Abstract
NKp30 is a stimulatory receptor on human NK cells implicated in tumor immunity, and is capable of promoting or terminating dendritic cell maturation. To gain a better understanding of NKp30 biology, we have investigated the expression and function of rat NKp30 (rNKp30). We generated stable transfectants of rNKp30 in RNK16 cells, a rat NK lymphoma line, and used a novel panel of mAb against rNKp30 to study this receptor. Using agonistic rNKp30 mAb, we demonstrated that rNKp30 mediates robust IFN-γ production and cytolytic responses from rNKp30-transfected RNK16 cells. We determined by flow cytometry that rNKp30 is expressed by a subset of primary NK cells isolated from the blood and spleen, and to a lesser extent also on liver NK cells. Stimulation of rNKp30 on primary NK cells led to IFN-γ production. Liver NK cells expressed low levels of NKp30 and had reduced rNKp30-mediated IFN-γ responses. During an alloimmune response in vivo, the proportion of the rNKp30+ NK cell subset in the peripheral blood significantly increased, suggesting that rNKp30 may play an important role during alloactivation. Thus, our data demonstrate that NKp30 is indeed expressed in rodents and is a functional stimulatory receptor in a subset of rat NK cells.
Keywords: Cellular activation, Cell surface, molecules, Innate immunity, NK cells, Transplantation
Introduction
Natural killer (NK) cells are lymphocytes of the innate immune system that are important during immune responses against viruses and tumors [1, 2]. Virally infected and transformed cells can be recognized and killed by NK cells without prior sensitization. In addition, NK cells can influence adaptive immunity through the secretion of cytokines, such as IFN-γ and TNF-α, and through interactions with dendritic cells (DC) [3–10].
NK cell activity is controlled by multiple stimulatory and inhibitory surface receptors. Stimulatory receptors are characterized by short cytoplasmic tails that lack signaling motifs, but instead contain a positively charged residue in the transmembrane domain that associates with immunoreceptor tyrosine-based activating motif (ITAM)-containing signaling adapter molecules, such as CD3ζ, DAP12, and FcεRIγ. The ligands identified for activation receptors include viral proteins, and stress or transformation-induced molecules. In contrast, inhibitory receptors bind MHC class I and are characterized by a long cytoplasmic tail that contains immunoreceptor tyrosine-based inhibitory motifs (ITIM). Engagement of inhibitory receptors can override NK cell activation to prevent targeting of normal self cells.
The natural cytotoxicity receptors (NCR), NKp30 (CD337), NKp46 (CD335) and NKp44 (CD336), are immunoglobulin (Ig)-like NK cell activation receptors that have been shown on human NK cells to be key receptors in tumor immunity and possibly viral immunity [11, 12]. Blocking the NCR inhibits NK cell cytotoxicity against multiple tumor cells in vitro [13–17]. However, tumor cell ligands for the NCR have yet to be identified. There is evidence that NKp44 and NKp46 detect viral ligands, such as influenza and sendai viral hemagglutinins, and that recognition may lead to killing of viral-infected cells [18]. NKp30 may also be important in viral immunity as the CMV protein pp65 appears to target NKp30 to evade NK cells [19]. Indeed, ligands that stimulate NKp30 remain undefined.
Furthermore, human NKp30 (hNKp30) is distinguished from other NCR and NK cell receptors through interactions with DC. NK-DC cross-talk can result in maturation or elimination of immature DC (iDC), and activation of NK cells [7, 20, 21]. In vitro evidence indicates that NKp30 is the major receptor involved in determining the fate of iDC during these interactions as NKp30 can mediate iDC killing, or maturation via TNF and IFN-γ [7, 20].
The importance of NKp30 in the human prompts investigation of NKp30 in vivo using rodent models. NKp30 is a pseudogene in inbred mouse strains as a result of two premature stop codons [22]. However, we and others have previously identified and cloned the rat homolog of NKp30 (rNKp30) [23, 24]. We report here the characterization of rNKp30 surface expression and function on NK cells. Our data show that NKp30 is expressed on a subset of NK cells in rat spleen, blood, and to a lesser extent in the liver. Activation of rNKp30 induces IFN-γ production and cytotoxicity. Interestingly, we show that the proportion of the rNKp30+ NK cell subset in the peripheral blood is significantly increased after alloactivation. Our data demonstrate that NKp30 is a functional NK cell activation receptor in the rat and this model can be used to study NKp30 during immune responses in vivo.
Results
Novel mAb detect rNKp30 on rNKp30 transfectants
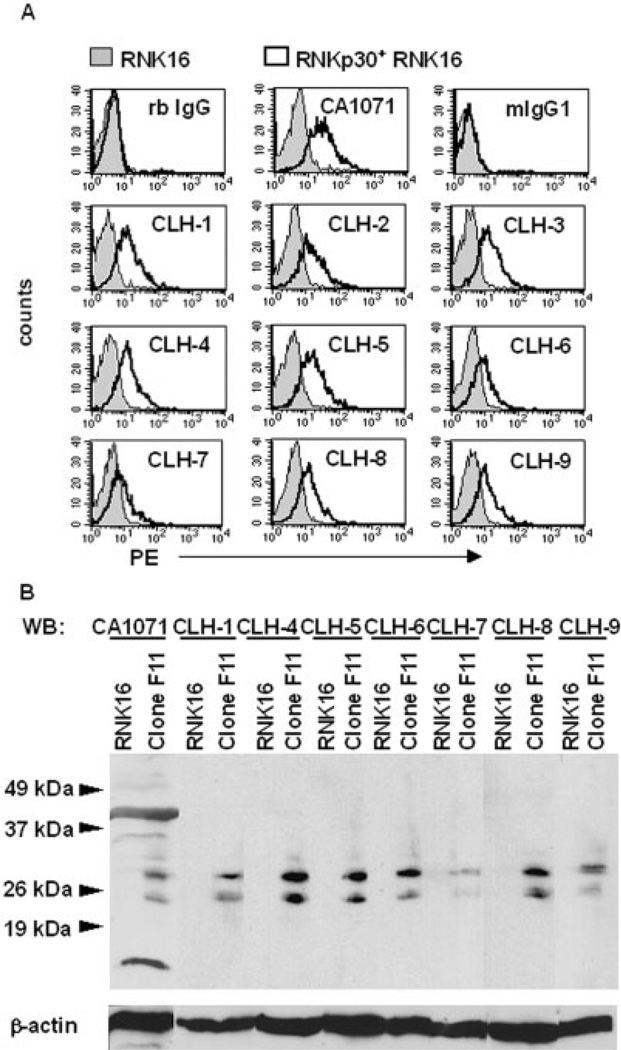
To study the expression and function of rNKp30, we generated nine mouse mAb against rNKp30 protein that were produced against an rNKp30-Fc chimera, and are designated CLH-1 through CLH-9. The rNKp30 mAb were screened by ELISA against the rNKp30 domain (not shown), and by flow cytometry for specific binding to rNKp30-transfected RNK16 cell lines (Fig. 1). CA1071, a rabbit (rb) anti-rat NKp30 polyclonal antibody (pAb) that we previously generated was used as a positive control for detection of rNKp30 expression [23]. All nine mAb and CA1071 bound, to varying degrees, to stably transfected rNKp30+ RNK16 cells (clone F11), but not to parental RNK16 cells (Fig. 1A).
Fig. 1.
Anti-rNKp30 mAb specifically detect rNKp30 on transfected cells. (A) Anti-rNKp30 pAb, CA1071, anti-rNKp30 mAb CLH-1 through CLH-9, and isotype control antibodies followed by PE-conjugated secondary antibodies were used to stain parental RNK16 cells (filled histograms) and stably transfected rNKp30+ RNK16 Clone F11 cells (open histograms). (B) Immunoblot analysis of lysates from parental RNK16 and rNKp30+ Clone F11 cells were performed and blots were probed with anti-rNKp30 antibodies as specified and reprobed for β-actin. Molecular masses (kDa) of a protein size ladder are indicated on the left by arrowheads. rNKp30 antibodies detect two prominent bands between 25 and 30 kDa specifically in the rNKp30+ RNK16 clone F11 lysates (n=3).
