, tethering of CENP-C and -T is not sufficient to recruit CENP-A [58].
Although the KMN network can directly bind microtubules through KNL-1 and through the Ndc80 complex, the CCAN may also play a more direct and dynamic role in microtubule binding. During typical kinetochore oscillations, CENP-H/I show dynamic enrichment at kinetochores coupled to growing versus shrinking microtubules [61]. In addition, a complex involving CENP-O (Mcm21R), -P, -Q, and -U (CENP-50) appears to play a role in regulating the quality of microtubule attachment to kinetochores and is essential for recovery from experimentally induced spindle damage in vivo [62]. Human CENP-Q can directly bind microtubules in vitro, so it is speculated to serve as the microtubule-binding component in the CENP-O sub-complex [61]. Phosphorylation of CENP-U by Aurora B is required for spindle damage recovery [62]. Because CENP-U is regulated by Aurora B, the role of the CENP-O sub-complex, and thereby the entire CCAN is not simply binding kinetochore components, but rather playing a more active role in generating correctly formed kinetochore-microtubule attachments.
The CENP-A deposition pathway
In order for centromeres to be stably inherited through many generations, new CENP-A nucleosomes must be assembled specifically at the site of the pre-existing centromere after each round of DNA replication. The CENP-A deposition pathway can be broken down into three basic stages that involve distinct protein complexes: initiation, deposition, and maintenance. At the correct moment in the cell cycle, the location of the centromere must be sensed and the underlying chromatin must be modified to, or maintained in, a permissive state for CENP-A deposition. Once this occurs, CENP-A-specific assembly factors associate with the centromere and allow for CENP-A deposition. Finally, through a chromatin remodeling and a maturation process, centromeric chromatin is fully stabilized (Fig. 3).
Fig. 3.
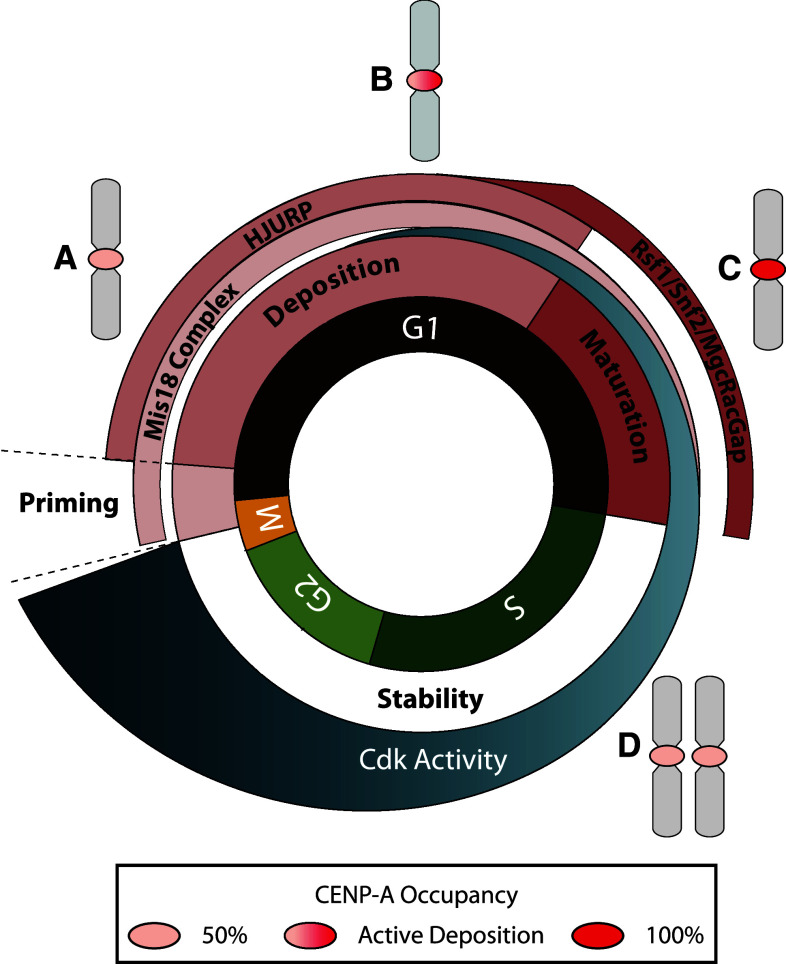
Overview of the cell cycle control mechanism of CENP-A deposition. The deposition of CENP-A is tightly regulated by the cell cycle. Chromosome schematics show the stepwise change in CENP-A protein levels at the centromere. a Starting at the exit from mitosis, each daughter centromere possesses one-half of the full complement of CENP-A nucleosomes (light pink oval). Cyclins are rapidly degraded following mitotic exit, and Cdk activity dramatically drops (dark blue gradient). Mis18BP1KNL2 can interact with centromeric chromatin in its unphosphorylated state and primes the centromere for CENP-A deposition. b Once Mis18BP1KNL2 associates with centromeres, HJURP is recruited and deposits newly synthesized CENP-A nucleosomes (light pink to red gradient oval). c In mid-to-late G1, the RSF complex (Rsf1-Snf2h) and MgcRacGap interact transiently with centromeres to stabilize newly assembly CENP-A nucleosomes and generate mature centromeric chromatin (red oval). By the S-phase transition, Cdk activity levels have increased above a threshold and are now unfavorable for Mis18 complex association with centromeres. Mis18BP1KNL2 is phosphorylated, releases from chromatin, and CENP-A deposition is inhibited. d During S-phase, CENP-A nucleosomes are parceled to each daughter centromere (two light pink ovals)
Initiation of CENP-A deposition: key players
In human cells, the deposition of CENP-A occurs during G1, after cells exit mitosis [63]. Therefore, initiation must occur prior to CENP-A loading. The earliest recognized step in the recruitment of new CENP-A nucleosomes is the association of the Mis18 complex, which localizes to centromeres in late anaphase, directly after mitotic exit [64]. Mis18 was first discovered in fission yeast by a temperature-sensitive screen for mutants that missegregated their chromosomes [65]. Along with Mis16 mutants (homolog of the human proteins RbAp48 and RbAp46), the Mis18 temperature-sensitive mutants resulted in the missegregation of chromosomes during mitosis due to a reduction of centromeric Cnp1, the CENP-A homolog in S. pombe [65]. This indicated that Mis16 and Mis18 were both required for the deposition of Cnp1-containing nucleosomes at the fission yeast centromere.
Through sequence homology searches, two human homologs of the Mis18 protein were discovered: Mis18α and Mis18β [64]. Pull downs of both Mis18α and Mis18β from chromatin fractions confirmed a physical interaction between the two Mis18 proteins as well as with the human homologs of Mis16, RbAp48 and RbAp46 [64]. In addition, an uncharacterized protein, termed Mis18 Binding Protein 1 (Mis18BP1KNL2), was also found to interact with chromatin-associated Mis18α and Mis18β [64]. Concurrently, an RNAi screen in C. elegans discovered a homolog of Mis18BP1KNL2 (named KNL-2) that was also found to be required for CENP-A centromeric localization; thus providing further evidence of the conserved nature of these proteins [66]. RNAi knockdown experiments in human cells showed Mis18α, Mis18β, and Mis18BP1KNL2 were dependent upon each other for localizing to the centromere [64]. In addition, knockdown of all three proteins as well as RbAp48/46 confirmed that the entire complex was required for the deposition of newly synthesized CENP-A at the centromere [64, 65]. In summary, the human Mis18 complex was found to consist of Mis18α, Mis18β, Mis18BP1KNL2, RbAp48 and RbAp46, which were all required to deposit new CENP-A at centromeres. However, no physical interaction between Mis18 and CENP-A has been found to date.
The Holliday junction recognition protein, HJURP, was found to specifically interact with CENP-A/H4 in its prenucleosomal form [38, 67, 68]. In human cells, HJURP is necessary and sufficient for the deposition of newly synthesized CENP-A nucleosomes in vivo and is able to assemble CENP-A nucleosomes onto plasmid DNA in vitro suggesting that HJURP is the assembly factor for CENP-A [17, 38, 68]. Cell cycle analysis of HJURP localization shows that HJURP is localized to centromeres in G1, which follows the late anaphase localization of the human Mis18 complex [38, 68]. Indeed, human Mis18α and Mis18BP1KNL2 are required for the centromeric localization of HJURP as siRNA knockdowns of either protein abolished centromeric localization of HJURP [17]. Studies in frogs and fission yeast showed the dependency of HJURP on the Mis18 complex was highly conserved [69–71]; however, no physical interaction has been observed between Mis18 and HJURP except in fission yeast.
Initiation of CENP-A deposition: temporal regulation
Since CENP-A deposition only occurs in G1, it is hypothesized that the proteins involved in CENP-A deposition must be regulated by the cell cycle. Progression through the cell cycle is orchestrated by the cyclical accumulation and destruction of the cyclin proteins and their interactions with the various cyclin dependent kinases (CDKs) [72]. Throughout G1 and S-phase, the cyclins accumulate, which results in increased Cdk activity. By the G2/M transition point, Cdk activity levels are maximal, ensuring that the vast majority of Cdk substrates are maintained in a phosphorylated state. Cdk activity levels remain elevated by the spindle assembly checkpoint until all sister chromatids are bi-oriented at the metaphase plate. After the SAC is satisfied, the cyclins are rapidly degraded by the anaphase promoting complex and the cell enters G1, with minimal Cdk activity.
