Abstract
Background
Altered expression of serotonin-1A (5-HT1A) receptors, both presynaptic in the raphe nuclei and in limbic and cortical target areas, has been implicated in mood disorders such as major depression and anxiety. Within the 5-HT1A receptor gene (HTR1A), a powerful dual repressor element (DRE) is regulated by two protein complexes: Freud-1/CC2D1A and a second, unknown repressor. Here we identify human Freud-2/CC2D1B, a Freud-1 homologue, as the second repressor.
Methods
Freud-2 distribution was examined using Northern and Western blot, RT-PCR, immunohistochemistry/immunofluorescence; Freud-2 function was examined by electrophoretic mobility shift, reporter assay and Western blot.
Results
Freud-2 RNA was widely distributed in brain and peripheral tissues. Freud-2 protein was enriched in the nuclear fraction of human prefrontal cortex and hippocampus, but was weakly expressed in the dorsal raphe nucleus. Freud-2 immunostaining was co-localized with 5-HT1A receptors, neuronal and glial markers. In prefrontal cortex, Freud-2 was expressed at similar levels in control and depressed male subjects. Recombinant hFreud-2 protein bound specifically to 5′ or 3′ human DRE adjacent to the Freud-1 site. Human Freud-2 showed strong repressor activity at the human 5-HT1A or heterologous promoter in human HEK293 5-HT1A-negative cells and neuronal SK-N-SH cells, a model of post-synaptic 5-HT1A receptor-positive cells. Furthermore siRNA knockdown of endogenous hFreud-2 expression de-repressed 5-HT1A promoter activity and increased levels of 5-HT1A receptor protein in SK-N-SH cells.
Conclusion
Human Freud-2 binds to the 5-HT1A DRE and represses the human 5-HT1A receptor gene to regulate its expression in non-serotonergic cells and neurons.
Keywords: 5-HT1A receptor, transcription factor, raphe, polymorphism, anxiety, major depressive disorder, serotonin receptors
Introduction
The serotonin-1A receptor (5-HT1A) is expressed presynaptically as an autoreceptor in raphe nuclei and postsynaptically in the limbic system including lateral septum, hippocampus, amygdala, and entorhinal cortex (1; 2) and is implicated in regulation of the serotonin system and of mood and emotion. Reductions in 5-HT1A receptor expression or activity are observed in patients with anxiety, major depression or suicide victims (3–6). Down-regulation of postsynaptic 5-HT1A receptors in the hippocampus and prefrontal cortex is implicated in schizophrenia, major depression, and type I bipolar disorder (7; 8). Genetic rescue studies indicate that early post-natal restoration of forebrain 5-HT1A receptors restores normal anxiety-like behaviour in 5-HT1A−/− mice (9). Moreover, postsynaptic 5-HT1A receptors are a potential target of antidepressant drugs (10), implicating the level of expression of postsynaptic 5-HT1A receptors in the pathophysiology and treatment of mood disorders.
In order to identify the mechanisms of transcriptional regulation of the 5-HT1A gene, others and we have investigated the 5-HT1A promoter region. The 5-HT1A proximal promoter contains a series of GC-rich MAZ/Sp1-binding sequences that drive strong expression in all cell types, which is silenced by upstream repressor elements (11; 12). In the rat 5-HT1A promoter we identified a 31-bp dual repressor element (DRE) that is located between −1555/−1524 bp from the translation initiation codon that strongly silences the promoter (13). The human 5-HT1A gene contains two tandem imperfect repeats of the DRE (−1624 to −1570 bp) with 71% nucleotide identity to the rat 5-HT1A DRE and displaying similar silencer activity (14; 15). The DRE is composed of a 5′ 14-bp element (FRE) and adjacent 3′-element (TRE). In post-synaptic 5-HT1A-expressing neuronal cells or 5-HT1A-negative cells, two protein complexes bound to the DRE, and deletion of the entire DRE was required to de-repress the gene. However, in raphe cells a single complex bound the DRE and mutation of the FRE blocked this complex and completely de-repressed the 5-HT1A promoter. By yeast one-hybrid screen we identified a novel transcription factor named Freud-1 (FRE Under Dual repression binding protein-1)/CC2D1A (Coiled-coil/C2-Domain-1A) that interacts with FRE and represses the 5-HT1A promoter in raphe RN46A cells (16). However the identity of the second protein complex that binds to the 5-HT1A-TRE has remained unknown.
In this study we report a new Freud-1 homologue, Freud-2/CC2D1B, which binds to the 5-HT1A-DRE at distinct sites that overlap with the Freud-1 site. Freud-2 negatively regulates 5-HT1A gene transcription via the DRE, and depletion of Freud-2 increased 5-HT1A transcription and receptor expression in a post-synaptic 5-HT1A-expressing cell model. Freud-2 staining was enriched in hippocampus and prefrontal cortex, but very sparse in the dorsal raphe nucleus. These data indicate that in the brain, unlike Freud-1, Freud-2 regulates post-synaptic rather than presynaptic 5-HT1A expression.
Materials and Methods
PCR and Plasmids
A 2.6-kb fragment of human Freud-2 cDNA including the complete coding sequence was amplified using specific primers 5-CCGGAATTCC GGATGCCAGG GCCAAGACCT CG-3′, 5′-CCGCTCGAGC GGCAGGCCCC GAGGCTCCAG GACC-3′ from a human brain cDNA library (Clontech). PCR products were gel-purified, subcloned in pGEMT-easy vector (Promega, Madison, WI) and subcloned into EcoRI/XhoI-cut pcDNA3 (Invitrogen, Burlington, Ontario, Canada) or pGEX-4T-1 (Amersham Bioscience). Human 5-HT1A promoter constructs were described previously (14). All constructs were purified by maxiprep kit (Sigma), quantified spectrophotometrically, and verified by DNA sequence analysis.
Freud-2 protein expression
Escherichia coli BL21cells were transformed with pGEX-4T-1-Freud-2 to express GST-Freud-2, and induced with 0.1 M isopropyl-β-D-thiogalactopyranoside. Bacteria were pelleted (3000xg, 4°C, 20 min) and recombinant proteins purified from pellets using B-PER GST Fusion Protein Purification Kit (Pierce, IL, USA) and stored at −80°C.
Cell culture and transfection
Human embryonic kidney (HEK) 293 cells and SK-N-SH human neuroblastoma cells were cultured and transfected as described previously (13; 14; 17). HEK293 cells were transfected by calcium phosphate co-precipitation (18), with 20μg luciferase reporter construct and 5μg pCMVβgal per 10-cm plate. For co-transfections total DNA was 25μg: 10μg of luciferase plasmid, indicated amounts of other constructs or empty vector, and 0.1 μg pCMVβgal to normalize transfection efficiency. SK-N-SH cells were subcultured into 6-well plates and transfected at 50–60% confluency with 1:1.5 ratio of plasmid:lipofectamine 2000 reagent (Invitrogen) using 7.5–10 μg/plate of luciferase and Freud-2 plasmids or empty vector with 2 μg/plate pCMVβgal. For reporter assays, triplicate samples after 48h of transfection were washed three times with ice-cold PBS and extracted with 150μl reporter lysis buffer (Promega). Supernatants were collected, assayed for luciferase activity using Spectramax M2 (Molecular Devices) luminometer and analyzed using Softmax Pro 4.8 software. Activities from at least three independent experiments was based on the ratio of luciferase/βgalactosidase activity and normalized to vector-transfected extracts. Data are presented as mean± SEM. Statistical significance was evaluated using two tailed unpaired t test.