To confirm the specificity of anti-rNKp30 antibodies, lysates were prepared from parental RNK16 and rNKp30-transfected RNK16 clone F11 cells, separated by SDS-PAGE, and immunoblotted for rNKp30. Immunoblots probed with either CA1071 or anti-rNKp30 mAb (CLH-1, CLH-4–9) demonstrated two prominent bands between 25–30 kDa in clone F11 lysates, but not in the parental RNK16 lysates (Fig. 1B). We have previously determined that these bands correspond to different glycosylated forms of rNKp30 [23]. Equal protein loading was confirmed by probing for β-actin. Taken together, these results indicate that the anti-rNKp30 mAb we have generated specifically detect rNKp30 in transfected cells.
rNKp30 activation induces IFN-γ secretion and cytotoxicity
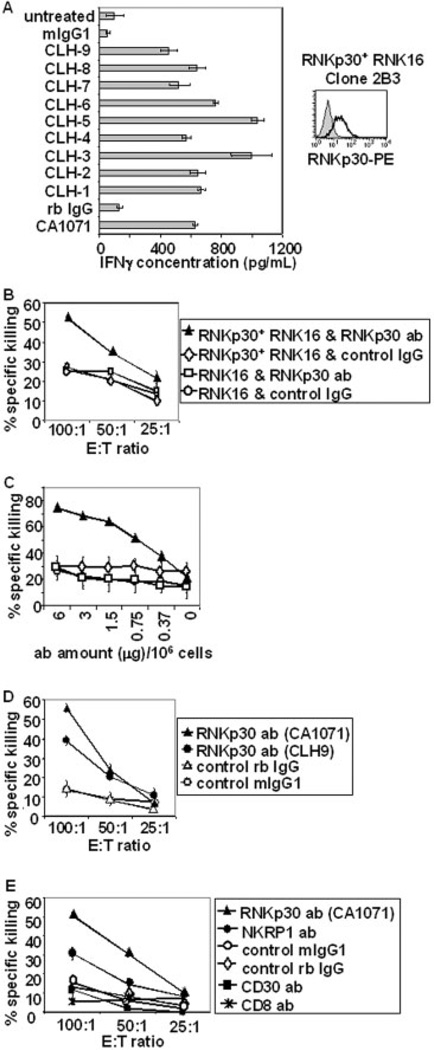
To determine the function of rNKp30 and to test if it could promote Th1 responses, rNKp30-transfected RNK16 cells were utilized to assess IFN-γ secretion following rNKp30 activation. rNKp30+ RNK16 clone 2B3 expressed rNKp30, as detected by flow cytometry (Fig. 2, right), and was used in functional assays. Clone 2B3 was cultured with immobilized anti-rNKp30 antibodies for 24 h, and the supernatants were tested for IFN-γ by ELISA. The anti-rNKp30 antibodies CA1071 and CLH-1–9 all induced significant levels of IFN-γ secretion (400–1100 pg/mL) from rNKp30+ RNK16 cells (Fig. 2A). In contrast, the isotype control antibodies, mIgG1 and rb IgG, did not induce IFN-γ secretion. Similarly, no IFN-γ production was detected in supernatants from parental RNK16 cells stimulated with rNKp30 antibodies (data not shown). By comparison, PMA and ionomycin treatment induced 1800 ± 300 pg/ mL IFN-γ from rNKp30+ RNK16 cells (data not shown).
Fig. 2.
rNKp30 activation in rNKp30+ RNK16 transfectants induces IFN-γ production and cytolytic activity. (A) rNKp30+ RNK16 clone 2B3 cells express rNKp30 as detected by flow cytometry (right). Anti-rNKp30 antibody, CA1071 (open histogram), followed by PE-conjugated secondary antibodies, and not isotype control antibodies (filled histogram), positively stained clone 2B3 cells. On the left, clone 2B3 cells were treated with immobilized antibodies (10 µg/mL) as indicated on the y-axis for 24 h (n=3). Harvested supernatants were analyzed for IFN-γ by ELISA. (B) rNKp30+ RNK16 clone 2B3 and parental RNK16 cells were analyzed for cytolytic activity in 4-h redirected lysis assays against [3H]thymidine-labeled P815 targets (n=8). On the left, rNKp30+ RNK16 cells stimulated with rNKp30 antibody, CA1071 (black triangles), showed increased cytotoxicity with increasing E:T ratios. Target cells were resistant to killing when control IgG or parental RNK16 cells were used (white diamonds, white squares, and white circles). (C) Redirected lysis assays were performed with titrated amounts of rNKp30 antibodies (CA1071) or control antibodies at an E:Tof 100:1 (n=2). Symbols used in (B) and (C) represent the same effector cells and antibody treatments. (D) Redirected lysis assays were performed to compare the anti-rNKp30 antibodies, CA1071 pAb (black triangles) and CLH-9 mAb (black circles). Both anti-rNKp30 antibodies induce killing as compared to control antibodies (white triangles and white circles). A symbol legend for (D) is boxed on the right. (E) Antibodies specific for activation receptors triggered cytotoxicity from rNKp30+ RNK16 cells. Antibodies (2 µg/106 effector cells) against rNKp30 (black triangles), NKRP1A (black circles), CD30 (black squares), CD8 (asterisks), and control mIgG1 (white circles) and control rabbit IgG (white diamonds) were analyzed in redirected killing assays (n=2). A symbol legend for (E) is boxed on the right.
To determine if rNKp30 activation could trigger cytolytic activity, parental and rNKp30+ RNK16 cells were analyzed in redirected lysis assays using anti-rNKp30 pAb CA1071, and P815 target cells, which are resistant to RNK16 cell killing. Only rNKp30+ RNK16 cells treated with CA1071 (black triangle) induced specific killing (Fig. 2B). Treatment of rNKp30+ RNK16 cells with prebleed IgG (white diamonds) or parental RNK16 cells treated with anti-rNKp30 antibody (white squares) and prebleed IgG (white circles) showed background levels of activity against P815 targets. Data shown are representative of eight experiments performed with either rNKp30+ RNK16 clone 2B3 or clone F11. Direct killing assays against YAC-1 lymphoblastoma targets were also performed to ensure functional killing activity of all effector cells (not shown). We also titrated the amount of anti-rNKp30 antibody in redirected lysis assays using a constant E:T ratio of 100:1. Cytolytic activity of rNKp30+ RNK16 clone 2B3 cells was dependent on the dose of CA1071 utilized (Fig. 2C). In addition, the mAb CLH-9 (black circles) induced elevated cytotoxicity, compared to control antibodies, in redirected lysis assays (Fig. 2D). There was no significant difference in killing with the parental RNK16 cell line when treated with CLH-9 or control antibodies (not shown).
To further demonstrate that cytotoxicity was induced by rNKp30 activation and was not the result of cell-to-cell contact mediated by an antibody that binds both the NK cell and FcR+ target, redirected lysis assays using other antibodies that bind RNK16 cells were used. CD30 and CD8 are surface molecules expressed by rNKp30 + RNK16 cells at similar intensities as rNKp30 as determined by flow cytometry (not shown). Only antibodies against NK activation receptors, rNKp30 (black triangles) and NKRP1A (black circles), the latter receptor to a lesser extent, induced specific killing by rNKp30+ RNK16 cells (Fig. 2E). Thus, activation directly through rNKp30 can trigger cytolytic activity.