In many organisms, it has been shown that the deposition of newly synthesized CENP-A nucleosomes can occur via a replication-independent process unlike the assembly of canonical H3.1 nucleosomes, which is concurrent with DNA synthesis [73]. Indeed, in human cells, CENP-A is not available for deposition during DNA replication. CENP-A mRNA and protein levels are not maximal until the end of S-phase, after the majority of centromeres have already completed replication [73, 74]. Instead, human cells and D. melanogaster embryos load new CENP-A nucleosomes in G1 only after mitotic exit [63, 75]. In D. melanogaster S2 cells, deposition occurs slightly earlier during mitosis [76]. In an independent genome-wide RNAi screen in D. melanogaster, depletion of cyclin A and Rca1, an inhibitor of the Chd1-APC complex, caused a direct loss of CID at centromeres [77, 78]. Therefore, although the timing between systems differs slightly, data suggest that CENP-A deposition is regulated by cell cycle progression.
As stated above, starting at anaphase onset, the level of Cdk activity drops dramatically, and this dearth of Cdk activity coincides with the deposition of newly synthesized CENP-A [63, 75]. This suggests that there may be a Cdk-controlled mechanism that directly prevents CENP-A deposition in S-phase when Cdk activity is high, but allows for CENP-A assembly into centromeric chromatin after the exit from mitosis, when Cdk activity is low (Fig. 3). Many of the proteins required for the deposition of CENP-A at centromeres share the localization pattern of associating with centromeres after the exit from mitosis when Cdk activity is low [38, 64, 65, 68].
Recently, deposition of CENP-A has been found to be regulated by Cdk1 and Cdk2 activity [79]. While Cdks are highly active in late G1 through G2, Mis18BP1KNL2 is unable to associate with centromeric chromatin. Once Cdk activity sharply declines after anaphase onset, there is a change in Mis18BP1KNL2 phosphorylation status, and the complex is able to associate with the centromere and recruit downstream CENP-A deposition factors [79]. When Cdk1 and Cdk2 are artificially inhibited at other points of the cell cycle, the Mis18 complex and its downstream effectors aberrantly associate with the centromere and deposit newly synthesized CENP-A outside of G1 [79]. This indicates that not only is centromere location spatially regulated by the Mis18 complex, but Cdk phosphorylation of Mis18BP1KNL2 also temporally regulates CENP-A deposition. While the localization of the Mis18 complex is affected by the inhibition of CDKs, it is conceivable that CDK phosphorylation may regulate multiple players in this pathway.
Initiating CENP-A deposition: spatial regulation
CENP-A deposition must be spatially regulated to occur only at the designated centromere locus. This may be achieved by coupling the CENP-A deposition machinery to the constitutive centromere (Fig. 4). Recent experiments have identified a physical interaction between the CCAN protein, CENP-C, and the Mis18 complex [69, 80]. While centromeric localization of CENP-C was shown to be dependent upon the presence of CENP-A nucleosomes, several studies in D. melanogaster cells determined that CENP-C is required for efficient deposition of CENP-A [77, 81, 82]. In egg extracts, Xenopus Mis18BP1KNL2 (M18BP1) was dependent upon CENP-C to localize to metaphase centromeres and thereby initiate CENP-A deposition [69]. Recent studies in mouse cells suggest a similar interaction between Mis18BP1KNL2 and CENP-C [80]. Of the various CCAN proteins assayed, only CENP-C co-localized with Mis18BP1KNL2 when it was targeted to a chromatin domain outside of the endogenous mouse centromeres [80]. As in Xenopus, this colocalization was found to be mediated through a physical interaction with Mis18BP1KNL2 and a C-terminal portion of CENP-C [69, 80].
Fig. 4.
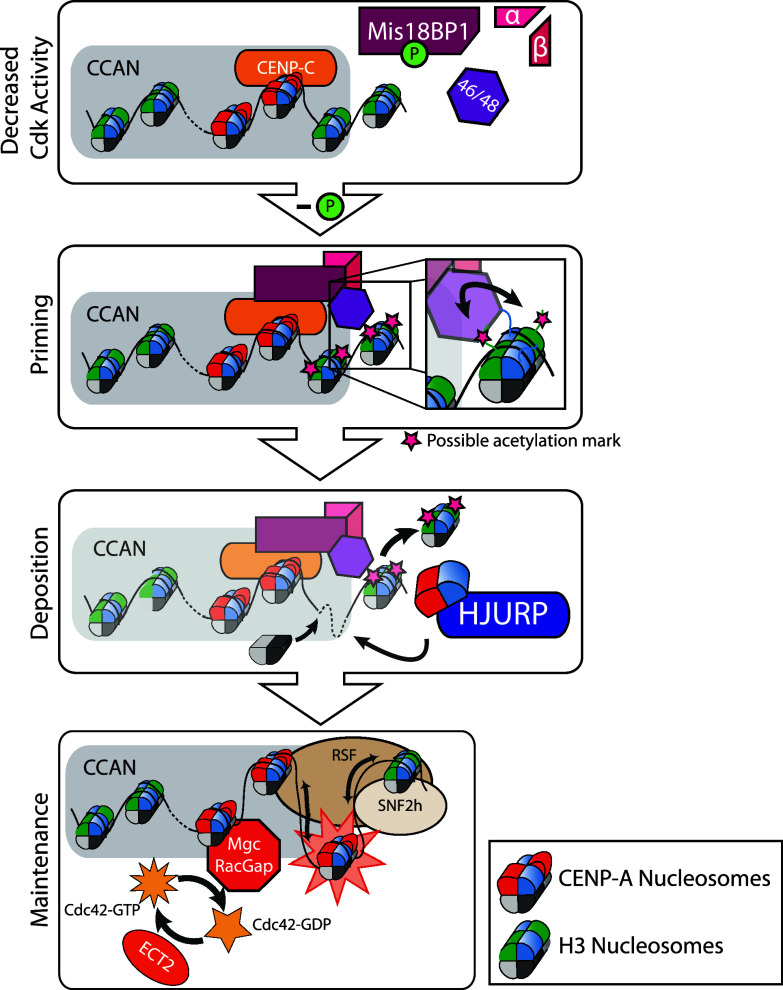
The deposition of newly synthesized CENP-A occurs during G1. The schematic shows a current model of CENP-A deposition into centromeric chromatin during the G1 phase of the cell cycle. Cdk activity decreases after the exit from mitosis, which allows the association of the Mis18 complex with centromeric chromatin, likely through an interaction with CENP-C. It is theorized that the Mis18 complex recruits chromatin modifying activity to centromeric histones in order to prime chromatin for the assembly of newly synthesized CENP-A by HJURP. Finally, the association of Rsf1/Snf2 and MgcRacGap with the centromere enables the establishment of fully stable CENP-A nucleosomes through chromatin remodeling and GTP-GDP of Cdc42 cycling
This proposes a model of reinforcement whereby new CENP-A deposition is reinforced at an existing centromere by the presence of the CCAN protein, CENP-C, which directs the localization of the CENP-A assembly factors, the Mis18 complex and HJURP. Several observations support this idea of a feed-forward mechanism to ensure continual enrichment of newly synthesized CENP-A at active centromeres. Overexpression of CENP-A (CID) in flies results in its mislocalization throughout chromatin; however, only a subset of regions containing the mislocalized CENP-A develop into active centromeres that recruit CCAN proteins and downstream kinetochore components [83, 84]. It may be that the recruitment of CCAN proteins at non-centromeric loci is minimal, possibly due to limiting protein levels. Therefore, the continual recruitment of new CENP-A to non-endogenous CENP-A foci may be limited and centromeric CENP-A is continually replenished because of its recruitment of CCAN proteins.
The above data make a very compelling case for the recruitment of new CENP-A nucleosomes by the CCAN; however, CENP-C may not be the sole mechanism of Mis18 recruitment. Xenopus has two isoforms of Mis18BP1KNL2; one is recruited from metaphase through G1, while the other isoform localizes like human Mis18BP1KNL2 in G1 alone. In CENP-C depletions, only the metaphase localization was compromised, while the G1 localization remained intact [69]. Targeting CENP-C to ectopic locations does not initiate CENP-A deposition outside of the centromere as would be expected if CENP-C was sufficient for Mis18 recruitment [58]. Therefore, Mis18 recruitment may prove to be more complex than simple recognition of CENP-C by Mis18BP1KNL2 and instead may proceed through multiple mechanisms.