Electrophoretic mobility shift assay
Complementary human DRE oligonucleotides were labelled with [α-32P]-dCTP using Klenow DNA polymerase (13) and probe (60,000–100,000 cpm/sample) incubated with recombinant proteins in 25-μl reaction containing EMSA buffer (20 mM HEPES, 0.2 mM EDTA, 0.2mM EGTA, 100 mM KCl, 5% glycerol, and 2 mM DTT, pH 7.9) and 2 μg poly(d(I-C)) at 22°C. Unlabeled double-stranded DREs (Table I) were used as competitor. For supershift assay, 2 μl of purified (Pierce) polyclonal rabbit antibody (Cedarlane, Hornby, ON) against a C-terminal peptide of human Freud-2 (CDGRKPTGGKLF) was used in 25-μl reaction (20 min, 37°C), probe added and incubated (20 min, 37°C). DNA/protein complexes were separated on a 5% polyacrylamide gel at 4°C dried and exposed to film overnight at −80°C with an intensifying screen.
Table 1. 5-HT1A DRE primers for competition assay.
The human 5-HT1A 5′ and 3′ DRE nucleotide sequences and location (relative to translation initiation site) of forward primers used for competition assays are aligned. The minimal consensus sequence in common among primers that competed for Freud-2 binding is underlined.
| Sequence Name (Location) | DNA sequence |
|---|---|
| 5′-DRE (−1624/−1598) | AGATGGCACTCTAAAACATTTGCCAGA |
| 17-bp 5′ (−1624/−1608) | AGATGGCACTCTAAAAC |
| 16-bp 5′ (−1613/−1598) | TAAAACATTTGCCAGA |
| 3′-DRE (−1597/−1565) | AGGTGGCGACATAAAACCTCATTGCTTAGAACT |
| 19-bp 3′ (−1597/−1578) | AGGTGGCGACATAAAACCT |
| 12-bp 3′ (−1578/−1566) | CATTGCTTAGAA |
Northern blot analysis
Human brain MTN Blot and 12-lane MTN Blot (Clontech) or 10 μg of purchased human prefrontal cortex and kidney RNA (Clontech) run on formaldehyde gel and transferred to hybond-N (Amersham) were probed with 900-bp human Freud-2 cDNA fragment using Strip EZ DNA kit (Ambion). 50 μl of 25,000 cpm/μl of purified probe was incubated with human brain MTN Blot membranes overnight at 42°C. Membranes were washed twice with 2xSCC/0.1% SDS (10 min, 42°C) followed by 0.1%SCC/0.1% SDS (30 min, 65°C) and exposed to film overnight at −80°C with intensifier screen.
qRT-PCR
Total RNA (5 μg) treated with DNase I (TURBO DNA-free kit) was reversed-transcribed using cells-to-DNA™ II kit (Applied Biosystems, Foster City, CA, USA). Each reaction contained 10 μl Taqman Universal master mix, 0.87μg cDNA, 1.5μl Freud-2 FAM (Hs01054180-g1) and GAPDH JOY primers, and 7.6μl DNase-free water. Taqman reactions were carried out in triplicate using a RotorGene3000 cycler (Corbett Research, Sydney, Australia) at: 10 min, 95°C; 40 cycles, 95°C, 15 sec; 60°C, 60s. PCR products were verified by agarose gel electrophoresis, and non-reverse transcribed samples were run as negative control. Relative amounts of Freud-2 to GAPDH were obtained as 10(CtT-CtG), where CtT=target gene threshold cycle, CtG=GAPDH threshold cycle (19).
Western blot analysis
Tissue punches were homogenized in buffer A (10mM HEPES pH 7.9, 10mM KCl, 0.1mM EDTA, 1mM DTT, protease inhibitors, 0.1% Igepal CA-630) and centrifuged (11,000xg, 1 min). The pellet was resuspended in buffer C (20 mM HEPES, pH 7.9; 400mM NaCl; mM each of DTT, EDTA, EGTA; and protease inhibitors), incubated 15 min. with shaking and centrifuged (11,000xg). Supernatant protein (nuclear extract) was resolved on 12.5% SDS polyacrylamide gel, blotted on nitrocellulose membrane and incubated overnight at 4°C with affinity-purified primary rabbit anti-Freud-2 polyclonal antibody or preimmune serum (1:5000) washed and incubated with secondary horseradish peroxidase (HRP)-linked anti-rabbit antibody (Amersham Biosciences, Buckinghamshire, England). After incubation, blots were washed with PBS and developed using enhanced chemiluminescence detection (ECL; Perkin-Elmer Life Sciences Inc., Boston, MA) and exposed to film. Nuclear protein of SK-N-SH cells was extracted as previously described (14). 60 μg of extracts were separated by SDS-PAGE on an 8% polyacrylamide gel. Polyclonal rabbit anti-Freud-2 antibody was purified using Montage antibody purification PROSEP-G spin column (Millipore).
Immunohistochemistry/immunofluorescence
Frozen 30-μm sections were fixed in 4% paraformaldehyde in PBS (1h, 22°C), preincubated in 5% horse serum in PBS (30 min) and incubated with rabbit anti-Freud-2 polyclonal antibody (1:500; 24h at 4°C). Sections were washed in PBS, incubated in biotinylated horse anti-rabbit IgG (1:200; Vector Laboratories, Burlingame, CA) in PBS buffer (4h, 22°C), processed using Vectastain ABC immunoperoxidase kit (Vector, Burlingame, CA) for 24h at 4°C and stained using 3,3′-diaminobenzidine tetrahydrochloride (DAB; 0.05%, Sigma). For dual immunofluorescence, frozen 20-μm sections were incubated overnight with rabbit anti-Freud-2 antibody (1:500) and mouse monoclonal anti-GFAP (1:1000; Chemicon) anti-NeuN (1:1000; Chemicon), anti-5-HT1A (1:100; Immunostar), or anti-CNPase (1:1000; Covance) diluted in the incubation solution. After three washes in 0.1M Tris-HCl buffer (pH 7.6), sections were incubated for 90 min with Cy2- and Cy5-conjugated goat anti-mouse antibody (Jackson Immunochemicals; 1:200), washed and viewed using fluorescence microscopy.
siRNA transfection
Stealth siRNA targeting hFreud-2 [CC2D1BHSS153336] (5′-cccugcagcagaggcugaacaagua-3′) and stealth RNAi negative control duplexes were purchased from InVitrogen. HEK cells were transfected using lipofectamine 2000 (Invitrogen) with a final siRNA concentration of 100 nM. Transfection efficiency control was performed with Block-iT™ fluorescent oligo (Invitrogen). For luciferase assay, 5 μl specific Freud-2-SiRNA or RNAi negative control was co-transfected with 1.5 μg of human 5-HT1A luciferase construct (h5-HT1A) in HEK cells and incubated for 48–72 hr and assayed as described. For immunoblot, 60μg protein/lane blotted using rabbit 5-HT1A polyclonal antibody (Cedarlane laboratory (1:1000) and secondary HRP-linked rabbit antibody (1:2000; Amersham) (16).
Human subjects
The Institutional Review Board of the University of Mississippi Medical Center and University Hospitals of Cleveland approved all procedures in this study. Human brain specimens were obtained from autopsies conducted at the Cuyahoga County Coroner, Cleveland, OH, after obtaining written consent from legally-defined next-of-kin. Retrospective, informant-based psychiatric assessments were performed for all depressed and control subjects (20). Subjects included 5 male MDD subjects, 1 male with dysthymia, and 6 male control subjects without Axis I illness or neurological disorder. MDD subjects included four suicides, one accidental death and one death undetermined; control subjects included four heart disease, one asthma, one electrocution and one homicide. Post-mortem toxicology revealed diazepam in only one male depressed subject. The mean age (yrs.±SEM) was 49.7±6.60 (MDD) and 46.8±7.35 (control); mean post-mortem interval (hrs) was 16.1±2.80 (MDD) and 14.8±2.82 (control); mean brain pH was 6.7±0.11 (MDD) and 6.8±0.031 (control); there was no significant difference observed. The mean age of onset of depression was 38.0±4.93 years and the average duration of illness was 7.0±4.93 years.