rNKp30 is a functional activation receptor expressed on a subset of splenic NK cells
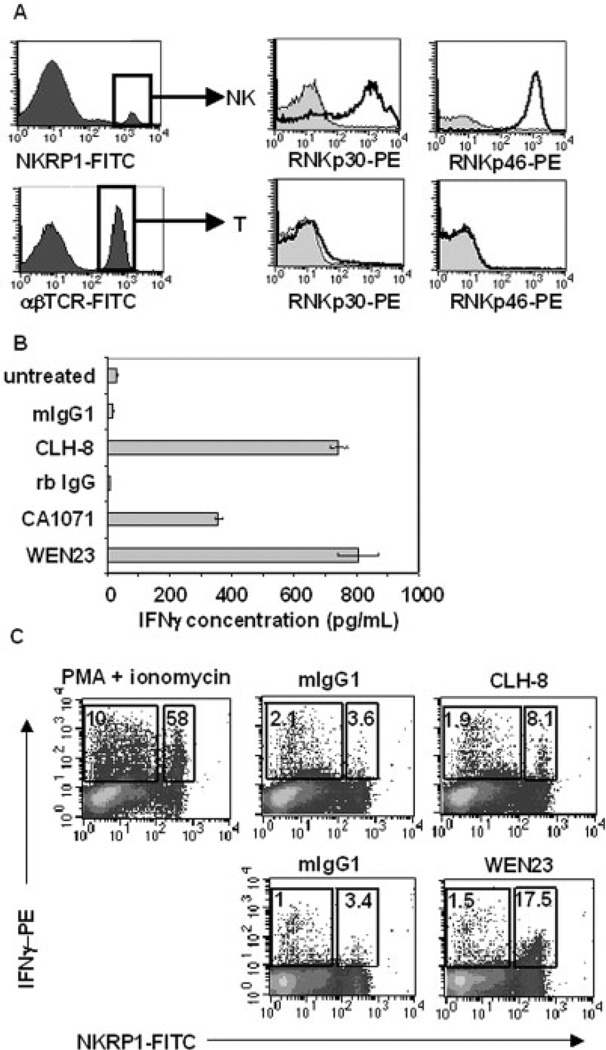
Previously, we had determined that splenic NK cells express rNKp30 transcripts [23]. To confirm that primary NK cells express rNKp30 protein, Fischer 344 (F344) rat splenic NK cells were stained with an anti-rNKp30 mAb (CLH-9) and analyzed by flow cytometry. Gates were established for NK cells on the NKRPlAhi βTCR− PI− population (Fig. 3A, top row). A subset of splenic NK cells clearly expressed rNKp30 (Fig. 3A, top row, middle). Both the CLH-9 or CLH-8 mAb yielded nearly identical results and representative staining of CLH-9 is shown. Interestingly, rNKp30 expression differs as compared to another NCR, rat NKp46 (rNKp46). RNKp46 was expressed on all NK cells (Fig. 3A, top row, right), as detected by an anti-rNKp46 mAb (WEN23), which is consistent with a previous report [25]. rNKp30 and rNKp46 were not expressed on T cells (Fig. 3A, bottom row).
Fig. 3.
rNKp30 is expressed on a major subset of F344 spleen NK cells and activation induces IFN-γ production. (A) rNKp30 and rNKp46 expression were analyzed on F344 rat spleen NK cells (NKRPl-FITChi, αβTCR-PerCP−) and T cells (αβTCR-FITC+) by flow cytometry. NKp30 expression was detected using CLH-9-biotin (middle, open histograms) and NKp46 expression was identified with WEN23-biotin (right, open histograms). NCR histograms were overlayed on isotype controls (filled histograms). (B) Splenocytes or PBMC (2.5 × 106/mL, 5 × 105 cells/ well) were cultured in media alone (untreated), or with immobilized antibodies (12 µg/mL). Splenocytes were treated with antibodies against rNKp30 (CLH-8 or CA1071), or control antibodies (mIgG1 or rb IgG) for 11 h. As a control for NK-mediated IFN-γ secretion, PBMC were treated with the anti-rNKp46 mAb, WEN23. Harvested supernatants were assessed for IFN-γ production by ELISA (n=5). (C) Treated mononuclear cells were permeabilized and analyzed for IFN-γ with a PE-conjugated anti-IFN-γ mAb, and costained with a FITC-conjugated anti-NKRPl antibody (n=3). Each dotplot contains a small box (right) indicating the percent of IFN-γ+ NK cells, and a large box (left) gating on the fraction of non-NK cells expressing IFN-γ. (Top row) Splenocytes were treated with PMA (10 ng/mL) and ionomycin (0.5 µg/mL) (left), mIgG1 isotype control (12 µg/mL) (middle), and anti-rNKp30 mAb CLH-8 (12 µg/mL) (right). (Bottom row) PBMC were treated with mIgG1 isotype control antibodies (12 µg/mL) or anti-rNKp46 mAb WEN23 (12 µg/mL). rNKp30 and rNKp46 activation induced increased IFN-γ production from a subset of NK cells.
Since rNKp30 activation induced IFN-γ production in the rNKp30-transfected RNK16 system, we asked if endogenous rNKp30 on primary cells was functional to induce cytokine responses. Spleen mononuclear cells were left untreated, or incubated with plate-bound anti-rNKp30 or control antibodies for 11–12 h, and harvested supernatants were analyzed for IFN-γ by ELISA. Anti-rNKp30 antibodies, CLH-8 and CA1071, induced 742 ± 28 pg/mL and 370 ± 10 pg/mL IFN-γ, respectively, from splenocytes (Fig. 3B). Isotype control antibodies, mIgG1 and rb IgG, induced less than 20 pg/mL IFN-γ (Fig. 3B). Additionally we demonstrate that activation of rNKp46 induces 806 ± 66 pg/mL of IFN-γ from PBMC stimulated with platebound anti-rNKp46 mAb (WEN23, Fig. 3B). PMA and ionomycin treatment resulted in abundant levels of IFN-γ (3673 ± 5 pg/mL) (not shown). ELISA data shown are representative of three to six experiments.
To confirm that NK cells were the source of IFN-γ after rNKp30 activation, intracellular staining was performed. Splenocytes were treated with PMA and ionomycin or antibodies and brefeldin A was added to retain secreted proteins. Permeabilized cells were analyzed for NKRP1A and IFN-γ expression by flow cytometry, and data representative of three experiments is shown. As a positive control, PMA and ionomycin induced IFN-γ production from 58% of NK cells and 10% of non-NK cells (top row, left, Fig. 3C). Our data demonstrate that the source of robust IFN-γ production from rNKp30-activated splenocytes was a subset of NK cells (CLH-8 treatment, top row, right). There was no increase in IFN-γ staining over background on non-NK cell population treated with CLH-8. Since NK cells are the source of rNKp30-mediated IFN-γ production, we determined the range of IFN-γ produced from rNKp30+ NK cells from six experiments to be 11–47 ng IFN-γ/106 rNKp30+ splenic NK cells (n = 6). Activation of rNKp46 from PBMC, which is expressed by all NK cells, consistently induced IFN-γ from a larger subset of NK cells compared to activation by rNKp30 (Fig. 3C, bottom row, right) (n = 3).