In replenishing CENP-A at centromeric sites, the overall architecture of a post mitotic chromosome may be the most efficient substrate for CENP-A deposition. Human CENP-A deposition occurs immediately following mitosis, at a time when chromatin is highly condensed. Human condensin I and II are multiunit complexes that function to condense chromosomes during mitosis [85]. Experiments in several organisms suggest that chromatin condensation may be important for the efficient deposition of CENP-A nucleosomes. At the budding yeast point centromere, condensin depletion results in a decrease of Cse4 localization [86]. Experiments using Xenopus egg extracts demonstrated that condensin II is required for efficient CENP-A deposition [87]. SMC2 depletion by siRNA in human cells leads to a decrease in the recruitment of new CENP-A nucleosomes to centromeres [88]. Condensin association with chromosomes peaks in anaphase, placing it directly prior to the association of the Mis18 complex with centromeres in late anaphase [64, 89]. While these experiments suggest a connection between CENP-A deposition and condensin, the exact function of condensin remains unclear. The function of condensin may be to simply maintain the three-dimensional structure of the centromere required to facilitate CENP-A deposition.
Priming the centromere for CENP-A deposition
The exact function of the Mis18 complex at centromeres largely remains a black box in the understanding of the centromere lifecycle. HJURP requires the activity of the Mis18 complex at centromeres for its recruitment [17, 69], but no direct interaction has been observed between the Mis18 complex proteins and HJURP or CENP-A in human cells. Therefore, current research in the field centers on the hypothesis that the Mis18 complex primes centromeres for CENP-A deposition by recruiting chromatin modifying activity to the centromere in order to generate or maintain a permissive state for the recruitment and deposition of CENP-A [64, 65].
In fission yeast, temperature-sensitive mutants of Mis16 and Mis18 show a significant increase in the acetylation of centromeric histone H3 and H4 at the inner centromere repeats (cnt1 and imr1) [65]. The human Mis18 complex also affects the post-translational modification status of the centromere, although where fission yeast Mis18 seems to inhibit acetylation, the human Mis18 complex promotes acetylation. Cells depleted for members of the Mis18 complex, lose the deposition of newly synthesized CENP-A at centromeres; however, treating those cells concurrently with the HDAC inhibitor, trichostatin A (TSA), rescues CENP-A deposition [64]. Global inhibition of HDAC activity with TSA would increase the acetylation status of the genome and theoretically the centromere as well. Consistent with these observations, targeting histone acetyltransferase activity of P300 or PCAF to a human artificial chromosome is sufficient to induce CENP-A deposition [51]. An increase in centromeric H3 acetylation can be seen in early G1, which correlates with the localization of the Mis18 complex at centromeres. However the identity of the endogenous histone acetyltransferase responsible for this activity is not known. Since an artificial increase in centromere acetylation seems to bypass the requirement for the Mis18 complex in CENP-A deposition, this argues that the human Mis18 complex functions by affecting the centromeric histone acetylation.
The Mis18 complex may also affect CENP-A deposition by altering epigenetic modifications of DNA. The DNA methyltransferases, DNMT3A and DNMT3B, interact with centromeric chromatin through interactions with Mis18α and CENP-C [90, 91]. Conditional knockout studies in mouse embryonic fibroblasts found that Mis18α loss resulted in a reduction of centromeric DNA methylation [91]. Disrupting DNMT3A/B decreased the methylation of centromeric DNA and lead to a significant decrease in the level of CENP-A at the centromere. It is not clear whether DNA methylation is part of the recruitment mechanism of HJURP or whether DNA methylation creates a permissive chromatin environment for CENP-A deposition by altering transcription.
Further evidence to support the role of the Mis18 complex in affecting the histone modification state of centromeric chromatin is the fact that the proteins of the Mis18 complex have several ties to chromatin remodeling and modifying complexes. Mis18BP1KNL2 contains a SANT (Swi3-Ada2-NCoR-TFIIIB) domain as well as a SANT-Associated (SANTA) domain [64, 92]. The SANTA domain was found in silico as a domain that characteristically is present in proteins which also contain a SANT domain [92]. In the human proteome, the SANTA domain has been identified exclusively in Mis18BP1KNL2. The function of the SANTA domain is currently unknown, although, the conserved hydrophobic residues are proposed to be involved in protein–protein interactions, possibly mediating Mis18BP1KNL2 interactions with its various binding partners at the centromere [92].
As its full name implies, the Swi3-Ada2-NCoR-TFIIIB (SANT) domain is found in a variety of chromatin remodeling and modifying complexes including the remodeler, SWI/SNF, and the SAGA histone acetyltransferase (HAT) complex [93]. SANT domains are made up of roughly 50 amino acids that form three alpha helices in a helix-turn-helix motif similar to the DNA binding domain in c-Myb [94]. The Myb domain in the proto-oncogene, c-Myb, has been shown to bind DNA in a sequence specific manner [95]. However, analysis of the crystal structure of the SANT domain in Xenopus ISWI shows that the amino acid residues responsible for the sequence specific DNA interactions in c-Myb are not conserved in the ISWI SANT domain [94].
SANT domains also mediate protein–protein interactions to recruit and activate additional binding partners in order to generate fully functional chromatin modifying complexes such as HDAC3 in the SMRT and N-CoR co-repressor complexes as well as the HAT activity of SAGA [96–98]. In addition, RbAp48 or RbAp46 are common to several known histone modifying and remodeling complexes [99, 100]. The presence of RbAp48 and RbAp46 as well as the domain architecture of Mis18BP1KNL2 give credence to the hypothesis that the Mis18 complex is capable of recruiting chromatin modifying activity to centromeric chromatin. However, direct evidence for the recruitment of these types of factors to centromeres during early G1 is lacking.
Deposition of CENP-A
The ultimate goal of the centromere specification pathway is the deposition of new CENP-A nucleosomes. Nucleosome assembly is facilitated by the activity of histone chaperone proteins [101, 102]. Known histone H3 variants, such as H3.1 and H3.3, utilize unique chaperone proteins in order to facilitate distinct timing and location of deposition [101, 102]. Despite vast differences in centromere organization between budding yeast and humans, these organisms all employ a related chaperone, known as HJURP (Holliday junction recognition protein) in humans and Scm3 in yeast, in order to achieve deposition of newly synthesized CENP-A nucleosomes.
CENP-A-histone H4 and HJURP form a prenucleosomal complex that localizes to centromeres in G1 during new CENP-A deposition [38, 68, 87, 103]. This complex is required for new CENP-A deposition and is sufficient to determine the site of centromere formation [17]. Deletion of Scm3 in budding or fission yeast leads to chromosome loss or missegregation due to defects in Cse4/Cnp1 recruitment [22, 104, 105]. A fly homolog for HJURP has not been identified; however, the localization of the CAL1 protein and its requirement in CenH3/CID deposition suggest that it may act as a functional homolog [76].
Although Scm3 and HJURP serve similar functions the entirety of their similarity is located within a small, 50 amino acid, region of homology within their N-termini [9]. HJURP is a much larger, 83-kD protein in humans compared to the 26-kD Scm3 protein of S. cerevisiae. Several regions of HJURP have been identified that are conserved among the higher eukaryotic forms of the protein, but none of these domains have yet been ascribed functional roles. Although HJURP was originally identified as a protein that recognizes synthetic holiday junctions (thus termed Holliday junction recognition protein) [67], a requirement for complex DNA structures has not been identified in CENP-A deposition. The differences between HJURP and Scm3 may reflect differences in the mechanism by which they are recruited to the centromere.
Centromere organization between yeast and humans differs greatly, and correlates with differences in HJURP versus Scm3 recruitment to centromeres. In humans and fission yeast, which both harbor regional centromeres; the Mis18 homologs are required for the recruitment of HJURP/Scm3 to centromeres (Fig. 4) [17, 65]. This is in contrast to budding yeast, which have a point centromere, and do not possess a Mis18 homolog. Instead, the S. cerevisiae Scm3 binds AT-rich DNA, which may serve as a possible recruitment mechanism [106]. Even among regional centromeres there are differences in the mechanism by which Mis18 recruits HJURP/Scm3. A direct physical interaction between Mis18 and Scm3 has been observed in fission yeast; however, despite several analyses of protein purifications, human HJURP and Mis18 have not been observed to associate [38, 64, 68, 107]. As such, the complexity of the Mis18 complex seems to mirror that of the centromere. A single Mis18 protein is present in S. pombe while two homologs are found in higher eukaryotes [64]. Moreover, the Mis18BP1KNL2 subunit appears to be specific to higher eukaryotes, as an S. pombe homolog has not been identified. It will be interesting to understand how the evolution of this important complex in centromere specification contributes to the differences in centromere organization.