Results
Molecular cloning of human Freud-2
To identify human Freud-1 homologues, we screened the GenBank database and identified Freud-2/CC2D1B, a distinct gene located on chromosome 1p32 (NP_115825) that encodes an 858-aa protein with 50% overall amino acid identity to hFreud-1 (Supplemental Fig. 1). Freud-2 contains highest conservation in a number of known domains such as four DM14 domains which extend from amino acids 170–237, 270–332, 385–433, 537–594; predicted helix-loop-helix (648–673); and a conserved protein kinase C (PKC) conserved region (C2 domain; 705–799). The DM-14 domain is highly conserved among species homologues of Freud/CC2D1, although its function is unknown. In Freud-1, the C2 domain is important for DNA binding and essential for its repressor activity (16). The helix-loop-helix domain is slightly shifted or expanded compared to that of Freud-1 and may mediate protein interactions and DNA binding. Several phosphorylation sites are conserved, including the calcium calmodulin-dependent kinase T642 site (T689 in Freud-2) that may mediate calcium sensitivity in Freud-1. A series of proline-directed protein kinase sites located in between DM14-2 and -3 in Freud-1 appear between DM14-3 and -4 in Freud-2. The conservation of important functional domains suggested that like Freud-1, Freud-2 might also bind to DNA and repress transcription.
Freud-2 RNA and protein expression
To verify the expression of Freud-2 in vivo, we examined the tissue distribution of Freud-2 mRNA by Northern blot analysis of human tissues (Fig. 1). A single 3.5-kb mRNA was highly expressed in several peripheral tissues particularly skeletal muscle, kidney and liver (Fig. 1A). In brain, Freud-2 RNA was also highly expressed and enriched in corpus callosum (Fig. 1B), which may reflect its expression in glial cells (see below) and in prefrontal cortex (Fig. 1C). By quantitative RT-PCR, Freud-2 RNA was approximately two-fold enriched in prefrontal cortex compared to dorsal raphe nucleus (Fig. 1D). Freud-2 protein expression was determined using a specific antibody generated against full-length Freud-2, which does not cross-react with Freud-1 (data not shown). By Western blot analysis of nuclear extracts from human postmortem brain tissue using anti-Freud-2, a major 120-kDa Freud-2 species was detected that was not present using preimmune serum (Fig. 2). The apparent molecular weight of the major Freud-2 isoform is larger than the predicted molecular weight of 89-kDa, but migrated with recombinant purified Freud-2 protein (data not shown) and may reflect post-translational modification. Interestingly, Freud-2 protein was enriched in PFC and hippocampus, but was only weakly detected in dorsal raphe nuclei (Fig. 2B). The presence of Freud-2 in the nuclear fraction is consistent with a possible role as a transcription factor.
Fig. 1.
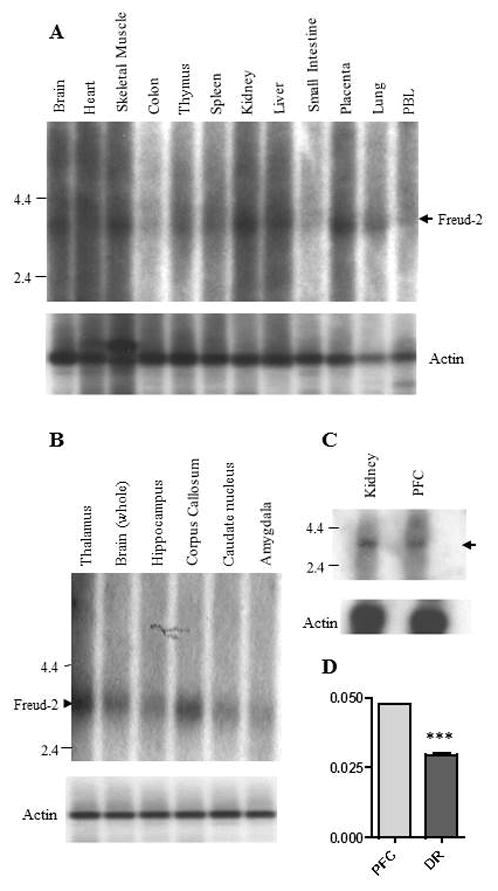
Tissue distribution of human Freud-2 RNA. RNA prepared from the indicated human tissues (Clontech) (A, C) or brain regions (B, C) was hybridized to labeled human Freud-2 cDNA for Northern blot analysis. A major Freud-2 RNA species of approximately 3.5-kb was identified (arrowhead) in most tissues; molecular size markers are shown. Below, the blots were reprobed with labeled beta-actin cDNA to control for RNA loading. D. Quantitative RT-PCR. Freud-2 RNA levels relative to GAPDH standard were quantified in total RNA from PFC and dorsal raphe using Taqman procedure for real-time PCR; Relative RNA levels are shown as mean ± S.E. of triplicate determinations. ***P<0.0001 by unpaired t-test.
Fig. 2.
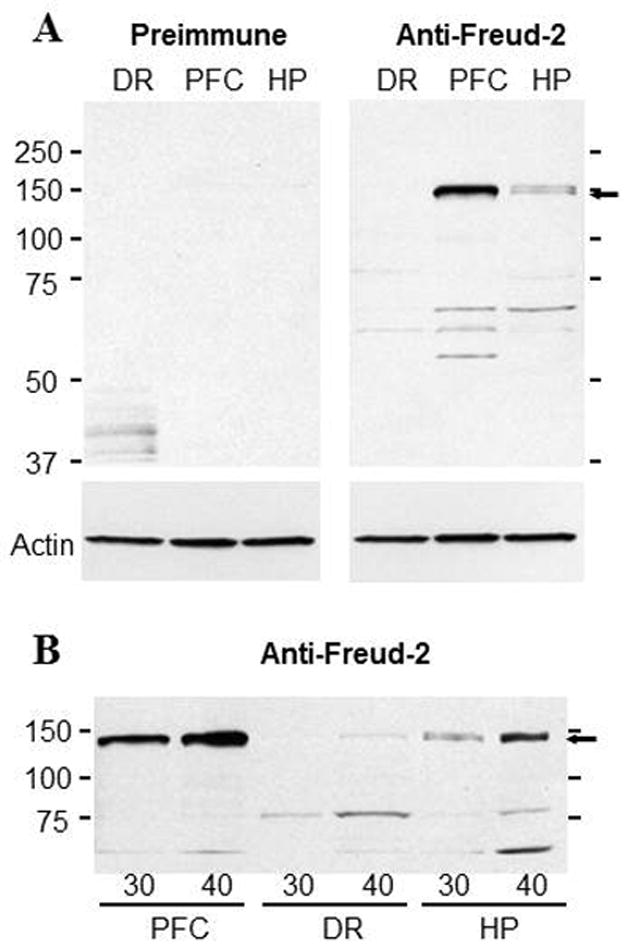
Freud-2 protein is enriched in human prefrontal cortex and hippocampus, but weakly detected in raphe. Nuclear fractions from individual samples of human prefrontal cortex (PFC), dorsal raphe (DR) and hippocampus (HP) were isolated and probed using anti-Freud-2 antiserum (Anti-Freud-2) or preimmune serum as negative control; a representative blot of three replicates is shown. Blots were reprobed for beta-actin as loading control. A major 120-kDa Freud-2 protein species was identified as a doublet. B. Detection of Freud-2 in raphe. Increasing amount of protein was loaded as indicated (30 or 40 μg) and probed using anti-Freud-2; the 120 kDa band was weakly detected in dorsal raphe tissue.