Blood and liver NK cells express different levels of rNKp30
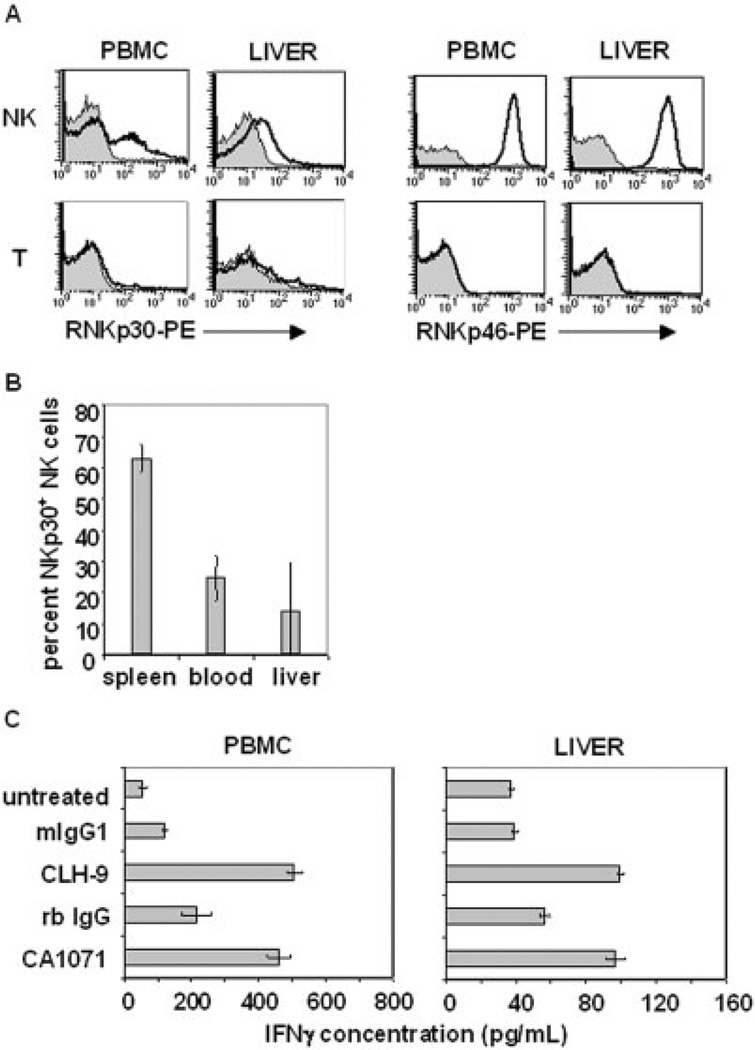
We examined normal peripheral blood and liver NK cells for rNKp30 expression. The liver was of interest as rNKp30 was initially cloned from a rejecting liver allograft [23]. NK cells were gated as previously shown in Fig. 3A. Similar to splenocytes, peripheral blood NK cells also contained a subset of rNKp30+ cells (Fig. 4A, upper left). Interestingly, liver rNKp30+ NK cells demonstrate low levels of rNKp30 as compared to the spleen and blood (Fig. 4A, upper left). As seen with splenic T cells, blood and liver T cells do not express rNKp30 or rNKp46 (Fig. 4A, bottom row). A greater proportion of splenic NK cells express rNKp30+ (63.1 ± 4.4%), compared to the proportion of rNKp30 + NK cells in the peripheral blood (24.6 ± 7.2%) and in the liver (14 ± 15.5%) (percentages indicate the mean of three experiments ± SD) (Fig. 4B). The expression of rNKp30 on liver NK cells was variable and the percent of liver NK cells expressing rNKp30 ranged from 5% to 32%. Interestingly, the median intensity of rNKp30 expression is highest on splenic NK cells, intermediate on blood NK cells, and the lowest on liver NK cells (Figs 3A, 4A). The decreased surface expression in the blood and liver is specific to rNKp30 as blood and liver NK cells maintain high NKRP1A (not shown) and rNKp46 levels (Fig. 4A, upper right), indicating there was no global down-regulation of surface receptors.
Fig. 4.
Peripheral blood and liver NK cells express rNKp30. (A) F344 rat PBMC and liver mononuclear cells were analyzed by flow cytometry for rNKp30 (left columns, open histograms) or rNKp46 (right columns, open histograms) using CLH-9 or WEN23 mAb, respectively. Isotype controls (filled histograms) and PE-conjugated secondary reagents were used. Gates were set on NK cells (NKRP1-FITChi αβTCR-PerCP−, upper panels) and T cells (αβTCR-FITC+, lower panels). Representative data from three to four separate experiments are shown. (B) The proportion of rNKp30+ cells in the total NK cell population was quantitated (n=3). rNKp30+ NK cells comprise 63.1 ± 4.4% of spleen NK cells, 24.6 ± 7.2% of peripheral blood NK cells, and 14 ± 15.5% of liver NK cells. (C) PBMC (left) and liver mononuclear cells (right) were cultured in media alone (untreated), or treated with platebound rNKp30 antibodies (CLH-9 or CA1071), or control antibodies (mIgG1 or rb IgG) for 24 h and supernatants were assessed for IFN-γ by ELISA.
To test rNKp30 function on blood and liver NK cells, mononuclear cells were treated with agonistic rNKp30 antibodies and assessed for IFN-γ secretion. PBMC produced significant IFN-γ levels when stimulated with either CLH-9 or CA1071 (Fig. 4C, left). In these assays, 12.8–14.4 ng IFN-γ/106 rNKp30+ NK cells was produced, which is at the lower end of the IFN-γ range produced from splenic rNKp30+ NK cells. Normal liver mononuclear cells also responded to anti-rNKp30 agonistic antibodies and produced IFN-γ, albeit at low levels (Fig. 4C, right). Although liver NK cells could produce significant amounts of IFN-γ in response to PMA and ionomycin (3.7 ± 0.34 ng/106 NK cells), in response to rNKp30 activation, only 0.08–1.2 ng IFN-γ/106 rNKp30+ liver NK cells was produced. Our data indicate that liver NK cells express low levels of rNKp30 and that rNKp30-mediated responses are reduced in the liver compared to the blood and spleen.
rNKp30 expression is increased during allo-activation
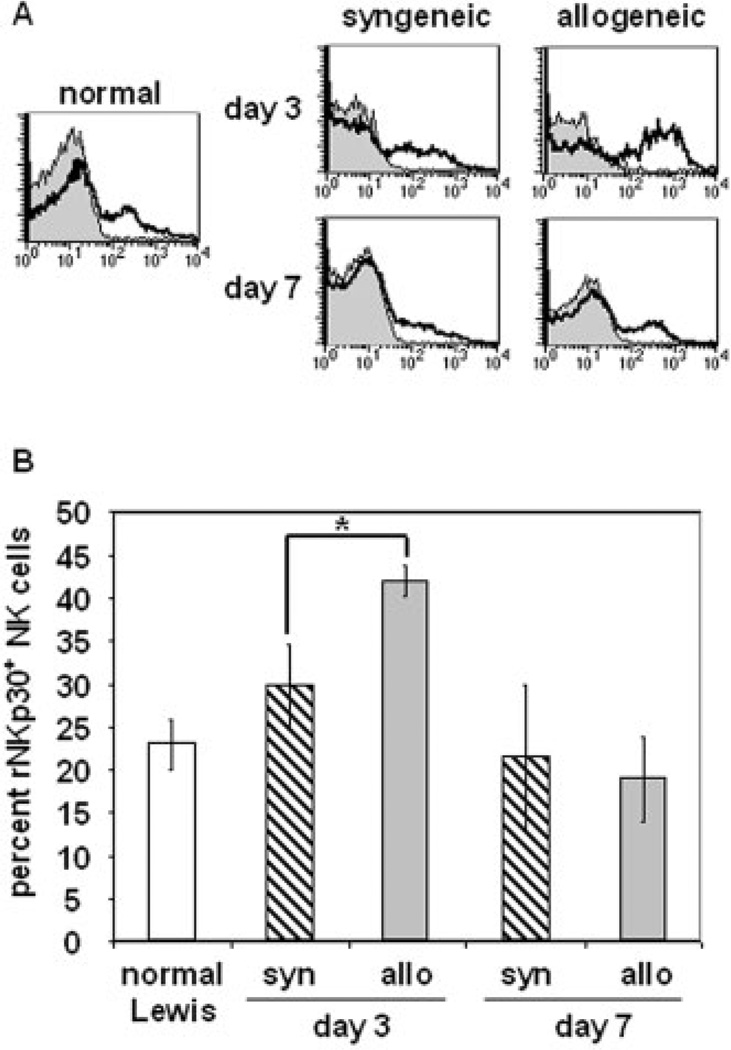
To examine rNKp30 expression during an in vivo immune response, we utilized a rat orthotopic liver transplant model where a dark agouti rat (DA) liver (RTlAa) was grafted into a Lewis recipient (RT1A1) [4]. In this fully MHC-mismatched combination, acute rejection occurs 9–13 days post-transplant as a result of cell-mediated processes. Depletion of NK cells prior to allogeneic liver transplantation resulted in a prolonged mean survival time and a significant decrease in serum IFN-γ levels [4]. To examine NKp30 expression following transplantation, PBMC were isolated from recipients of syngeneic and allogeneic liver grafts on days 3 and 7 post transplant, and from normal rats. rNKp30 expression was determined using the CLH-9 mAb and representative histogram plots of CLH-9 staining (open histograms) and isotype control staining (filled histograms) on all peripheral blood NK cells is shown (Fig. 5A). The profile of rNKp30 expression on PBMC from a normal Lewis recipient (Fig. 5A, left) is comparable to the rNKp30 expression profile of F344 rat PBMC (see Fig. 4A, B). In the PBMC of syngeneic graft recipients there was no significant difference in rNKp30 expression on days 3 or 7 post transplant (Fig. 5A, middle column). However, rNKp30 expression significantly increased on day 3 in allograft recipients and subsequently returned to normal levels by day 7 (Fig. 5A, right column). The increased proportion of the rNKp30+ NK subset is not a reflection of total rNKp30 + NK cell numbers increasing in the blood since the number of all NK cells in the blood actually decreased on day 3, likely due to NK cell recruitment to liver allografts [4]. It is intriguing that the increase in rNKp30 expression on day 3 in allograft recipients coincides with peak serum IFN-γ levels after alloactivation in this model [4]. These data suggest that early on after transplantation NKp30+ NK cells may be contributing to alloimmune responses.