The classical role of histone chaperones is to facilitate the deposition of histones into nucleosomes. The deposition of CENP-A nucleosomes is a conserved function of the HJURP/Scm3 protein. In vitro chromatin assembly assays, using recombinant proteins, show that human HJURP and budding yeast Scm3 are both sufficient to assemble CENP-A into nucleosomes [17, 108–111]. In each of these cases, HJURP/Scm3 assembles an octameric nucleosome that wraps DNA in a left-handed manner, similar to canonical H3 nucleosomes. In cells, the retargeting of HJURP to non-centromeric loci is sufficient to lead to the incorporation of CENP-A into chromatin [17]. CENP-A point mutants that affect the CENP-A dimerization interface are able to bind HJURP but cannot be stably assembled into chromatin by HJURP [27]. Heterotypic nucleosomes that contain one copy of CENP-A and histone H3 have been observed as a small fraction of human CENP-A nucleosomes [20]. Since histone H3 uses a similar dimerization interface, mutations in this region would also be expected to eliminate the formation heterotypic octameric nucleosomes. However, to date no function has been assigned to heterotypic CENP-A-H3 nucleosomes. These data suggest Scm3/HJURP proteins assemble octameric CENP-A nucleosomes at centromeres. Nonetheless, data exists that depicts other sub-octameric forms of centromeric nucleosomes. It is possible that the structure of the CENP-A nucleosome is dynamic throughout the cell cycle, and perhaps may change through downstream remodeling events.
Recent crystal structures provide interesting insight into the interaction between HJURP/Scm3 and the CENP-A/histone H4 heterodimer in budding yeasts and humans. The CENP-A binding domain (CBD) of HJURP includes the Scm3 homolog domain and forms a long alpha helix followed by a short beta sheet. In all structures, the long alpha helix, within the Scm3 homology domain of HJURP/Scm3, interacts with the CENP-A CATD [112–114]. The CBD of HJURP extends into the region of CENP-A self association and precludes CENP-A heterotetramer formation; therefore, the HJURP/CENP-A/H4 complex forms a heterotrimer which contains a single copy of each protein [27, 112]. Residues outside of the CATD domain also interact with the previously identified TLTY box recognition domain of HJURP/Scm3, although it is unlikely that these resides contribute to specificity [27, 103, 112]. These structures show that although the centromeres of these organisms are divergent, the specific interaction between HJURP/Scm3 and CENP-A/H4 heterodimers is conserved between budding yeast and man. Moreover, the formation of the HJURP/CENP-A/H4 prenucleosomal complex excludes tetramer formation and DNA interaction suggesting a step-wise conformational change is required for incorporating CENP-A to centromeres.
Centromeric chromatin maturation
A growing amount of evidence supports the idea that CENP-A nucleosomes are not fully stable after their initial deposition in early G1, but require additional changes through remodeling complexes and GTP cycling to become fully mature, stable centromeric nucleosomes (Fig. 4) [107, 115]. This maturation process occurs after the deposition of newly synthesized CENP-A by HJURP and does not affect CENP-A nucleosomes already present at the centromere. The RSF complex (Rsf1 and SNF2 h) associates with the centromere in mid G1 and confers stability to newly deposited CENP-A nucleosomes [115]. In addition, MgcRacGap and Ect2 GTP cycling activity is recruited to centromeric chromatin in late G1 and is also required to stabilize new CENP-A nucleosomes [107]. These events occur asynchronously and transiently at only a subset of centromeres during late G1. While the function of this maturation process is not completely understood at this time, RSF and MgcRacGap seem to help generate centromeric chromatin that is sufficiently stable to support its roles during the cell cycle, such as serving as the kinetochore platform during mitosis.
S-phase: maintaining centromere identity through DNA synthesis
Replication of the genome necessitates that CENP-A nucleosomes be distributed to each newly synthesized sister chromatid (Fig. 5). This is not unique to the centromeric epigenetic mark, but must also be the case for the stable propagation of many histone modifications that provide epigenetic regulation. Replication of general chromatin includes the incorporation of new histone H3.1-containing nucleosomes as well as the re-incorporation of histones from pre-existing nucleosomes [101, 102]. As the replication fork passes through chromatin, pre-existing nucleosomes are distributed among the daughter strands. Then, newly synthesized histones are deposited into both strands to make up for the dilution of histones that occurs during the replication process. How CENP-A nucleosomes are stably transited across the replication fork and whether this mechanism is similar to that used by H3-containing nucleosomes is not known.
Fig. 5.
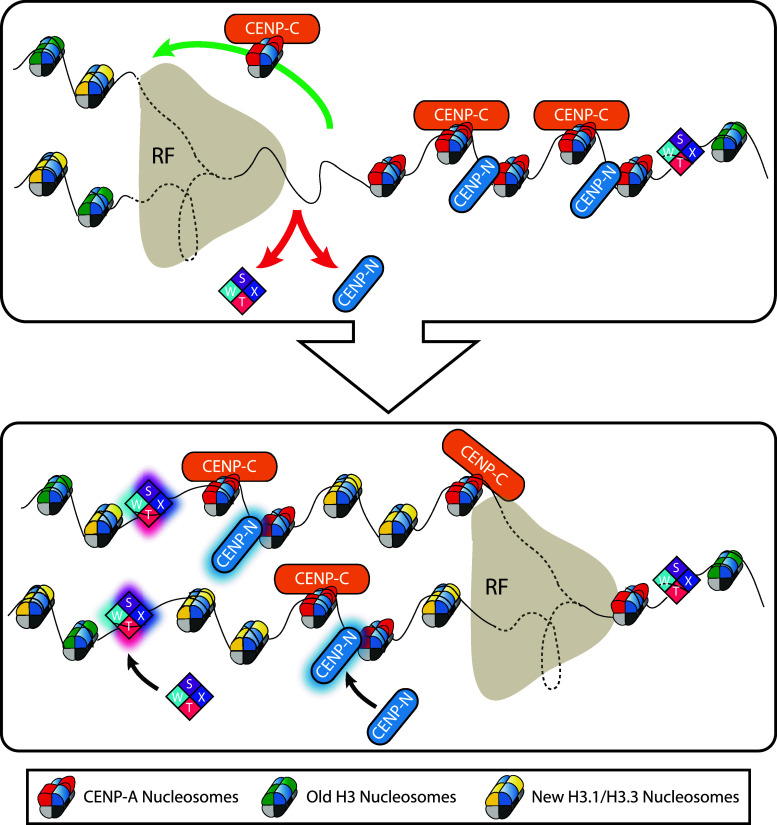
Replication of centromeric chromatin and the S-phase dynamics of the most CENP-A-chromatin proximal CCAN proteins as the replication fork (RF) passes. Centromeric chromatin is replicated in S-phase concurrently with general chromatin. S-phase dynamics of the most CENP-A-chromatin proximal CCAN proteins: CENP-T/W/S/X complex, CENP-C, and CENP-N are also shown. Red arrows symbolize dissociation from the centromere. The green arrow symbolizes replication fork passage. Black arrows symbolize loading of new CENPs. a Existing CENP-A nucleosomes are allotted to each daughter strand, but no new CENP-A nucleosomes are added during S-phase. New H3.1/H3.3 nucleosomes may serve as placeholders during replication coupled dilution of existing CENP-A nucleosomes at the centromere (yellow nucleosomes). CENP-C is stably associated with centromeres in S-phase and likely tracts with CENP-A nucleosomes across the replication fork. b As the replication fork passes, CENP-T/W/S/X complexes are turned over every cell cycle, and load during late S-phase. CENP-N localization is dynamic throughout the cell cycle, but loads to maximal levels during S-phase
Availability of CENP-A may help to preclude CENP-A deposition during S-phase. Overexpression of CENP-A can lead to the misincorporation of CENP-A into non-centromeric chromatin, suggesting that high levels of CENP-A can overtax the mechanisms that restrict CENP-A deposition to centromeres [83, 84, 116]. While canonical histone levels increase by early S-phase to allow for the massive deposition of new nucleosomes, CENP-A mRNA levels do not rise until mid S-phase leading to the accumulation of CENP-A protein and its chaperone HJURP in G2, after replication has finished [68, 73, 74]. As such, CENP-A protein is not available for deposition during replication, and G2 centromeres contain only half of the amount of CENP-A molecules that were present in the parental centromere prior to replication.
CENP-A nucleosomes are stably retained through S-phase [63]. Each CENP-A domain parses its CENP-A nucleosomes between the two daughter strands. Immunofluorescence studies of replicated chromatin fibers show that daughter strands have the same number of CENP-A blocks, but the blocks are one half of the intensity of pre-S-phase centromeres [117]. Since no new CENP-A nucleosomes are deposited until G1, cells may leave gaps in centromeric loci resulting in only partial nucleosome protection. Alternatively, H3.1/3.3-containing nucleosomes (or an alternative complex) may occupy these gaps. Recent data suggest that histone H3.3 nucleosomes may serve to fill in the gaps left by CENP-A distribution at the centromere during DNA replication, and these H3.3 nucleosomes are then exchanged in G1 concurrent with CENP-A deposition [117]. It is unclear whether the deposition of H3.3 at the centromere is through the replication machinery or dependent on centromeric transcription (see below). Regardless, it suggests that active displacement of a “placeholder” through a chromatin remodeling event may be necessary in order to make room for newly synthesized CENP-A nucleosomes in G1.
An alternative model is that old CENP-A nucleosomes are split between the two daughter chromosomes generating either a “hemisome” consisting of only one copy of each histone or a heterotypic nucleosome containing one copy of CENP-A and one copy of H3.1/3.3. There is some evidence to support the existence of a hemisome-like particle via crosslinking studies as well as atomic force microscopy, in which the height of the CENP-A nucleosome particles are one half the size of the canonical H3 nucleosomes [23, 24]. This would dictate that CENP-A hemisomes would be restricted to the G2 phase of the cell cycle.