Freud-2 protein was further localized by immunohistochemistry using anti-Freud-2 (Fig. 3) compared to preimmune serum, which lacked specific staining (data not shown). Consistent with Western blot data, Freud-2 immunoreactivity was enriched in the grey matter of the prefrontal cortex (BA10, Fig. 3A), and appeared to stain pyramidal neurons, as well as interneurons and glia (Fig. 3B). Freud-2 was also detected in dentate gyrus of the hippocampus (data not shown), which expresses 5-HT1A receptor RNA (8). In the dorsal raphe (DR) nucleus Freud-2 staining was sparsely distributed in cell bodies (Fig. 3C), consistent with the low level of Freud-2 staining in Western blot. Thus Freud-2 displays a predominant distribution in cells of the prefrontal cortex compared to presynaptic serotonergic raphe cells.
Fig. 3.
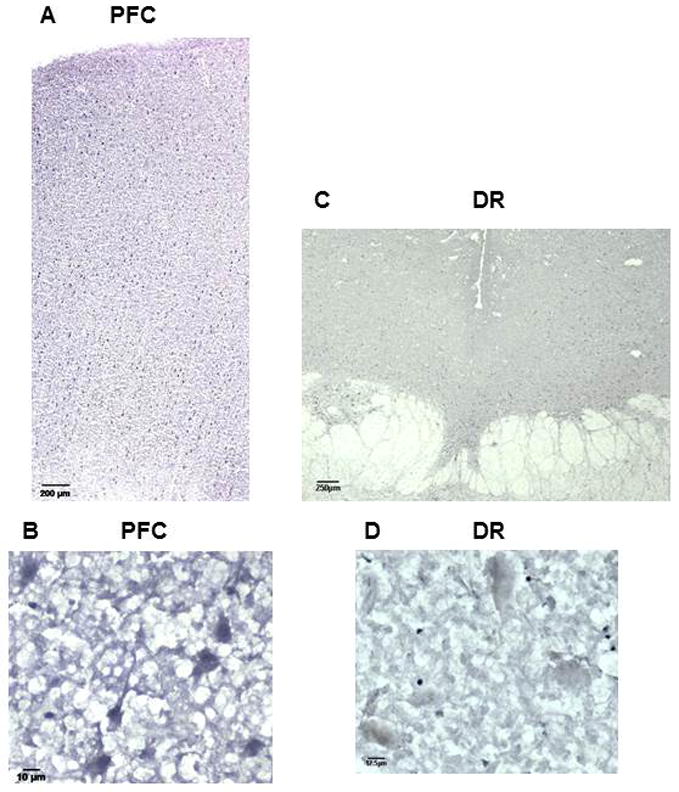
Distribution of Freud-2 immunoreactivity in human prefrontal cortex and dorsal raphe nuclei. Frozen sections of post-mortem human prefrontal (PFC; sections A and B) and dorsal raphe nuclei (DR; sections C and D) were incubated with anti-Freud-2 antibody and processed for immunohistochemistry using anti-Freud-2; no specific staining was observed using preimmune serum (not shown). Sections B and D are at high magnification (40x). Freud-2 staining is enriched in grey matter of prefrontal cortex, but sparse in dorsal raphe nuclei (DR).
The cell types expressing Freud-2 were identified by dual immunofluorescence (Fig. 4). Freud-2 immunoreactivity was strongly detected in cells of the human prefrontal cortex (PFC) with a primarily nuclear localization, and was colocalized with both astrocyte (GFAP) and neuronal (NeuN) markers (Fig. 4A). In dorsal raphe (Fig. 4B), Freud-2 staining was also colocalized with glial (CNPase) and neuronal markers (NeuN). In both raphe and prefrontal cortex Freud-2 was colocalized with 5-HT1A immunostaining (Fig. 4C), although Freud-2 was also detected in 5-HT1A-negative cells, suggesting that like Freud-1, Freud-2 may regulate other genes. Thus Freud-2 is expressed in the nuclei of subsets of neurons and glia in both prefrontal cortex and raphe. The sparse population of stained glial cells in the raphe nuclei may contribute to the higher background staining in the DR (Fig. 3C).
Fig. 4.
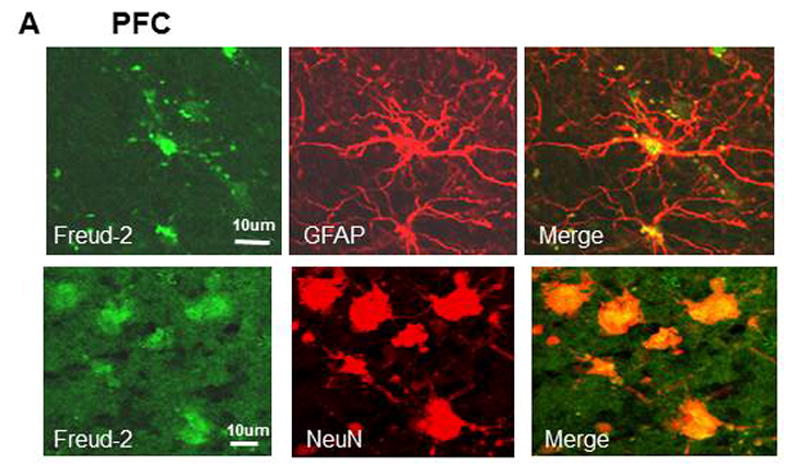
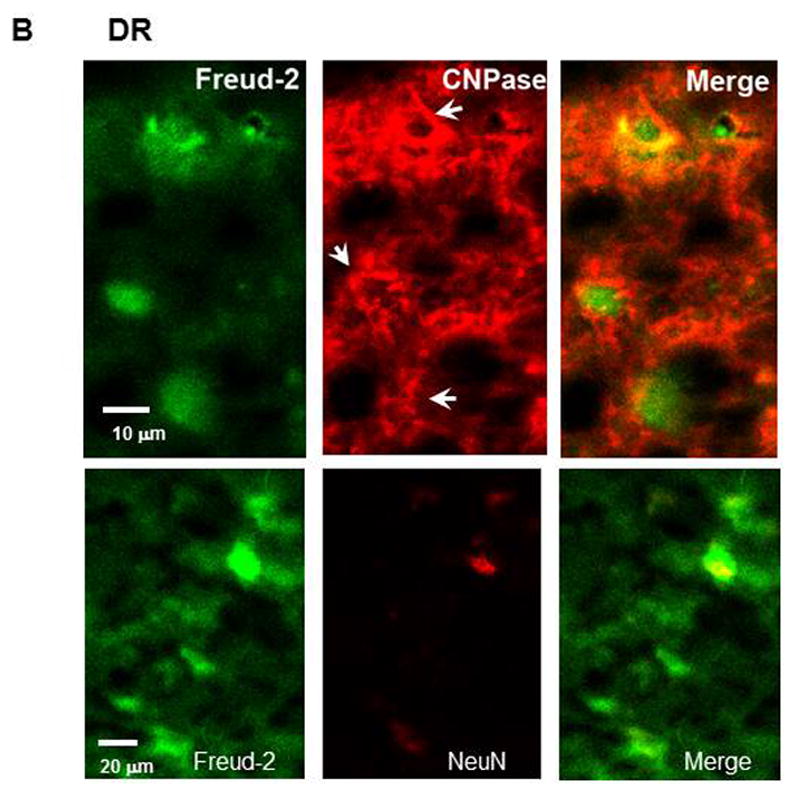
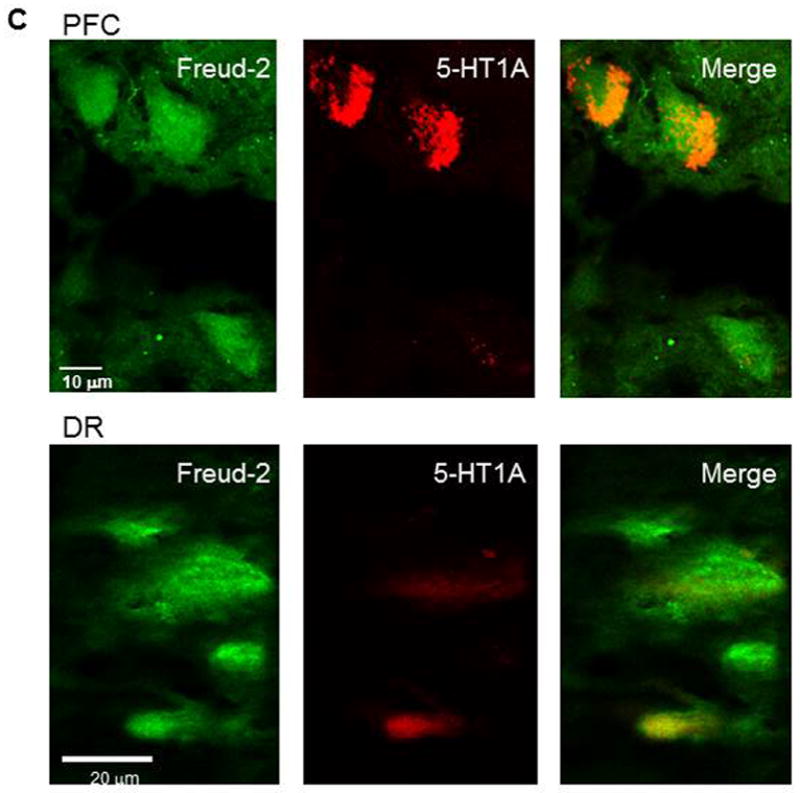
Colocalization of Freud-2 with glial and neuronal markers and 5-HT1A receptor in human PFC and DR. Brain sections from human post-mortem prefrontal cortex (A) or dorsal raphe (B) were probed using anti-Freud-2, GFAP or CNPase (glial) or NeuN (neuronal) antibodies and processed for immunofluorescence of each marker (Freud-2 in green, other markers in red), and merged. Freud-2 was colocalized with GFAP and NeuN indicating its presence in glial and neuronal cells. C. Freud-2 colocalization with 5-HT1A receptor. Sections from PFC or DR were co-stained with anti-Freud-2 and anti-5-HT1A antibodies, and colocalization shown in the merged sections.