Fig. 5.
The percent of rNKp30+ NK cells in the peripheral blood increases during allo-activation. PBMC were isolated from normal Lewis animals or from recipients of syngeneic (Lewis to Lewis) and allogeneic (DA to Lewis) liver grafts 3 and 7 days post transplant. rNKp30 expression on peripheral blood NK cells (NKRPlhi αβTCR−) was examined by flow cytometry. (A) NK cells from normal Lewis rat PBMC (left), and peripheral blood NK cells from syngeneic graft recipients (middle column) and allograft recipients (right column) were stained with the CLH-9 mAb (open histograms), and compared to staining with the isotype control antibody (filled histograms). Representative data are shown. (B) The mean percentage of rNKp30+ NK cells in the peripheral blood NK cell population was quantified and is reported ± SEM. The mean percent of rNKp30+ NK cells in normal Lewis rats is 23 ± 3% (white bar) (n=2). In syngeneic transplant recipients (striped bars), the mean percent of rNKp30+ NK cells is 29.9 ± 4.7% on day 3 (syn day 3, n=3), and 21.5 ± 8.5% on day 7 (syn day 7, n=2). In the allogeneic transplant group (gray bars), the mean percent of rNKp30+ NK cells increases to 41.9 ± 1.76% by day 3 (allo day 3, n=6), and decreases to 19 ± 4.9% by day 7 (allo day 7, n=3). Fisher's PLSD analysis determined significance as p≤ 0.05 (*p=0.0155).
The percentages of rNKp30+ NK cells in the peripheral blood of normal animals and in liver transplant recipients is shown in Fig. 5B. Approximately, 23 ± 3% of normal Lewis rat blood NK cells express rNKp30 (normal Lewis, n = 2). Following syngeneic transplantation, the mean percentage of blood rNKp30 + NK cells on day 3 is 29.9 ± 4.7% (syn day 3, n = 3). In recipients of allografts on day 3 (allo day 3, n=6), there is a marked and statistically significant (p = 0.0155) increase to 41.9 ± 1.76% in the percentage of blood NK cells expressing rNKp30. However, the increase in the proportion of rNKp30+ NK cells is transient since the mean percent returns to normal levels of 19 ± 4.93% by day 7 in allogeneic recipients (allo day 7, n = 3).
During the alloimmune response, host NK cells infiltrate liver allografts [4]. To determine whether rNKp30+ NK cells are recruited to allografts, we examined graft-infiltrating NK cells by flow cytometry on several days following transplantation. Although host NKRP1+ NKp46+ NK cells were found, we did not detect any significant increase in the percentage or expression intensity of surface rNKp30 on these NK cells compared to syngeneic and normal animals (not shown). It is possible that the rNKp30+ NK cell subset may not traffic into allografts. However, previous data from our laboratory showed that rNKp30 RNA is significantly increased in liver allografts compared to syngeneic and normal livers, suggesting that the NKp30+ NK subset is being recruited to the liver [23]. These data would suggest that surface NKp30 expression may be modulated on NK cells homing to the liver.
Discussion
We have demonstrated that NKp30 is a functional NK activation receptor in the rat. Our data show that rNKp30 can mediate important NK effector functions such as cytotoxicity and IFN-γ secretion. The increasing rNKp30+ NK cell subset in the peripheral blood of liver allograft recipients indicate that rNKp30 may be important during alloimmune responses. A subsequent key issue is to determine the true targets of NKp30 killing and the role of the receptor in the immune system. Rat NKp30, like its human homolog, may affect DC and influence antigen presentation. rNKp30-mediated IFN-γ responses could lead to multiple effects, such as maturation of APC [7], up-regulation of chemokines like IFN-γ-inducible protein-10 (IP-10) and monokine induced by IFN-γ, which recruit T and NK cells, and Th1 polarization of T cells.
Our analysis of rNKp30 reveals that it is expressed on a subset of splenic and blood NK cells, and to a lesser extent on liver NK cells. It remains to be determined if rNKp30 expression characterizes a functionally distinct NK cell subset. Furthermore, our data show that the spleen, blood, and liver express different percentages and levels of rNKp30, which is in stark contrast to other NK activation receptors, such as NKRP1A and NKp46, which are constitutively and brightly expressed in all three organs. The different amounts of rNKp30 expression and function in the spleen, blood, and liver suggest either preferential homing of rNKp30+ NK cells to certain organs and/or organ-specific regulation of receptor expression. There is precedence for the existence of NK cell subsets possessing different functional capabilities and homing patterns. For example, CD56bright human NK cells are more effective at producing cytokines, whereas CD56dim NK cells are better killers [2, 26]. Similarly, distinguishable homing pathways have been reported for the CD56+ NK subsets such that the lymph node recruits a higher proportion of CD56bright NK cells, while the spleen contains a higher proportion of CD56dim NK cells [27]. NK subsets in the rodent models are less well characterized and to date, no rodent homolog to the human CD56 subsets have been found.
NK cells localized in different compartments exhibit different phenotypes. An oligonucleotide array analysis reported 150 genes were expressed differentially between blood and hepatic rat NK cells [28]. For example, liver NK cells expressed higher levels of IP-10 and CCR5 than blood NK cells. Another report demonstrated that NK cells recruited to the liver by macrophage inflammatory protein-1α were required for IFN-γ-mediated mCMV resistance [6]. This suggested that resident liver NK cells were insufficient to mount an effective anti-viral response, perhaps due to impaired expression of activation receptors and/or a lower potential to produce IFN-γ. This trend would be consistent with our data indicating that the liver has reduced rNKp30-mediated responses compared to the spleen and blood. However, evidence suggests that hepatic NK cells have increased cytotoxic functions compared to splenic and blood NK cells [28, 29]. It is clear that NK cells localized in different organs require further characterization.
It has been shown that NKp30 surface expression can be modulated. It is suggested that humans infected with CMV or HIV, and patients with acute myeloid leukemia (AML) have impaired NCR surface expression, but the mechanism(s) remains unknown [30–32]. Our data would suggest that NK cells in the liver have low rNKp30 surface expression. The liver may express or secrete ligands for rNKp30 and upon ligand engagement, rNKp30 may be internalized. It has been shown that ligation of another activation receptor, NKG2D, can lead to reduced NKG2D surface expression and impaired function [33, 34]. Additionally, cytokines can modulate NK receptor expression. In vitro evidence demonstrated that transforming growth factor-β2 (TGF-αβ1) inhibited hNKp30, but not hNKp46 expression [35]. HNKp30 expression can be up-regulated on peripheral blood, tonsil and lymph node NK cells after exposure to IL-2 [27]. It is possible that molecules expressed in an organ-specific manner and/or during an immune response affect NKp30 expression.
Studying NKp30 in a rodent model facilitates in vivo experimentation and studies of controlled disease models. Our current understanding of human NKp30 has been acquired from in vitro investigations. However, in vivo disease models will be required to answer questions about NKp30 more definitively. We have initiated studies examining NKp30 during an in vivo alloimmune response.
NK cells play an important role in solid-organ transplantation. It was previously demonstrated in CD28−/− mice that NK cells were required for acute rejection of cardiac transplants [36]. In addition, NK cells mediate cardiac allograft vasculopathy during chronic rejection [37]. We formerly showed that NK cell depletion prolonged the mean survival time of liver allograft recipients, and confirmed that NK cells produce abundant amounts of IFN-γ [4]. We now demonstrate a marked and transient increase in the proportion of rNKp30+ NK cells in the total blood NK cell pool early during the alloimmune response
At least three possibilities exist for this increase in rNKp30 expression. One is that the rNKp30 receptor is up-regulated on blood NK cells after alloactivation. Another possibility is that rNKp30 ligation may induce increased proliferation of rNKp30+ NK cells as is seen upon ligation of Ly49H on NK cells [38]. A third possibility is that mobilization of the rNKp30+ NK cell subset during an immune response is unique. Chemokines expressed by the allograft may be preferentially recruiting the rNKp30+ NK cell subset into the peripheral blood. Interestingly, there is evidence that NK cells activated through different activation or inhibitory Ly49 receptors respond differently to chemokines [39]. Other evidence suggests that TGF- βl can up-regulate the chemokine receptor CXCR3 and down-regulate NKp30 [35, 40]. Thus, the cytokine environment may influence recruitment and/or NK receptor expression. Further examination of the relationship between NK and chemokine receptors in grafts may help elucidate if and how cells are selectively recruited.