Another level of complexity lies in the distribution of various CCAN components during S-phase. The CCAN is a large complex of centromere proteins (CENPs) that are constitutively localized at the centromere throughout the cell cycle [20]. While these proteins are continually present at centromeres, their localization is dynamic, with varied exchange profiles [118, 119]. CENP-N has a rapid exchange rate throughout the cell cycle, allowing it to dissociate as the replication fork passes (Fig. 5). However, during late S-phase, centromeric levels of CENP-N increase as the exchange rate drops [119]. This suggests that CENP-N is free to diffuse before replication, but afterwards loads onto centromeric chromatin in a more stable manner. This may enable CENP-N to mark centromeres that have completed replication. The CENP-T/W/S/X complex remains stably associated with centromeric DNA throughout the cell cycle, but becomes dynamic in late S-phase and completely turns over, suggesting that the CENP-T/W/S/X complex is disrupted by the replication fork and reassembles after it has passed [45]. In contrast, CENP-C is highly dynamic throughout the cell cycle but becomes stably associated with centromeres during S-phase and mitosis [118]. CENP-C interacts directly with CENP-A nucleosomes, meaning that CENP-C may pass the replication fork in a similar manner. How CENP-A nucleosomes transit the replication fork during S-phase is an extremely interesting yet poorly understood aspect of centromere inheritance, and it is fascinating to consider the possible roles of the various CCAN proteins in this process.
Ensuring centromere specificity: removal of CENP-A from non-centromeric loci
The directed recruitment of CENP-A deposition machinery to centromeres is the major method to ensure stable centromere identity. However, overexpression of CENP-A in several systems leads to the misincorporation of CENP-A nucleosomes into other sites within the genome [83, 84, 116]. This means that while CENP-A is deposited specifically at centromeres; it retains the ability to localize throughout the rest of the genome. Mislocalization of CENP-A throughout general chromatin by overexpression, or to specific non-centromeric loci via targeted deposition, can cause chromosome missegregation errors [17, 18, 83, 84]. Therefore, in order for a cell to ensure the formation of only one centromere per chromosome, it must also employ a mechanism that removes non-centromeric CENP-A. Experimental evidence strongly suggests that this occurs via ubiquitin-mediated proteasome degradation, a mechanism that seems to be conserved from yeast to humans (Fig. 6).
Fig. 6.
The removal of non-centromeric CENP-A through the proteasome pathway. a Centromeric CENP-A is protected from degradation by binding partners that compete with the binding of CENP-A specific E3 ubiquitin ligases, such as Psh1. HJURP binding to unassembled heterodimers or specific interactions between centromeric CENP-A nucleosomes and the CCAN, inhibit the degradation of CENP-A. b Non-centromeric CENP-A is removed from ectopic locations in a cell cycle independent manner. This may occur as a natural consequence of histone exchange during chromatin remodeling and metabolism across chromosome arms or via a targeted degradation event by a specific E3 ubiquitin ligase, such as Psh1. The CENP-A specific E3 ubiquitin ligase has yet to be found in humans; however, research indicates that a CENP-A degradation can occur through the ubiquitin-mediated proteasome pathway
In both budding yeast and flies, specific E3 ubiquitin ligases have been linked to CENP-A degradation. The S. cerevisiae E3 ligase, Psh1, specifically signals for degradation of Cse4 as compared to histone H3 [120, 121]. Deletion of Psh1 prevents Cse4 from being ubiquitinated and increases the association of Cse4 at non-centromeric loci [120, 121]. Psh1 is a major buffer to the effects of Cse4 overexpression, which are lethal in the absence of Psh1 [120, 121]. The major E3 ubiquitin ligase responsible for CenH3/CID degradation in D. melanogaster is the SCF complex component, Ppa [122]. Ubiquitin ligase activity, associated with CENP-A stability, has been identified in humans. Infection by HSV-1 hijacks the proteasome and targets CENP-A for degradation through a viral, RING finger domain-containing protein, ICP0 [123]. ICP0 is required and sufficient to cause CENP-A degradation after infection [123]. However, a host ubiquitin ligase that is coupled to targeted CENP-A degradation remains to be discovered in humans.
Both Ppa and Psh1 recognize the CATD domain of CENP-A [121, 122]. This is the same region recognized by both the CENP-A chaperone HJURP/Scm3 and the CCAN protein CENP-N in human cells [37, 68, 103]. Consistent with these observations, centromeric pools of Cse4 in budding yeast are resistant to proteolytic degradation [124]. Scm3 binding to Cse4, prevents its ubiquitination by Psh1 in vitro, and turning off Scm3 expression in vivo accelerates Cse4 degradation in budding yeast [120]. Likewise, the knockdown of HJURP reduces CENP-A protein levels in human cells [38, 68].
Depletion of Swi/Snf activity in budding yeast causes the accumulation of Cse4 at non-centromeric loci suggesting that non-centromeric CENP-A nucleosomes are sensitive to destabilization by chromatin remodelers [125]. These data suggesting that accessibility of CENP-A to degradation is limited by its interaction with either the HJURP chaperone complex or with the CCAN upon incorporation into centromeres. Misincorporated CENP-A lacks these interactions and is therefore removed by during remodeling and subjected to proteasome degradation.
Influences of heterochromatin in centromere specification
Regional centromeres are consistently organized such that a central CENP-A-containing region is flanked by pericentric heterochromatin. In humans, pericentric heterochromatin contain nucleosomes that are trimethylated on lysine 9 of histone H3 (H3K9me3), while H3K9me2 chromatin is interspersed with CENP-A in the centromere region (Fig. 2). Pericentric heterochromatin is a repressive chromatin structure, where the H3K9 trimethylation acts as a signal for the recruitment of the chromodomain protein, HP1 [126, 127].
Heterochromatin formation in S. pombe is required for de novo centromere formation [128]. Pericentromeric regions are established through the RNAi pathway. H3K9me3 and Chp1 mediate binding of the RITS complex, which in turn recruits RNA-dependent polymerase complex (RDRC) mediated transcription of double stranded RNA. Dicer then processes the double stranded RNA to generate siRNAs that help facilitate centromere silencing. In a positive-feedback loop, the methyltransferase Clr4 is locally recruited by the RNAi pathway and reinforces the trimethylation mark on H3K9 [129]. In plasmid based de novo centromere formation assays, when heterochromatin formation is inhibited on flanking regions, de novo CENP-A nucleosome deposition cannot occur. However, when the siRNA requirement is bypassed by direct targeting of the Clr4 methyltransferase, CENP-A nucleosomes are deposited demonstrating that the key requirement for de novo CENP-A deposition is the activity of Clr4 [130].
Neocentromere formation in experimental systems appears to prefer sequences that are in close proximity to heterochromatin, perhaps reflecting the need for pericentric heterochromatin in de novo centromere formation. When neocentromeres are induced by the removal of endogenous centromere sequences on chromosome one in S. pombe, neocentromeres most often arise near telomeric regions where H3K9 methylation is present [15]. Likewise, in D. melanogaster, overexpressed CID/CenH3 forms islands throughout the length of the chromosomes, but it appears that ectopic centromere formation is biased to regions where heterochromatin and euchromatin are in close apposition [131]. It seems that this boundary element may make a more permissive structure for de novo CID/CenH3 deposition. It is not clear if these same modifications influence neocentromere formation in humans, as the consistent localization of H3K9me3 regions with neocentromeres studies has so far not been observed [132].
Non-coding RNAs, transcription, and the centromere
Mounting evidence suggests that RNA polymerase II (RNAPII) mediated transcription through CENP-A-containing chromatin is a conserved feature of centromeric regions across several species. Transcripts have been identified from the central domain of S. pombe centromeres as well as from maize centromere sequences [133, 134]. In human cells, centromeric alpha-satellite repeat transcripts have been detected in several different cell lines [135, 136].
The process of transcription seems to be at odds with the highly stable character of CENP-A-containing chromatin, as CENP-A nucleosomes do not appear to turn over except for deposition of newly synthesized CENP-A during G1. However, centromeric character is not mutually exclusive with gene expression. Immunofluorescence studies of stretched centromeric chromatin fibers show that centromeres contain H3K4me2, a mark of open or permissive chromatin, and do not contain several H3 modifications implicated in transcription silencing [39]. In addition, human neocentromeres can form in chromosome regions containing actively transcribed genes, and CENP-A nucleosome deposition in gene coding regions of human artificial chromosomes (HACs) does not diminish gene expression [10, 50, 137]. Recent studies demonstrate that modest amounts of transcription across the alpha-satellite centromere region of a HAC are compatible with centromere function; however, driving high levels of transcription leads to a loss of kinetochore function and a destabilization of CENP-A chromatin, [138] suggesting that the levels of transcription may be key to the stability of centromeres.