Freud-2 binding to human 5-HT1A DRE
We hypothesized that Freud-2 may bind to the 5-HT1A DRE based on the amino acid similarity between Freud-1 and Freud-2 (Supplemental Fig. 1) and the partially-overlapping sequences of 5-HT1A DREs (13; 14). To examine Freud-2 binding to the DRE, EMSA was done using labeled 5-HT1A DREs incubated with purified recombinant GST-Freud-2 fusion protein (Fig. 5). GST-Freud-2, but not GST alone, bound to labeled 5′- or 3′-DRE as a single complex, which was competed by unlabelled DRE oligonucleotides indicating that Freud-2 protein binds specifically to both 5′ and 3′ 5-HT1A DRE elements. Antibody to GST also partly reduced GST-Freud-2 interaction with DRE. To localize the site within the DRE that Freud-2 recognizes, competition EMSA was done in which GST-Freud-2 was incubated with labeled 3′ or 5′ 5-HT1A-DRE and competed with unlabelled segments of the DRE (Table I, Fig. 6). A single 3′-DRE-Freud-2 complex was detected, which was competed as effectively with the 19-bp primers as with the complete 3′-DRE, but was not competed with the adjacent 12-bp portion (Fig. 6A), indicating that Freud-2 binds specifically to the 5′ half of the 3′DRE (Table-1). A polyclonal antibody raised against Freud-2 C-terminal peptide (Anti-cF2) super-shifted the protein-DNA complex (upper arrowhead), while antibody alone did not form a complex with the probe (Fig. 6A). These results confirm the presence of Freud-2 in the complex. Analysis of Freud-2 interactions with the 5-HT1A 5′-DRE revealed that both 16- and 17-bp primers competed for Freud-2 binding to the 5′ DRE (Fig. 6B). Alignment of these sequences in Table I reveals that recombinant Freud-2 binds specifically to a minimal consensus sequence of 5′-TAAAAC-3′, conserved between 5′ and 3′ DREs, and competing oligonucleotides.
Fig. 5.
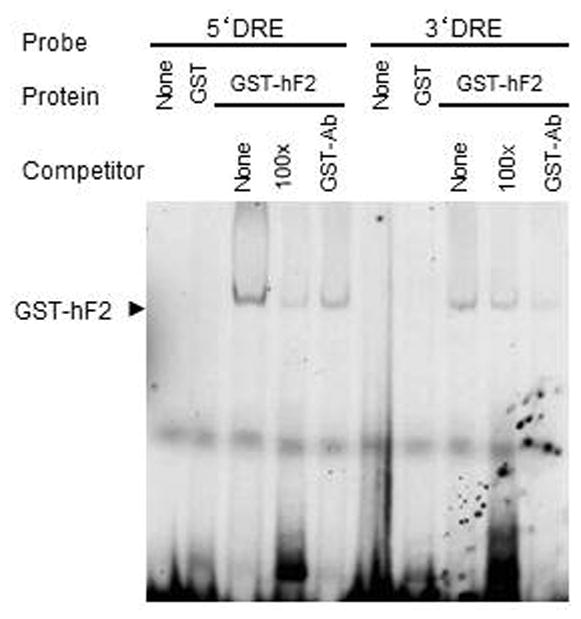
Specific binding of Freud-2 to human DRE sequences. Electrophoretic mobility shift assay (EMSA) was done using bacterially expressed purified recombinant GST-Freud-2 fusion protein (GST-hF2) or GST alone with labelled 5′or 3′DRE from the human 5-HT1A promoter. For competition, unlabelled 5′or 3′DRE (cold) were used at 100-fold molar excess. Antibody to GST (GST-Ab, 2 μl/sample) was added as indicated. A single band (arrow) was detected which was competed with excess unlabeled 5′ or 3′ DRE, indicating that Freud-2 protein binds both 5′ and 3′ DRE from human 5-HT1A promoter.
Fig. 6.
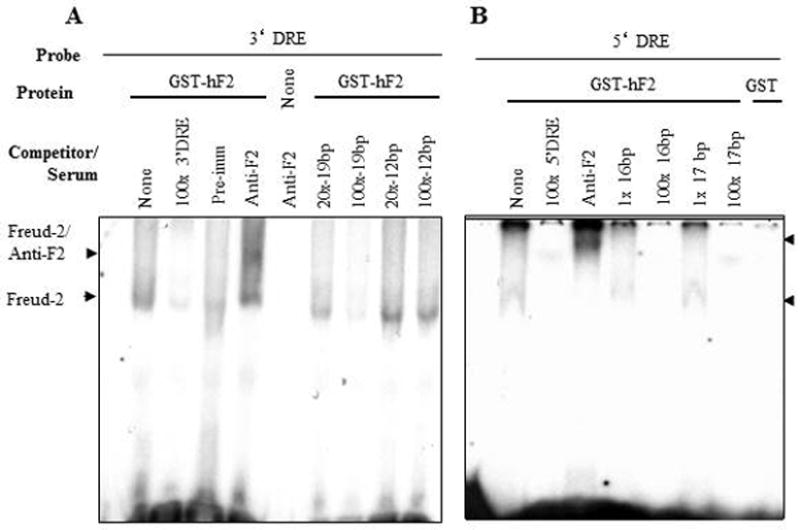
Binding specificity of Freud-2/DRE complexes. EMSA was done using purified recombinant GST-Freud-2 (GST-hF2), GST or no protein incubated with labelled 5-HT1A 3′ or 5′ DRE (A or B, respectively). A specific complex (lower arrowhead) was observed for GST-hF2 but not GST, and this complex was competed with the indicated molar excess of unlabelled primers (see Table I). The 3′ 19-bp and 5′ 16- and 17-bp primers effectively competed (at 100x), indicating that human Freud-2 specifically binds to sequences in common between these primers (Table I). To confirm the presence of Freud-2 in the complex, antiserum (2 μl) to Freud-2 C-terminal (Anti-F2) was added and a supershifted complex was observed in the presence of GST-hF2 (upper arrowhead).