Our study has shown that rNKp30 is a functional and unique NK activation receptor on a subset of NK cells. Further studies that characterize NK cell subsets, and studies to understand the regulation of NK receptors during homeostasis and during an immune response are warranted. Finally, additional investigation of rNKp30 in vivo will elucidate important roles for this receptor.
Materials and methods
Animals
Male Fischer 344 (5–12 weeks), and Lewis rats (11–12 weeks) were purchased from either Charles River Laboratories (Wilmington, MA) or Harlan (Indianapolis, IN) and Dark Agouti rats (DA) (11–12 weeks) were purchased from Harlan. All animals were housed in accordance with institutional animal care. The Institutional Animal Care and Use Committee (IACUC) at Stanford University approved all animal protocols.
Mononuclear cell preparation
Mononuclear cells were isolated from the blood, spleen, and liver using Ficoll-hypaque (Amersham Pharmacia, Piscataway, NJ). Isolation of liver mononuclear cells was performed as previously described [4, 41]. Briefly, livers were perfused through the portal vein with PBS to remove residual blood. Livers were then perfused with a collagenase solution of 10% FBS (Cellgro), 0.05% collagenase (Sigma, St. Louis, MO), 5000 U DNaseI (Sigma) in Hanks' buffered salt solution (HBSS, Invitrogen, Carlsbad, CA) and incubated for 10 min at room temperature. The perfused liver tissue was minced, incubated for 30 min at 37°C, and filtered through a nylon mesh (100-µm pore size) to produce a cell suspension.
Cell transfection
The spontaneous rat NK lymphoma line RNK16 was a generous gift from Dr. Mary Nakamura (UCSF, CA) and was maintained in complete RPMI (RPMI with 10% FBS, 1% penicillin/ streptomycin) with 50 µM 2-mercaptoethanol (2-ME) (Sigma). RNK16 cells were transfected by electroporation with a linearized pCDNA3.1a construct (Invitrogen) containing an rNKp30 cassette. Transfection and selection were carried out essentially as described by Ryan et al. [42] using electroporation at 120 mV, 960 µF with a BioRad (Hercules, CA) electroporator. Cells were plated at 105 cells/mL in complete RPMI with 10 mM HEPES (Invitrogen) and 1.0 mg/mL neomycin (Invitrogen). Stably transfected clonal populations of RNK16 cells uniformly expressing rNKp30 (clones 2B3, F10, and F11) as determined by flow cytometry were used for expression and functional studies.
Generation of anti-rat NKp30 mAb
Mouse anti-rat NKp30 mAb were generated against an rNKp30-Fc fusion protein. The immunogen was produced by cloning the extracellular domain of rNKp30 fused to the human IgG1 Fc domain (a generous gift from Dr. David Cosman, Amgen, Seattle, WA) into a pCDNA3.1a construct. The rNKp30-Fc construct was stably transfected into 293F cells maintained in serum-free media (Invitrogen). BALB/c mice were immunized with purified fusion protein and hybridomas were generated (Zymed Laboratories, South San Francisco, CA). Hybridoma supernatants were screened by ELISA and further screened by flow cytometry for specific staining of rNKp30-transfected RNK16 cells. All of the mAb (CLH-1, CLH-2, CLH-3, CLH-4, CLH-5, CLH-6, CLH-7, CLH-8, CLH-9) are mouse IgG1,κ isotype. mAb were purified from ascites fluids by affinity chromatography against NKp30-Fc bound to Sulfo-link coupling gel (Pierce, Rockford, IL) and biotinylated using EZ-Link Sulfo-NHS-LC-Biotin (Pierce).
Flow cytometry
To screen the rNKp30 mAb and to examine the expression of rNKp30 on rNKp30-transfected RNK16 cells, we labeled parental RNK16 cells with 10 µM 5-chloromethylfluorescein diacetate (CMFDA) according to manufacturer instructions (Molecular Probes, Eugene, OR) and mixed the CMFDA-labeled parental RNK16 cells with rNKp30+ RNK16 Clone F11 cells at a 1:1 ratio. The mixed cell populations were stained with anti-rNKp30 mAb (0.25–0.5 µg/106 cells), rabbit anti-rNKp30 pAb CA1071 [23] or isotype control antibodies for 30 min on ice. Cells were washed with cold FACS buffer (PBS, 1% FBS, 0.1% sodium azide). RPE-conjugated donkey anti-rabbit or anti-mouse F(ab′)2 antibodies (1:200; Jackson Immunoresearch, West Grove, PA) were used as secondary reagents. For primary cell staining, biotinylated mAb that detect rNKp30 or rNKp46 (WEN23 mAb, a generous gift from Dr. Erik Dissen, University of Oslo, Oslo, Norway), or isotype controls were used and followed by streptavidin-RPE (1:400) (BD Pharmingen, La Jolla, CA). Where indicated, the following antibodies, NKRP1A-FITC (clone 10/78, Serotec, Raleigh, NC), αβTCR-PerCP (clone R73, BD Pharmingen), αβTCR-FITC (clone R73, Serotec), and isotype controls (Dako, Carpinteria, CA) were also used to stain primary cells. Cells were washed, resuspended in 0.5 µg/mL PI, and analyzed using a FACScan flow cytometer and CellQuest software (Becton Dickinson, San Jose, CA). The percent of rNKp30+ NK cells were calculated after subtracting background staining from isotype controls, and were reported as the mean percent ± SD for normal animals.
To detect intracellular cytokines, splenocytes or PBMC were plated at 2.5 × 106 cells/mL in 96-well flat-bottom plates coated with immobilized anti-rNKp30 (CLH-8), anti-rNKp46 (WEN23) or control antibodies (10–12 µg/mL), or the cells were treated with PMA (10 ng/mL) and ionomycin (0.5 µg/ mL). Cells were cultured in RPMI complete with 50 µM 2-ME, 10 U/mL recombinant human IL-2 (NCI, Bethesda, MD), 1 mM sodium pyruvate (Invitrogen) and 10 mM HEPES for 11–12 h. Brefeldin A (10 µg/mL, Epicentre, Madison, WI) was added for the last 6.5 h. Cells were washed, stained with FITC-conjugated anti-NKRPlA antibody or isotype controls as above, and fixed with CytoFix (BD Pharmingen). Following permeabilization with 0.4% saponin in FACS buffer, IFN-γ was detected with an RPE-conjugated anti-rat IFN-γ antibody (clone DB1; BD Pharmingen).
Western blotting
Washed parental RNK16 and rNKp30-transfected RNK16 cells were lysed in ice-cold lysis buffer (50 mM Tris pH 7.4,1% NP- 40, 0.5% DOC, 150 mM NaCl, 0.5 mM EDTA, 2 mM PMSF, 0.5 mg/mL aprotinin, 0.5 mg/mL leupeptin, 0.05 mg/mL pepstatin) for 30 min. Whole cell lysates (300 µg/lane) were resolved on a 12% SDS-PAGE gel under reducing conditions. Proteins were transferred to nitrocellulose membranes. After incubation with blocking solution (5% milk, TBS, 0.1% Tween 20), blots were probed overnight with either CA1071 pAb (1 µg/mL) or CLH-1–9 mAb (2 µg/mL). Bound antibodies were detected using either HRP-conjugated goat anti-rabbit (1:10 000) or HRP-conjugated donkey anti-mouse secondary antibodies (1:10 000) (Jackson Immunoresearch), followed by enhanced chemiluminescence (Amersham Pharmacia). Blots were reprobed with an anti-β actin mAb (Sigma) followed by the anti-mouse-HRP secondary antibody.