Recently Chan et al. [135] observed an accumulation of active RNAPII at human centromeres in metaphase. Analysis of a pseudodicentric chromosome showed that RNAPII only colocalized with the active neocentromere and was not found at the inactivated, alpha-satellite-containing region of the original centromere. Pulse labeling using FITC-rUTP showed nascent α-satellite transcripts colocalizing specifically with centromeres during mitosis that were abolished upon α-amanitin treatment. Disruption of these centromere transcripts in mitosis caused lagging chromosomes in the subsequent anaphase [135]. The increase in lagging chromosomes correlated with a measurable decrease in centromeric CENP-C levels [135]. CENP-C has been previously implicated as binding to centromeric RNA transcripts suggesting a connection between centromeric transcription and the CCAN [139].
While it is clear that centromeric transcription occurs, its function remains unclear. One hypothesis has been proposed where centromeric transcription is coupled to chromatin remodeling activity in order to facilitate the exchange of histone H3 nucleosomes for CENP-A nucleosomes [133]. SSRP1 (structure-specific recognition protein), a subunit of the FACT (facilitates chromatin transcription complex), was also found to localize to RNAPII foci in human cells [135]. FACT is a general chromatin remodeler that has been found to associate with human CENP-A along with another chromatin remodeler, chromo-helicase DNA-binding protein 1 (Chd1) [19, 20, 140, 141]. The fission yeast homolog, Hrp1, has already been implicated in efficient CENP-A deposition at centromeres [142]. Depletion of Hrp1 caused an increase in H3 nucleosomes in the inner repeats of the fission yeast centromere [133]. Like in humans, S. pombe central domain centromeres were transcribed by RNAPII to produce small amounts of non-coding RNA (ncRNA) [133]. In both species, Chd1 associates with actively transcribed centromeres and is present at a moment when histone exchange would occur. However, this remains highly speculative and is an area in need of intense study.
Perspective: centromeres in the treatment and genesis of human disease
Overexpression of centromere assembly pathway members, CENP-A and HJURP, has been observed in several types of cancer [67, 143, 144]. Interestingly, co-overexpression of CENP-A and HJURP is correlated in breast cancer and results in increased rates of chromosome missegregation in cultured cells [145]. The overexpression of HJURP alone or in combination with other genes is proving to be a significant biomarker for cancer prognosis in both breast cancer and high-grade gliomas [144, 146].
Ectopic centromere formation may contribute to genomic instability. Recent experiments show that deposition of CENP-A at non-centromeric loci may form active, stable, ectopic centromeres [17, 18]. If an ectopic centromere is formed on a chromosome arm, it results in chromosome breakage. Likewise, the amplification of alpha-satellite repeats has also been shown to result in chromosome missegregation and the formation of chromosome bridges during anaphase [147].
Examples of neocentromere formation have been observed in a variety of cancer cells including carcinoma, sarcoma, and leukemia. However, cases of lipomatous tumors have been consistently associated with neocentromere formation [148–150]. In all of these cases, the reported neocentromere supported the stable inheritance of a marker chromosome that contained an extra copy of chromosome region 12q14–15. This suggests that the amplification resulted from a period of genomic instability, and that neocentromere formation was able to stabilize a cancer promoting phenotype. The potential stability of neocentromeres in cancer is evident in an example of a lung carcinoma, where a neocentromere formed on the p arm of chromosome 9. The 9p neocentromeric chromosome was present in the primary lung tumor and in two separate metastatic tumors of the same individual [151]. This suggests that the neocentromere was present in the primary tumor and stable through the process of metastasis. Neocentromeres in cancer consistently result in the amplification of a region, either because the chromosome fragment is part of an inverted duplication or because the neocentromeric chromosome is present at multiple copies in each cell. These amplifications are likely to result in changes in dosage of many genes. Most instances of neocentromere formation in cancers have no consistent site of neocentromere location, with the exception of the lipomatous tumor mentioned previously. The true degree of neocentromere formation has not been studied systematically in tumors. It may be that neocentromere formation is a necessary step to allow the stable propagation of complex chromosomal rearrangements and gene amplifications seen in tumors, which may drive cancer progression.
The generation of human artificial chromosomes (HACs) has been an important and insightful tool in determining how centromeres are stably propagated. A long-standing goal has been to develop viable and efficient HACs as gene therapy vectors to treat human disease using endogenous centromere machinery in order to ensure their stable inheritance [152]. HACs have been derived from endogenous, cloned alpha-satellite repeats, but more recently, synthetic arrays are using alpha-satellite repeats combined with binding sites for the TET repressor in order to control centromere activity and the inheritance of therapeutic genes [50, 153]. Human artificial chromosomes are able to correct genetic deficiencies in cultured cells and reprogram induced pluripotent stem cells (iPS cells) [154–156]. Recent work demonstrating the sufficiency of HJURP and CENP-A to initiate de novo centromere formation may lead to the development of highly efficient, completely synthetic human artificial chromosomes that are independent of repetitive alpha-satellite DNA sequences; surely an exciting tool for prospective gene therapeutics [17, 18].
References
- 1.Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McClintock B. The stability of broken ends of chromosomes in Zea mays . Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colnaghi R, Carpenter G, Volker M, O’Driscoll M. The consequences of structural genomic alterations in humans: genomic disorders, genomic instability and cancer. Semin Cell Dev Biol. 2011;22:875–885. doi: 10.1016/j.semcdb.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 5.Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, Choo KH. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cottarel G, Shero JH, Hieter P, Hegemann JH. A 125-base-pair CEN6 DNA fragment is sufficient for complete meiotic and mitotic centromere functions in Saccharomyces cerevisiae . Mol Cell Biol. 1989;9:3342–3349. doi: 10.1128/mcb.9.8.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- 8.Maddox PS, Oegema K, Desai A, Cheeseman IM. “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans . Chromosome Res. 2004;12:641–653. doi: 10.1023/B:CHRO.0000036588.42225.2f. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marshall OJ, Chueh AC, Wong LH, Choo KH. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bassett EA, Wood S, Salimian KJ, Ajith S, Foltz DR, Black BE. Epigenetic centromere specification directs aurora B accumulation but is insufficient to efficiently correct mitotic errors. J Cell Biol. 2010;190:177–185. doi: 10.1083/jcb.201001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lo AW, Craig JM, Saffery R, Kalitsis P, Irvine DV, Earle E, Magliano DJ, Choo KH. A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J. 2001;20:2087–2096. doi: 10.1093/emboj/20.8.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, Poirier GG, Earnshaw WC. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- 14.Voullaire LE, Slater HR, Petrovic V, Choo KH. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am J Hum Genet. 1993;52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- 15.Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- 16.Williams BC, Murphy TD, Goldberg ML, Karpen GH. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila . Nat Genet. 1998;18:30–37. doi: 10.1038/ng0198-30. [DOI] [PubMed] [Google Scholar]
- 17.Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendiburo MJ, Padeken J, Fulop S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- 19.Obuse C, Yang H, Nozaki N, Goto S, Okazaki T, Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- 20.Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 21.Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, 3rd, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 22.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 23.Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y. Tetrameric organization of vertebrate centromeric nucleosomes. Proc Natl Acad Sci USA. 2010;107:20317–20322. doi: 10.1073/pnas.1009563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, Kimura H, Kurumizaka H. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 27.Bassett EA, DeNizio J, Barnhart-Dailey MC, Panchenko T, Sekulic N, Rogers DJ, Foltz DR, Black BE. HJURP is a molecular chaperone and CENP-A assembly factor that recognizes the exposed surface of the CATD. Dev Cell. 2012;22(4):749–762. doi: 10.1016/j.devcel.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Colmenares SU, Karpen GH. Assembly of Drosophila centromeric nucleosomes requires CID dimerization. Mol Cell. 2012;45:263–269. doi: 10.1016/j.molcel.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Black BE, Brock MA, Bedard S, Woods VL, Jr, Cleveland DW. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci USA. 2007;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 31.Panchenko T, Sorensen TC, Woodcock CL, Kan ZY, Wood S, Resch MG, Luger K, Englander SW, Hansen JC, Black BE. Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc Natl Acad Sci USA. 2011;108:16588–16593. doi: 10.1073/pnas.1113621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conde e Silva N, Black BE, Sivolob A, Filipski J, Cleveland DW, Prunell A. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J Mol Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 33.Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 37.Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc Natl Acad Sci USA. 2006;103:4186–4191. doi: 10.1073/pnas.0507947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishino T, Takeuchi K, Gascoigne KE, Suzuki A, Hori T, Oyama T, Morikawa K, Cheeseman IM, Fukagawa T. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 2012;148:487–501. doi: 10.1016/j.cell.2011.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marino-Ramirez L, Levine KM, Morales M, Zhang S, Moreland RT, Baxevanis AD, Landsman D (2011) The histone database: an integrated resource for histones and histone fold-containing proteins. Database (Oxford) bar048 [DOI] [PMC free article] [PubMed]