Freud-2 repression of human 5-HT1A expression
To test whether Freud-2 regulates 5-HT1A gene transcription, Freud-2 was cotransfected with human 5-HT1A promoter-luciferase constructs to assay transcriptional activity in either 5-HT1A-expressing human SK-N-SH neuroblastoma cells or HEK cells, which lack detectable 5-HT1A expression (Table 2). Compared to pGL3P, the activity of the DRE containing 5-HT1A constructs was low, consistent with basal repression observed at these elements (13; 14). In both cell types, transfection of human Freud-2 significantly reduced the transcriptional activity of DRE-containing 5-HT1A reporter constructs (Table 2, upper rows). To examine the activity of endogenous Freud-2 on 5-HT1A gene activity, Freud-2 protein level was decreased by cotransfection of Freud-2/CC2D1B-siRNA. Reduction of endogenous Freud-2 protein level induced a 1.6-fold derepression of transcriptional activity of human 5-HT1A gene in HEK-293 cells (Table 2, lower row).
Table 2. Repressor activity of Freud-2 at the 5-HT1A DRE.
Human embryonic kidney HEK-293 or 5-HT1A-expressing human neuroblastoma SK-N-SH cells were transiently co-transfected with vector (pcDNA3, Control) human Freud-2 expression plasmid (Freud-2) and either 5-HT1A-DRE-containing pGL3P luciferase reporter or vector (pGL3P, lacking DRE). For siRNA experiments, cells were treated with 5 μl of scrambled control (Control) or Freud-2 siRNA (F2-siRNA). Transcriptional activity is expressed as luciferase/β-galactosidase activity of cell extracts normalized to pGL3P (=1); % change is Freud-2 or Freud-2 siRNA relative to Control (100%). Data represent the mean ± SEM of three independent experiments. P <0.05 compared with Control by t-test.
| Cell type | Control (DRE/pGL3P) | Freud-2 (DRE/pGL3P) | Change (%) | p < |
|---|---|---|---|---|
| HEK-293 | 0.18 ± 0.01 | 0.096 ± 0.037 | −47% | 0.05 |
| SK-N-SH | 0.27 ± 0.06 | 0.068 ± 0.034 | −75% | 0.05 |
| Cell type | Control (DRE/pGL3P) | Freud-2 siRNA (DRE/pGL3P) | Change (%) | p < |
|---|---|---|---|---|
| HEK-293 | 0.11 ± 0.01 | 0.18 ± 0.02 | +64% | 0.05 |
To determine whether reduction in Freud-2 alters endogenous 5-HT1A expression, 5-HT1A-positive SK-N-SH cells were transiently transfected by either control (i.e., scrambled siRNA) or two different CC2D1B siRNAs (1–2) and the level of endogenous 5-HT1A protein was examined by Western blot (Fig. 7). Both siRNA-1 or siRNAs-1/2 combination reduced the expression of endogenous Freud-2 protein and increased the level of 5-HT1A protein, although siRNA1 alone was more effective in both cases. Thus, these experiments show that human Freud-2 represses 5-HT1A gene transcription in neuronal or non-neuronal cell types. Reduction of Freud-2 protein by specific CC2D1B siRNA increases the level of 5-HT1A protein in SK-N-SH cells, indicating that Freud-2 negatively regulates the basal level of 5-HT1A expression in these cells.
Fig. 7.
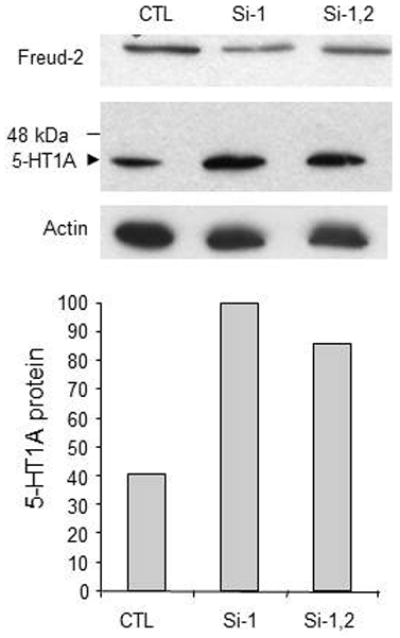
Depletion of Freud-2 increases 5-HT1A receptor expression in SK-N-SH cells. SKN-SH cells were treated with scrambled control siRNA (CTL) or siRNA to Freud-2 (Si-1, Si-2) and cell extracts were examined by Western blot using anti-Freud-2 (top) or anti-5-HT1A antibody (middle); the blot was probed for β-actin as loading control (bottom). Treatment with Freud-2 siRNA1 reduced Freud-2 protein and proportionately increased 5-HT1A receptor expression. The relative intensity of 5-HT1A protein normalized to si-1 (100) is plotted below. The data shown are representative of three independent experiments.
Freud-2 expression in depressed subjects
Since alteration in 5-HT1A receptor expression is implicated in depression, the level of Freud-2 protein was examined in PFC tissue from subjects with major depression and psychiatrically normal matched control subjects (Supplemental Fig. 2). In these preliminary studies, Freud-2 protein was reduced by 40% in the PFC of MDD subjects (relative optical density of Freud-2/beta-actin in control: 5.53 ± 2.26 vs. MDD: 3.37 ± 1.24). Furthermore, Freud-2 protein was decreased in four out of the six male MDD subjects relative to paired controls (Supplemental Fig. 2B). However, the decrease in Freud-2 protein failed to reach statistical significance (two-tailed Wilcoxon signed rank test: W = 15.00, P=0.1563, Supplemental Fig. 2C).
Discussion
Freud-2: novel repressor of post-synaptic 5-HT1A expression
In order to elucidate transcriptional mechanisms that regulate the brain serotonin system, we have initially focused on the identification of transcription factors that regulate the 5-HT1A receptor gene (21). The 5-HT1A is a key presynaptic regulator of serotonergic activity, and also a major post-synaptic receptor that mediates serotonin actions in anxiety and depression (6; 22). We previously identified the DRE as a powerful repressor element in both human and rat 5-HT1A genes (13; 14), and identified Freud-1/CC2D1A as a strong repressor at the DRE (15; 16). In this study we have identified the repressor function of Freud-2/CC2D1B, a homologue of Freud-1, which binds to the DRE to repress the human 5-HT1A receptor gene. Recombinant purified Freud-2 protein bound specifically to both 5′ and 3′-DRE, but recognized a consensus sequence (5′-TAAAAC, Table 1) that is adjacent and partly overlapping with the Freud-1 site defined at the rat 5-HT1A promoter (5′-CATAAAGCAAG) (13). These observations suggest that both Freud-1 and Freud-2 mediate repression of 5-HT1A receptor expression at the DRE.