Cytotoxicity assays
Redirected lysis assays were performed in which apoptosis was assessed using the JAM method to measure DNA fragmentation [43]. Effector cells were plated at 20 × 106 cells/mL and incubated with 0–6 µg/mL of antibodies for 20 min. Antibodies used were anti-rNKp30 pAb (CA1071) and anti-rNKp30 mAb (CLH-9), control prebleed rb IgG, anti-rat CD30 pAb, anti-NKRPlA (Clone 10/78), anti-CD8a (OX-8) mAb (Serotec), and control mIgG1 (MOPC31) (BD Pharmingen). Effector cells were plated as twofold serial dilutions in triplicate in 96-well round-bottom plates. Target FcR+ P815 mouse mastocytoma cells were cultured in complete DMEM and labeled with [3H] thymidine (5 µCi/mL, Perkin-Elmer, Boston, MA) for 8 h. Washed target cells were plated with and without effector cells for 4 h at 104 target cells/well. Cells were harvested onto a glass fiber filtermat (Wallac, Turku, Finland) using a Tomtec harvester 96 MACHII (Orange, CT) and a Wallacl205 betaplate reader measured radioactivity. Percent specific killing was calculated as follows: (cpm of spontaneous killing without effector cells – cpm of experimental killing) / cpm of spontaneous killing) × 100.
Measurement of IFN-γ in culture supernatants
RNK16 cell lines (0.5 × 106 cells/mL) were treated for 24 h with plate-bound antibodies (10–12 µg/mL) or PMA and ionomycin as above. Primary mononuclear cells (2.5 × 106 cells/mL or 5 × 105 cells/well) were plated in complete RPMI with 50 µM 2-ME, 1 mM sodium pyruvate (Invitrogen), and 10 mM HEPES (Invitrogen) with 10–100 U/ mL human IL-2 (NCI). Splenocytes were cultured for 11–12 h (IFN-γ levels peak near 11 h and remain high for 24 h), PBMC and liver mononuclear cells were cultured for 24 h, and supernatants were recovered. Supernatants were plated in triplicate, and IFN-γ levels were determined by ELISA using an rb anti-mouse/rat IFN-γ coating antibody and a mouse anti-mouse/rat IFN-γ-biotin (clone DB-1) antibody (Biosource International, Camarillo, CA). IFN-γ ELISA assays were carried out according to manufacturer protocols. ELISA data was reported as the mean (pg/mL) of the triplicate ± SD.
Orthotopic liver transplantation
Orthotopic liver transplantation was performed as previously described [4]. In the syngeneic group, Lewis animals were grafted with Lewis livers. In the allogeneic group, Lewis recipients were grafted with DA livers. Surgeries were performed under general anesthesia with isoflurane (Abbott Laboratories, North Chicago, IL). No immunosuppressive drugs were used in this study. Percentages of rNKp30+ NK cells were reported as the mean percent ± SEM. Statistical analysis of multiple groups was calculated using analysis of variance (ANOVA) with p≤0.05 determined as statistically significant. When significance was found Fisher's protected least significance difference (PLSD) analysis was conducted.
Acknowledgements
We are grateful to Dr. Karine Piard-Ruster, Guadalupe Rodriguez, and Manisha Bahl for their excellent technical assistance. We also thank Dr. Jay Campisi for statistical assistance, and Dr. Stacie Lambert for her review of our manuscript. This study was supported by NIH R01 A144095 and the Roche Organ Transplantation Research Foundation. Christine L. Hsieh was supported by NIH Immunology Training Grant 5 T32 AI07290.
Abbreviations
- DA
dark agouti rat
- iDC
immature DC
- NC
natural cytotoxicity receptor
- pAb
polyclonal antibody
- r
rat
- rNKp30
rat NKp30
References
- 1.Cerwenka A, Lanier LL. Natural killer cells, viruses and cancer. Nat. Rev. Immunol. 2001;1:41–49. doi: 10.1038/35095564. [DOI] [PubMed] [Google Scholar]
- 2.Raulet DH. Interplay of natural killer cells and their receptors with the adaptive immune response. Nat. Immunol. 2004;5:996–1002. doi: 10.1038/ni1114. [DOI] [PubMed] [Google Scholar]
- 3.Salazar-Mather TP, Hamilton TA, Biron CA. A chemokine-to-cytokine-to-chemokine cascade critical in antiviral defense. J. Clin. Invest. 2000;105:985–993. doi: 10.1172/JCI9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Obara H, Nagasaki K, Hsieh CL, Ogura Y, Esquivel CO, Martinez OM, Krams SM. IFN-gamma, produced by NK cells that infiltrate liver allografts early after transplantation, links the innate and adaptive immune responses. Am. J. Transplant. 2005;5:2094–2103. doi: 10.1111/j.1600-6143.2005.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin-Fontecha A, Thomsen LL, Brett S, Gerard C, Lipp M, Lanzavecchia A, Sallusto F. Induced recruitment of NK cells to lymph nodes provides IFN-gamma for T(H)1 priming. Nat. Immunol. 2004;5:1260–1265. doi: 10.1038/ni1138. [DOI] [PubMed] [Google Scholar]
- 6.Salazar-Mather TP, Orange JS, Biron CA. Early murine cytomegalovirus (MCMV) infection induces liver natural killer (NK) cell inflammation and protection through macrophage inflammatory protein 1alpha (MlP-1alpha)-dependent pathways. J. Exp. Med. 1998;187:1–14. doi: 10.1084/jem.187.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vitale M, Delia Chiesa M, Carlomagno S, Pende D, Arico M, Moretta L, Moretta A. NK-dependent DC maturation is mediated by TNFalpha and IFNgamma released upon engagement of the NKp30 triggering receptor. Blood. 2005;106:566–571. doi: 10.1182/blood-2004-10-4035. [DOI] [PubMed] [Google Scholar]
- 8.Gerosa F, Baldani-Guerra B, Nisii C, Marchesini V, Carra G, Trinchieri G. Reciprocal activating interaction between natural killer cells and dendritic cells. J. Exp. Med. 2002;195:327–333. doi: 10.1084/jem.20010938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piccioli D, Sbrana S, Melandri E, Valiante NM. Contact-dependent stimulation and inhibition of dendritic cells by natural killer cells. J. Exp. Med. 2002;195:335–341. doi: 10.1084/jem.20010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munz C, Steinman RM, Fujii S. Dendritic cell maturation by innate lymphocytes: coordinated stimulation of innate and adaptive immunity. J. Exp. Med. 2005;202:203–207. doi: 10.1084/jem.20050810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bottino C, Castriconi R, Moretta L, Moretta A. Cellular ligands of activating NK receptors. Trends Immunol. 2005;26:221–226. doi: 10.1016/j.it.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Moretta L, Bottino C, Pende D, Vitale M, Mingari MC, Moretta A. Human natural killer cells: Molecular mechanisms controlling NK cell activation and tumor cell lysis. Immunol. Lett. 2005;100:7–13. doi: 10.1016/j.imlet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Cantoni C, Bottino C, Vitale M, Pessino A, Augugliaro R, Malaspina A, Parolini S, et al. NKp44, a triggering receptor involved in tumor cell lysis by activated human natural killer cells, is a novel member of the immunoglobulin superfamily. J. Exp. Med. 1999;189:787–796. doi: 10.1084/jem.189.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pessino A, Sivori S, Bottino C, Malaspina A, Morelli L, Moretta L, Biassoni R, Moretta A. Molecular cloning of NKp46: a novel member of the immunoglobulin superfamily involved in triggering of natural cytotoxicity. J. Exp. Med. 1998;188:953–960. doi: 10.1084/jem.188.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pende D, Parolini S, Pessino A, Sivori S, Augugliaro R, Morelli L, Marcenaro E, et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J. Exp. Med. 1999;190:1505–1516. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sivori S, Parolini S, Marcenaro E, Castriconi R, Pende D, Millo R, Moretta A. Involvement of natural cytotoxicity receptors in human natural killer cell-mediated lysis of neuroblastoma and glioblastoma cell lines. J. Neuroimmunol. 2000;107:220–225. doi: 10.1016/s0165-5728(00)00221-6. [DOI] [PubMed] [Google Scholar]