- 44.Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, Cheeseman IM, Fukagawa T. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 45.Prendergast L, van Vuuren C, Kaczmarczyk A, Doering V, Hellwig D, Quinn N, Hoischen C, Diekmann S, Sullivan KF. Premitotic assembly of human CENPs -T and -W switches centromeric chromatin to a mitotic state. PLoS Biol. 2011;9:e1001082. doi: 10.1371/journal.pbio.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci USA. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cardinale S, Bergmann JH, Kelly D, Nakano M, Valdivia MM, Kimura H, Masumoto H, Larionov V, Earnshaw WC. Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol Biol Cell. 2009;20:4194–4204. doi: 10.1091/mbc.E09-06-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohzeki JI, Bergmann JH, Kouprina N, Noskov VN, Nakano M, Kimura H, Earnshaw WC, Larionov V, Masumoto H. Breaking the HAC barrier: histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J. 2012;31(10):2391–2402. doi: 10.1038/emboj.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marshall OJ, Marshall AT, Choo KH. Three-dimensional localization of CENP-A suggests a complex higher order structure of centromeric chromatin. J Cell Biol. 2008;183:1193–1202. doi: 10.1083/jcb.200804078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng C, Lu X, Hansen JC, Hayes JJ. Salt-dependent intra- and internucleosomal interactions of the H3 tail domain in a model oligonucleosomal array. J Biol Chem. 2005;280:33552–33557. doi: 10.1074/jbc.M507241200. [DOI] [PubMed] [Google Scholar]
- 54.Milks KJ, Moree B, Straight AF. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol Biol Cell. 2009;20:4246–4255. doi: 10.1091/mbc.E09-05-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM. CENP-C is a structural platform for kinetochore assembly. Curr Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 56.Screpanti E, De Antoni A, Alushin GM, Petrovic A, Melis T, Nogales E, Musacchio A. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki A, Hori T, Nishino T, Usukura J, Miyagi A, Morikawa K, Fukagawa T. Spindle microtubules generate tension-dependent changes in the distribution of inner kinetochore proteins. J Cell Biol. 2011;193:125–140. doi: 10.1083/jcb.201012050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schleiffer A, Maier M, Litos G, Lampert F, Hornung P, Mechtler K, Westermann S. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat Cell Biol. 2012;14(6):604–613. doi: 10.1038/ncb2493. [DOI] [PubMed] [Google Scholar]
- 60.Cheeseman IM, Hori T, Fukagawa T, Desai A. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol Biol Cell. 2008;19:587–594. doi: 10.1091/mbc.E07-10-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amaro AC, Samora CP, Holtackers R, Wang E, Kingston IJ, Alonso M, Lampson M, McAinsh AD, Meraldi P. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat Cell Biol. 2010;12:319–329. doi: 10.1038/ncb2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hori T, Okada M, Maenaka K, Fukagawa T. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol Biol Cell. 2008;19:843–854. doi: 10.1091/mbc.E07-06-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 65.Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 66.Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kato T, Sato N, Hayama S, Yamabuki T, Ito T, Miyamoto M, Kondo S, Nakamura Y, Daigo Y. Activation of holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 2007;67:8544–8553. doi: 10.1158/0008-5472.CAN-07-1307. [DOI] [PubMed] [Google Scholar]
- 68.Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, Allshire RC. Fission yeast Scm3: a CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Uhlmann F, Bouchoux C, Lopez-Aviles S. A quantitative model for cyclin-dependent kinase control of the cell cycle: revisited. Philos Trans R Soc Lond B Biol Sci. 2011;366:3572–3583. doi: 10.1098/rstb.2011.0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 76.Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 2011;7:e1002068. doi: 10.1371/journal.pgen.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grosskortenhaus R, Sprenger F. Rca1 inhibits APC-Cdh1(Fzr) and is required to prevent cyclin degradation in G2. Dev Cell. 2002;2:29–40. doi: 10.1016/s1534-5807(01)00104-6. [DOI] [PubMed] [Google Scholar]
- 79.Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell. 2011;22(1):52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 80.Dambacher S, Deng W, Hahn M, Sadic D, Frohlich J, Nuber A, Hoischen C, Diekmann S, Leonhardt H, Schotta G. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus. 2012;3(1):101–110. doi: 10.4161/nucl.18955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Orr B, Sunkel CE. Drosophila CENP-C is essential for centromere identity. Chromosoma. 2011;120:83–96. doi: 10.1007/s00412-010-0293-6. [DOI] [PubMed] [Google Scholar]
- 82.Goshima G, Kiyomitsu T, Yoda K, Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J Cell Biol. 2003;160:25–39. doi: 10.1083/jcb.200210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moreno-Moreno O, Torras-Llort M, Azorin F. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res. 2006;34:6247–6255. doi: 10.1093/nar/gkl902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cuylen S, Haering CH. Deciphering condensin action during chromosome segregation. Trends Cell Biol. 2011;21:552–559. doi: 10.1016/j.tcb.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 86.Yong-Gonzalez V, Wang BD, Butylin P, Ouspenski I, Strunnikov A. Condensin function at centromere chromatin facilitates proper kinetochore tension and ensures correct mitotic segregation of sister chromatids. Genes Cells. 2007;12:1075–1090. doi: 10.1111/j.1365-2443.2007.01109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bernad R, Sanchez P, Rivera T, Rodriguez-Corsino M, Boyarchuk E, Vassias I, Ray-Gallet D, Arnaoutov A, Dasso M, Almouzni G, Losada A. Xenopus HJURP and condensin II are required for CENP-A assembly. J Cell Biol. 2011;192:569–582. doi: 10.1083/jcb.201005136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Samoshkin A, Arnaoutov A, Jansen LE, Ouspenski I, Dye L, Karpova T, McNally J, Dasso M, Cleveland DW, Strunnikov A. Human condensin function is essential for centromeric chromatin assembly and proper sister kinetochore orientation. PLoS ONE. 2009;4:e6831. doi: 10.1371/journal.pone.0006831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tada K, Susumu H, Sakuno T, Watanabe Y. Condensin association with histone H2A shapes mitotic chromosomes. Nature. 2011;474:477–483. doi: 10.1038/nature10179. [DOI] [PubMed] [Google Scholar]
- 90.Gopalakrishnan S, Sullivan BA, Trazzi S, Della Valle G, Robertson KD. DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Hum Mol Genet. 2009;18:3178–3193. doi: 10.1093/hmg/ddp256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim IS, Lee M, Park KC, Jeon Y, Park JH, Hwang EJ, Jeon TI, Ko S, Lee H, Baek SH, Kim KI. Roles of Mis18alpha in epigenetic regulation of centromeric chromatin and CENP-A loading. Mol Cell. 2012;46(3):260–273. doi: 10.1016/j.molcel.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 92.Zhang D, Martyniuk CJ, Trudeau VL. SANTA domain: a novel conserved protein module in Eukaryota with potential involvement in chromatin regulation. Bioinformatics. 2006;22:2459–2462. doi: 10.1093/bioinformatics/btl414. [DOI] [PubMed] [Google Scholar]
- 93.Boyer LA, Langer MR, Crowley KA, Tan S, Denu JM, Peterson CL. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol Cell. 2002;10:935–942. doi: 10.1016/s1097-2765(02)00634-2. [DOI] [PubMed] [Google Scholar]
- 94.Horton JR, Elgar SJ, Khan SI, Zhang X, Wade PA, Cheng X. Structure of the SANT domain from the Xenopus chromatin remodeling factor ISWI. Proteins. 2007;67:1198–1202. doi: 10.1002/prot.21352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- 96.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sterner DE, Wang X, Bloom MH, Simon GM, Berger SL. The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J Biol Chem. 2002;277:8178–8186. doi: 10.1074/jbc.M108601200. [DOI] [PubMed] [Google Scholar]
- 98.You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc Natl Acad Sci USA. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 100.Philpott A, Krude T, Laskey RA. Nuclear chaperones. Semin Cell Dev Biol. 2000;11:7–14. doi: 10.1006/scdb.1999.0346. [DOI] [PubMed] [Google Scholar]
- 101.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 103.Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci USA. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 105.Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci USA. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xiao H, Mizuguchi G, Wisniewski J, Huang Y, Wei D, Wu C. Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol Cell. 2011;43:369–380. doi: 10.1016/j.molcel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lagana A, Dorn JF, De Rop V, Ladouceur AM, Maddox AS, Maddox PS. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat Cell Biol. 2010;12:1186–1193. doi: 10.1038/ncb2129. [DOI] [PubMed] [Google Scholar]