Our studies indicate that Freud-2 regulates the basal expression of 5-HT1A receptors. Transfection of Freud-2 cDNA reduced the activity of 5-HT1A DRE-luciferase constructs in neuronal 5-HT1A-positive SK-N-SH cells and 5-HT1A-negative non-neuronal HEK293 cells, indicating that Freud-2 may ubiquitously repress this gene. Oppositely, depletion of Freud-2 protein using siRNA increased reporter activity and 5-HT1A receptor protein in SK-N-SH cells, indicating that endogenous Freud-2 represses basal 5-HT1A receptor expression in this post-synaptic neuronal model. Both Freud-1 and Freud-2 are expressed widely in brain and appear to partially overlap in regulating 5-HT1A receptor expression. The strong expression of Freud-2 protein in serotonin target brain regions that express 5-HT1A receptors, such as PFC and hippocampus, suggests that Freud-2 may play a role in regulating the basal expression of post-synaptic 5-HT1A receptors in these regions. However, Freud-1 is also strongly expressed in raphe nuclei and plays a key role in the regulation of basal 5-HT1A autoreceptor expression in serotonergic raphe RN46A cells (16), while Freud-2 did not repress 5-HT1A transcription in these cells (data not shown). Unlike Freud-1, Freud-2 was weakly expressed in the raphe nuclei, consistent with a lack of Freud-2 repression of presynaptic 5-HT1A autoreceptors. Thus, our results indicate that unlike Freud-1, which regulates both pre- and post-synaptic 5-HT1A receptors, Freud-2 appears to preferentially regulate the level of expression of post-synaptic 5-HT1A receptors.
Our data show that like Freud-1, Freud-2 is also widely expressed in peripheral tissues that lack 5-HT1A receptors, suggesting that both Freud-1 and Freud-2 mediate redundant repression to silence 5-HT1A transcription in these tissues. In addition, REST/NRSF, a pan-neuronal repressor that silences neuronal genes in non-neuronal cells, represses the 5-HT1A receptor at a conserved RE-1 recognition site that is located adjacent to the 5-HT1A 3′-DRE (13; 14). Thus, the combination of Freud-1, Freud-2 and REST/NRSF appear to contribute to the silencing of 5-HT1A transcription in non-neuronal tissues. In addition, its broad tissue distribution suggests that like Freud-1, Freud-2 may act as a global repressor of other genes.
Potential roles of Freud-2 in vivo
The strong immunostaining of Freud-2 in forebrain regions and its repression of 5-HT1A expression suggest a role in mental illnesses such as depression or anxiety. Freud-2 protein levels were slightly but not significantly reduced in preliminary studies of depressed versus control PFC (Supplemental Fig. 2), suggesting that down-regulation of Freud-2 could be a compensatory adaptation following the down-regulation of 5-HT1A receptor expression in depression due to genotype (e.g. 5-HTT s/s genotype (23)) or environmental factors (e.g., chronic mild stress, (24) (25)). Interestingly, a recent study indicates that in male depressed suicide brains there is an increase in 5-HT1A RNA in BA10 (26), which could be mediated by Freud-2 down-regulation.
In imaging studies, region- and disorder-specific reductions in the density of cortical and hippocampal 5-HT1A receptors have been observed in depression (27–31) and anxiety disorders (3; 20; 32), while cortical 5-HT1A receptors are increased in anorexia or bulimia nervosa (33; 34). In post-mortem tissue, 5-HT1A RNA and protein levels are reduced in the hippocampus of major depression and bipolar I disorder patients compared to control subjects (7; 8) and 5-HT1A signaling is reduced in several brain regions (35). In 5-HT1A−/− mice, early post-natal rescue of 5-HT1A receptors in hippocampus and cortex restored anxiety phenotype to normal (9), and specific 5-HT1A rescue and activation in the dentate gyrus reduced conditioned freezing response to ambiguous stimuli (36). In mice, 5-HT1A receptors were required for fluoxetine-mediated hippocampal neurogenesis and anti-anxiety actions (10), although this effect appears to be strain-dependent (37). However, the regulation of Freud-2 and its role in reduction of 5-HT1A receptors in depression and anxiety disorders remains to be elucidated. Similarly, the expression of Freud-2 in cortical glia and corpus callosum is of interest since a reduction of glial cells has been observed in the prefrontal cortex of depressed suicides (38; 39) and is counteracted by electroconvulsive seizure (40–42). However the mechanisms involved and the role of Freud-2 in glial function remain unknown.
The structural and functional similarities between Freud-1 and Freud-2 suggests overlapping functions. Recently, a deletion mutation in the Freud-1/CC2D1A gene has been linked to non-syndromal mental retardation (43), implicating Freud-1 in cognitive development. This mutation truncates the protein, eliminating the C2 domain that is required for Freud-1 repressor activity (16), suggesting that the truncated mutant is non-functional or a dominant-negative protein (44). Similarly, Freud-2 may also participate in cognitive development, although it is unable to rescue the Freud-1 mutant phenotype.
Although Freud-2 represses the 5-HT1A gene and depletion of Freud-2 leads to up-regulation of 5-HT1A receptor expression, like Freud-1 Freud-2 may regulate additional genes. In particular, we recently characterized a DRE in the human dopamine-D2 receptor gene that is repressed by Freud-1 (45). The D2-DRE contains a consensus GATAAG sequence for Freud-1 binding, but also contains an adjacent TAAAAG sequence that is similar to the TAAAAC sequence we identified for Freud-2 binding to the 5-HT1A DRE. Further studies will be required to determine whether Freud-2 also regulates dopamine-D2 receptor expression.
In summary, we have identified Freud-2 as a novel transcriptional repressor, which, in combination with the homologue Freud-1, regulates the expression of 5-HT1A receptors in neuronal and nonneuronal cells.
Supplementary Material
Acknowledgments
We acknowledge the work of Drs James C. Overholser and George Jurjus, Lesa Dieter and Nicole Herbst in tissue collection and the retrospective psychiatric diagnoses. We are deeply appreciative of the assistance of the next-of-kin of the deceased and gratefully acknowledge the assistance of the Cuyahoga County Coroner’s Office, Cleveland, OH, USA. We thank Dr. Anastasia Rogaeva, Dr. Xiaoming Ou, Margaret Czesak and Heidi Fitzgibbon for technical assistance and guidance. This study was supported by a CIHR grant to P.R.A. and NIH grants MH67996 and RR17701 to M.C.A.
Footnotes
Financial Disclosures: None.