- 17.Neville MJ, Campbell RD. A new member of the Ig superfamily and a V-ATPase G subunit are among the predicted products of novel genes close to the TNF locus in the human MHC. J. Immunol. 1999;162:4745–4754. [PubMed] [Google Scholar]
- 18.Arnon TI, Lev M, Katz G, Chernobrov Y, Porgador A, Mandelboim O. Recognition of viral hemagglutinins by NKp44 but not by NKp30. Eur. J. Immunol. 2001;31:2680–2689. doi: 10.1002/1521-4141(200109)31:9<2680::aid-immu2680>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 19.Arnon TI, Achdout H, Levi O, Markel G, Saleh N, Katz G, Gazit R, et al. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat. Immunol. 2005;6:515–523. doi: 10.1038/ni1190. [DOI] [PubMed] [Google Scholar]
- 20.Ferlazzo G, Tsang ML, Moretta L, Melioli G, Steinman RM, Munz C. Human dendritic cells activate resting natural killer (NK) cells and are recognized via the NKp30 receptor by activated NK cells. J. Exp. Med. 2002;195:343–351. doi: 10.1084/jem.20011149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spaggiari GM, Carosio R, Pende D, Marcenaro S, Rivera P, Zocchi MR, Moretta L, Poggi A. NK cell-mediated lysis of autologous antigen-presenting cells is triggered by the engagement of the phosphatidylinositol 3-kinase upon ligation of the natural cytotoxicity receptors NKp30 and NKp46. Eur. J. Immunol. 2001;31:1656–1665. doi: 10.1002/1521-4141(200106)31:6<1656::aid-immu1656>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 22.Hollyoake M, Campbell RD, Aguado B. NKp30 (NCR3) is a pseudogene in 12 inbred and wild mouse strains, but an expressed gene in Mus caroli . Mol. Biol. Evol. 2005;22:1661–1672. doi: 10.1093/molbev/msi162. [DOI] [PubMed] [Google Scholar]
- 23.Hsieh CL, Ogura Y, Obara H, Ali UA, Rodriguez GM, Nepomuceno RR, Martinez OM, Krams SM. Identification, cloning, and characterization of a novel rat natural killer receptor, RNKP30: a molecule expressed in liver allografts. Transplantation. 2004;77:121–128. doi: 10.1097/01.TP.0000110423.27977.6F. [DOI] [PubMed] [Google Scholar]
- 24.Backman-Petersson E, Miller JR, Hollyoake M, Aguado B, Butcher GW. Molecular characterization of the novel rat NK receptor 1C7. Eur. J. Immunol. 2003;33:342–351. doi: 10.1002/immu.200310008. [DOI] [PubMed] [Google Scholar]
- 25.Westgaard IH, Berg SF, Vaage JT, Wang LL, Yokoyama WM, Dissen E, Fossum S. Rat NKp46 activates natural killer cell cytotoxicity and is associated with FcepsilonRIgamma and CD3zeta. J. Leukoc. Biol. 2004;76:1200–1206. doi: 10.1189/jlb.0903428. [DOI] [PubMed] [Google Scholar]
- 26.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 27.Ferlazzo G, Thomas D, Lin SL, Goodman K, Morandi B, Muller WA, Moretta A, Munz C. The abundant NK cells in human secondary lymphoid tissues require activation to express killer cell Ig-like receptors and become cytolytic. J. Immunol. 2004;172:1455–1462. doi: 10.4049/jimmunol.172.3.1455. [DOI] [PubMed] [Google Scholar]
- 28.Vermijlen D, Seynaeve C, Luo D, Kruhoffer M, Eizirik DL, Orntoft TF, Wisse E. High-density oligonucleotide array analysis reveals extensive differences between freshly isolated blood and hepatic natural killer cells. Eur. J. Immunol. 2004;34:2529–2540. doi: 10.1002/eji.200324712. [DOI] [PubMed] [Google Scholar]
- 29.Vermijlen D, Luo D, Froelich CJ, Medema JP, Kummer JA, Willems E, Braet F, Wisse E. Hepatic natural killer cells exclusively kill splenic/blood natural killer-resistant tumor cells by the perforin/ granzyme pathway. J. Leukoc. Biol. 2002;72:668–676. [PubMed] [Google Scholar]
- 30.Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 2004;104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- 31.De Maria A, Fogli M, Costa P, Murdaca G, Puppo F, Mavilio D, Moretta A, Moretta L. The impaired NK cell cytolytic function in viremic HIV-1 infection is associated with a reduced surface expression of natural cytotoxicity receptors (NKp46, NKp30 and NKp44) Eur. J. Immunol. 2003;33:2410–2418. doi: 10.1002/eji.200324141. [DOI] [PubMed] [Google Scholar]
- 32.Costello RT, Sivori S, Marcenaro E, Lafage-Pochitaloff M, Mozziconacci MJ, Reviron D, Gastaut JA, et al. Defective expression and function of natural killer cell-triggering receptors in patients with acute myeloid leukemia. Blood. 2002;99:3661–3667. doi: 10.1182/blood.v99.10.3661. [DOI] [PubMed] [Google Scholar]
- 33.Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 2002;419:734–738. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 34.Ogasawara K, Hamerman JA, Hsin H, Chikuma S, Bour-Jordan H, Chen T, Pertel T, et al. Impairment of NK cell function by NKG2D modulation in NOD mice. Immunity. 2003;18:41–51. doi: 10.1016/s1074-7613(02)00505-8. [DOI] [PubMed] [Google Scholar]
- 35.Castriconi R, Cantoni C, Della Chiesa M, Vitale M, Marcenaro E, Conte R, et al. Transforming growth factor {beta}1 inhibits expression of NKp30 and NKG2D receptors: Consequences for the NKmediated killing of dendritic cells. Proc. Natl. Acad. Sci. USA. 2003;100:4120–4125. doi: 10.1073/pnas.0730640100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maier S, Tertilt C, Chambron N, Gerauer K, Huser N, Heidecke CD, Pfeffer K. Inhibition of natural killer cells results in acceptance of cardiac allografts in CD28−/− mice. Nat. Med. 2001;7:557–562. doi: 10.1038/87880. [DOI] [PubMed] [Google Scholar]
- 37.Uehara S, Chase CM, Kitchens WH, Rose HS, Colvin RB, Russell PS, Madsen JC. NK cells can trigger allograft vasculopathy: the role of hybrid resistance in solid organ allografts. J. Immunol. 2005;175:3424–3430. doi: 10.4049/jimmunol.175.5.3424. [DOI] [PubMed] [Google Scholar]
- 38.Dokun AO, Kim S, Smith HR, Kang HS, Chu DT, Yokoyama WM. Specific and nonspecific NK cell activation during virus infection. Nat. Immunol. 2001;2:951–956. doi: 10.1038/ni714. [DOI] [PubMed] [Google Scholar]
- 39.Inngjerdingen M, Rolstad B, Ryan JC. Activating and inhibitory Ly49 receptors modulate NK cell chemotaxis to CXC chemokine ligand (CXCL) 10 and CXCL12. J. Immunol. 2003;171:2889–2895. doi: 10.4049/jimmunol.171.6.2889. [DOI] [PubMed] [Google Scholar]
- 40.Inngjerdingen M, Damaj B, Maghazachi AA. Expression and regulation of chemokine receptors in human natural killer cells. Blood. 2001;97:367–375. doi: 10.1182/blood.v97.2.367. [DOI] [PubMed] [Google Scholar]
- 41.Ogura Y, Martinez OM, Villanueva JC, Tait JF, Strauss HW, Higgins JPT, Tanaka K, et al. Apoptosis and Allograft Rejection in the Absence of CD8+ T Cells. Transplantation. 2001;71:1827–1834. doi: 10.1097/00007890-200106270-00020. [DOI] [PubMed] [Google Scholar]
- 42.Ryan JC, Niemi EC, Nakamura MC. Functional analysis of natural killer cell receptors in the RNK-16 rat leukemic cell line. In: Campbell KS, Colonna M, editors. Natural Killer Cell Protocols. Totowa: Humana Press; 1999. pp. 283–295. [DOI] [PubMed] [Google Scholar]
- 43.Matzinger P. The JAM test. A simple assay for DNA fragmentation and cell death. J. Immunol. Methods. 1991;145:185–192. doi: 10.1016/0022-1759(91)90325-a. [DOI] [PubMed] [Google Scholar]