- 108.Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kingston IJ, Yung JS, Singleton MR. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J Biol Chem. 2011;286:4021–4026. doi: 10.1074/jbc.M110.189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shivaraju M, Camahort R, Mattingly M, Gerton JL. Scm3 is a centromeric nucleosome assembly factor. J Biol Chem. 2011;286:12016–12023. doi: 10.1074/jbc.M110.183640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, Li G, Xu RM. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cho US, Harrison SC. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc Natl Acad Sci USA. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou Z, Feng H, Zhou BR, Ghirlando R, Hu K, Zwolak A, Miller Jenkins LM, Xiao H, Tjandra N, Wu C, Bai Y. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011;472:234–237. doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K. Active establishment of centromeric CENP-A chromatin by RSF complex. J Cell Biol. 2009;185:397–407. doi: 10.1083/jcb.200903088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- 117.Dunleavy EM, Almouzni G, Karpen GH. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G phase. Nucleus. 2011;2:146–157. doi: 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S. Dynamics of inner kinetochore assembly and maintenance in living cells. J Cell Biol. 2008;180:1101–1114. doi: 10.1083/jcb.200710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hellwig D, Emmerth S, Ulbricht T, Doring V, Hoischen C, Martin R, Samora CP, McAinsh AD, Carroll CW, Straight AF, Meraldi P, Diekmann S. Dynamics of CENP-N kinetochore binding during the cell cycle. J Cell Sci. 2011;124:3871–3883. doi: 10.1242/jcs.088625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell. 2010;40:444–454. doi: 10.1016/j.molcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell. 2010;40:455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Moreno-Moreno O, Medina-Giro S, Torras-Llort M, Azorin F. The F box protein partner of paired regulates stability of Drosophila centromeric histone H3, CenH3(CID) Curr Biol. 2011;21:1488–1493. doi: 10.1016/j.cub.2011.07.041. [DOI] [PubMed] [Google Scholar]
- 123.Lomonte P, Sullivan KF, Everett RD. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J Biol Chem. 2001;276:5829–5835. doi: 10.1074/jbc.M008547200. [DOI] [PubMed] [Google Scholar]
- 124.Collins KA, Furuyama S, Biggins S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 125.Gkikopoulos T, Singh V, Tsui K, Awad S, Renshaw MJ, Scholfield P, Barton GJ, Nislow C, Tanaka TU, Owen-Hughes T. The SWI/SNF complex acts to constrain distribution of the centromeric histone variant Cse4. EMBO J. 2011;30:1919–1927. doi: 10.1038/emboj.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 127.Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 128.Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev. 2010;20:134–141. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, Simmer F, Urano T, Hamilton GL, Allshire RC. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science. 2009;324:1716–1719. doi: 10.1126/science.1172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Olszak AM, van Essen D, Pereira AJ, Diehl S, Manke T, Maiato H, Saccani S, Heun P. Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat Cell Biol. 2011;13:799–808. doi: 10.1038/ncb2272. [DOI] [PubMed] [Google Scholar]
- 132.Alonso A, Hasson D, Cheung F, Warburton PE. A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin. 2010;3:6. doi: 10.1186/1756-8935-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Choi ES, Stralfors A, Castillo AG, Durand-Dubief M, Ekwall K, Allshire RC. Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J Biol Chem. 2011;286:23600–23607. doi: 10.1074/jbc.M111.228510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Topp CN, Zhong CX, Dawe RK. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc Natl Acad Sci USA. 2004;101:15986–15991. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chan FL, Marshall OJ, Saffery R, Won Kim B, Earle E, Choo KH, Wong LH. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc Natl Acad Sci USA. 2012;109:1979–1984. doi: 10.1073/pnas.1108705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, Choo KH. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KH, Wong LH. LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet. 2009;5:e1000354. doi: 10.1371/journal.pgen.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bergmann JH, Jakubsche JN, Martins NM, Kagansky A, Nakano M, Kimura H, Kelly DA, Turner BM, Masumoto H, Larionov V, Earnshaw WC. Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J Cell Sci. 2012;125:411–421. doi: 10.1242/jcs.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Du Y, Topp CN, Dawe RK. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 2010;6:e1000835. doi: 10.1371/journal.pgen.1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Izuta H, Ikeno M, Suzuki N, Tomonaga T, Nozaki N, Obuse C, Kisu Y, Goshima N, Nomura F, Nomura N, Yoda K. Comprehensive analysis of the ICEN (interphase centromere complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 2006;11:673–684. doi: 10.1111/j.1365-2443.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- 141.Okada M, Okawa K, Isobe T, Fukagawa T. CENP-H-containing complex facilitates centromere deposition of CENP-A in cooperation with FACT and CHD1. Mol Biol Cell. 2009;20:3986–3995. doi: 10.1091/mbc.E09-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Walfridsson J, Bjerling P, Thalen M, Yoo EJ, Park SD, Ekwall K. The CHD remodeling factor Hrp1 stimulates CENP-A loading to centromeres. Nucleic Acids Res. 2005;33:2868–2879. doi: 10.1093/nar/gki579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–3516. [PubMed] [Google Scholar]
- 144.Hu Z, Huang G, Sadanandam A, Gu S, Lenburg ME, Pai M, Bayani N, Blakely EA, Gray JW, Mao JH. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res. 2010;12:R18. doi: 10.1186/bcr2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mishra PK, Au WC, Choy JS, Kuich PH, Baker RE, Foltz DR, Basrai MA. Misregulation of Scm3p/HJURP causes chromosome instability in Saccharomyces cerevisiae and human cells. PLoS Genet. 2011;7(9):e1002303. doi: 10.1371/journal.pgen.1002303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.de Tayrac M, Aubry M, Saikali S, Etcheverry A, Surbled C, Guenot F, Galibert MD, Hamlat A, Lesimple T, Quillien V, Menei P, Mosser J. A 4-gene signature associated with clinical outcome in high-grade gliomas. Clin Cancer Res. 2011;17:317–327. doi: 10.1158/1078-0432.CCR-10-1126. [DOI] [PubMed] [Google Scholar]
- 147.Gisselsson D, Hoglund M, Mertens F, Mandahl N. Variable stability of chromosomes containing amplified alpha-satellite sequences in human mesenchymal tumours. Chromosoma. 1999;108:271–277. doi: 10.1007/s004120050378. [DOI] [PubMed] [Google Scholar]
- 148.Sirvent N, Forus A, Lescaut W, Burel F, Benzaken S, Chazal M, Bourgeon A, Vermeesch JR, Myklebost O, Turc-Carel C, Ayraud N, Coindre JM, Pedeutour F. Characterization of centromere alterations in liposarcomas. Genes Chromosomes Cancer. 2000;29:117–129. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1014>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 149.Forus A, Bjerkehagen B, Sirvent N, Meza-Zepeda LA, Coindre JM, Berner JM, Myklebost O, Pedeutour F. A well-differentiated liposarcoma with a new type of chromosome 12-derived markers. Cancer Genet Cytogenet. 2001;131:13–18. doi: 10.1016/s0165-4608(01)00516-7. [DOI] [PubMed] [Google Scholar]
- 150.Italiano A, Maire G, Sirvent N, Nuin PA, Keslair F, Foa C, Louis C, Aurias A, Pedeutour F. Variability of origin for the neocentromeric sequences in analphoid supernumerary marker chromosomes of well-differentiated liposarcomas. Cancer Lett. 2009;273:323–330. doi: 10.1016/j.canlet.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 151.Italiano A, Attias R, Aurias A, Perot G, Burel-Vandenbos F, Otto J, Venissac N, Pedeutour F. Molecular cytogenetic characterization of a metastatic lung sarcomatoid carcinoma: 9p23 neocentromere and 9p23-p24 amplification including JAK2 and JMJD2C. Cancer Genet Cytogenet. 2006;167:122–130. doi: 10.1016/j.cancergencyto.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 152.Kazuki Y, Oshimura M. Human artificial chromosomes for gene delivery and the development of animal models. Mol Ther. 2011;19:1591–1601. doi: 10.1038/mt.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Iida Y, Kim JH, Kazuki Y, Hoshiya H, Takiguchi M, Hayashi M, Erliandri I, Lee HS, Samoshkin A, Masumoto H, Earnshaw WC, Kouprina N, Larionov V, Oshimura M. Human artificial chromosome with a conditional centromere for gene delivery and gene expression. DNA Res. 2010;17:293–301. doi: 10.1093/dnares/dsq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kim JH, Kononenko A, Erliandri I, Kim TA, Nakano M, Iida Y, Barrett JC, Oshimura M, Masumoto H, Earnshaw WC, Larionov V, Kouprina N. Human artificial chromosome (HAC) vector with a conditional centromere for correction of genetic deficiencies in human cells. Proc Natl Acad Sci USA. 2011;108:20048–20053. doi: 10.1073/pnas.1114483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kazuki Y, Hiratsuka M, Takiguchi M, Osaki M, Kajitani N, Hoshiya H, Hiramatsu K, Yoshino T, Kazuki K, Ishihara C, Takehara S, Higaki K, Nakagawa M, Takahashi K, Yamanaka S, Oshimura M. Complete genetic correction of ips cells from Duchenne muscular dystrophy. Mol Ther. 2010;18:386–393. doi: 10.1038/mt.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hiratsuka M, Uno N, Ueda K, Kurosaki H, Imaoka N, Kazuki K, Ueno E, Akakura Y, Katoh M, Osaki M, Kazuki Y, Nakagawa M, Yamanaka S, Oshimura M. Integration-free iPS cells engineered using human artificial chromosome vectors. PLoS ONE. 2011;6:e25961. doi: 10.1371/journal.pone.0025961. [DOI] [PMC free article] [PubMed] [Google Scholar]