References
- 1.Pompeiano M, Palacios JM, Mengod G. Distribution and cellular localization of mRNA coding for 5-HT1A receptor in the rat brain: correlation with receptor binding. Journal of Neuroscience. 1992;12:440–453. doi: 10.1523/JNEUROSCI.12-02-00440.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert PR, Zhou QY, Van Tol HH, Bunzow JR, Civelli O. Cloning, functional expression, and mRNA tissue distribution of the rat 5-hydroxytryptamine1A receptor gene. Journal of Biological Chemistry. 1990;265:5825–5832. [PubMed] [Google Scholar]
- 3.Lanzenberger RR, Mitterhauser M, Spindelegger C, Wadsak W, Klein N, Mien LK, et al. Reduced serotonin-1A receptor binding in social anxiety disorder. Biol Psychiatry. 2007;61:1081–1089. doi: 10.1016/j.biopsych.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan GM, Oquendo MA, Simpson N, Van Heertum RL, Mann JJ, Parsey RV. Brain serotonin1A receptor binding in major depression is related to psychic and somatic anxiety. Biol Psychiatry. 2005;58:947–954. doi: 10.1016/j.biopsych.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Pitchot W, Hansenne M, Pinto E, Reggers J, Fuchs S, Ansseau M. 5-Hydroxytryptamine 1A receptors, major depression, and suicidal behavior. Biol Psychiatry. 2005;58:854–858. doi: 10.1016/j.biopsych.2005.05.042. [DOI] [PubMed] [Google Scholar]
- 6.Albert PR, Lemonde S. 5-HT1A Receptors, Gene Repression, and Depression: Guilt by Association. Neuroscientist. 2004;10:575–593. doi: 10.1177/1073858404267382. [DOI] [PubMed] [Google Scholar]
- 7.Gray L, Scarr E, Dean B. Serotonin 1a receptor and associated G-protein activation in schizophrenia and bipolar disorder. Psychiatry Res. 2006;143:111–120. doi: 10.1016/j.psychres.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Lopez-Figueroa AL, Norton CS, Lopez-Figueroa MO, Armellini-Dodel D, Burke S, Akil H, et al. Serotonin 5-HT1A, 5-HT1B, and 5-HT2A receptor mRNA expression in subjects with major depression, bipolar disorder, and schizophrenia. Biol Psychiatry. 2004;55:225–233. doi: 10.1016/j.biopsych.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 9.Gross C, Zhuang X, Stark K, Ramboz S, Oosting R, Kirby L, et al. Serotonin1A receptor acts during development to establish normal anxiety-like behaviour in the adult. Nature. 2002;416:396–400. doi: 10.1038/416396a. [DOI] [PubMed] [Google Scholar]
- 10.Santarelli L, Saxe M, Gross C, Surget A, Battaglia F, Dulawa S, et al. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science. 2003;301:805–809. doi: 10.1126/science.1083328. [DOI] [PubMed] [Google Scholar]
- 11.Parks CL, Shenk T. The serotonin 1a receptor gene contains a TATA-less promoter that responds to MAZ and Sp1. Journal of Biological Chemistry. 1996;271:4417–4430. doi: 10.1074/jbc.271.8.4417. [DOI] [PubMed] [Google Scholar]
- 12.Storring JM, Charest A, Cheng P, Albert PR. TATA-driven transcriptional initiation and regulation of the rat serotonin 5-HT1A receptor gene. J Neurochem. 1999;72:2238–2247. doi: 10.1046/j.1471-4159.1999.0722238.x. [DOI] [PubMed] [Google Scholar]
- 13.Ou XM, Jafar-Nejad H, Storring JM, Meng JH, Lemonde S, Albert PR. Novel dual repressor elements for neuronal cell-specific transcription of the rat 5-HT1A receptor gene. J Biol Chem. 2000;275:8161–8168. doi: 10.1074/jbc.275.11.8161. [DOI] [PubMed] [Google Scholar]
- 14.Lemonde S, Rogaeva A, Albert PR. Cell type-dependent recruitment of trichostatin A-sensitive repression of the human 5-HT1A receptor gene. J Neurochem. 2004;88:857–868. doi: 10.1046/j.1471-4159.2003.02223.x. [DOI] [PubMed] [Google Scholar]
- 15.Rogaeva A, Albert PR. The mental retardation gene CC2D1A/Freud-1 encodes a long isoform that binds conserved DNA elements to repress gene transcription. Eur J Neurosci. 2007;26:965–974. doi: 10.1111/j.1460-9568.2007.05727.x. [DOI] [PubMed] [Google Scholar]
- 16.Ou XM, Lemonde S, Jafar-Nejad H, Bown CD, Goto A, Rogaeva A, Albert PR. Freud-1: A novel calcium-regulated repressor of the 5-HT1A receptor gene. J Neuroscience. 2003;23:7415–7425. doi: 10.1523/JNEUROSCI.23-19-07415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Czesak M, Lemonde S, Peterson EA, Rogaeva A, Albert PR. Cell-specific repressor or enhancer activities of Deaf-1 at a serotonin 1A receptor gene polymorphism. J Neurosci. 2006;26:1864–1871. doi: 10.1523/JNEUROSCI.2643-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charest A, Wainer BH, Albert PR. Cloning and differentiation-induced expression of a murine serotonin1A receptor in a septal cell line. J Neuroscience. 1993;13:5164–5171. doi: 10.1523/JNEUROSCI.13-12-05164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Czesak M, Burns AM, Lenicov FR, Albert PR. Characterization of rat rostral raphe primary cultures: Multiplex quantification of serotonergic markers. J Neurosci Methods. 2007;164:59–67. doi: 10.1016/j.jneumeth.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Szewczyk B, Albert PR, Burns AM, Czesak M, Overholser JC, Jurjus GJ, et al. Gender-specific decrease in NUDR and 5-HT1A receptor proteins in the prefrontal cortex of subjects with major depressive disorder. Int J Neuropsychopharmacol. 2008 doi: 10.1017/S1461145708009012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Francois B, Czesak M, Steubl D, Albert PR. Transcriptional regulation at a HTR1A polymorphism associated with mental illness. Neuropharmacology. 2008;55:977–985. doi: 10.1016/j.neuropharm.2008.06.046. [DOI] [PubMed] [Google Scholar]
- 22.Gross C, Hen R. The developmental origins of anxiety. Nat Rev Neurosci. 2004;5:545–552. doi: 10.1038/nrn1429. [DOI] [PubMed] [Google Scholar]
- 23.David SP, Murthy NV, Rabiner EA, Munafo MR, Johnstone EC, Jacob R, et al. A functional genetic variation of the serotonin (5-HT) transporter affects 5-HT1A receptor binding in humans. J Neurosci. 2005;25:2586–2590. doi: 10.1523/JNEUROSCI.3769-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez JF, Liberzon I, Vazquez DM, Young EA, Watson SJ. Serotonin 1A receptor messenger RNA regulation in the hippocampus after acute stress. Biol Psychiatry. 1999;45:934–937. doi: 10.1016/s0006-3223(98)00224-8. [DOI] [PubMed] [Google Scholar]
- 25.Griffin WC, 3rd, Skinner HD, Birkle DL. Prenatal stress influences 8-OH-DPAT modulated startle responding and [3H]-8-OH-DPAT binding in rats. Pharmacol Biochem Behav. 2005;81:601–607. doi: 10.1016/j.pbb.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 26.Anisman H, Du L, Palkovits M, Faludi G, Kovacs GG, Szontagh-Kishazi P, et al. Serotonin receptor subtype and p11 mRNA expression in stress-relevant brain regions of suicide and control subjects. J Psychiatry Neurosci. 2008;33:131–141. [PMC free article] [PubMed] [Google Scholar]
- 27.Sargent PA, Kjaer KH, Bench CJ, Rabiner EA, Messa C, Meyer J, et al. Brain serotonin1A receptor binding measured by positron emission tomography with [11C]WAY-100635: effects of depression and antidepressant treatment. Arch Gen Psychiatry. 2000;57:174–180. doi: 10.1001/archpsyc.57.2.174. [DOI] [PubMed] [Google Scholar]
- 28.Drevets WC, Frank E, Price JC, Kupfer DJ, Greer PJ, Mathis C. Serotonin type-1A receptor imaging in depression. Nucl Med Biol. 2000;27:499–507. doi: 10.1016/s0969-8051(00)00119-0. [DOI] [PubMed] [Google Scholar]
- 29.Bhagwagar Z, Rabiner EA, Sargent PA, Grasby PM, Cowen PJ. Persistent reduction in brain serotonin1A receptor binding in recovered depressed men measured by positron emission tomography with [11C]WAY-100635. Mol Psychiatry. 2004;9:386–392. doi: 10.1038/sj.mp.4001401. [DOI] [PubMed] [Google Scholar]
- 30.Shively CA, Friedman DP, Gage HD, Bounds MC, Brown-Proctor C, Blair JB, et al. Behavioral depression and positron emission tomography-determined serotonin 1A receptor binding potential in cynomolgus monkeys. Arch Gen Psychiatry. 2006;63:396–403. doi: 10.1001/archpsyc.63.4.396. [DOI] [PubMed] [Google Scholar]
- 31.Moses-Kolko EL, Price JC, Thase ME, Meltzer CC, Kupfer DJ, Mathis CA, et al. Measurement of 5-HT1A receptor binding in depressed adults before and after antidepressant drug treatment using positron emission tomography and [11C]WAY-100635. Synapse. 2007;61:523–530. doi: 10.1002/syn.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neumeister A, Bain E, Nugent AC, Carson RE, Bonne O, Luckenbaugh DA, et al. Reduced serotonin type 1A receptor binding in panic disorder. J Neurosci. 2004;24:589–591. doi: 10.1523/JNEUROSCI.4921-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tiihonen J, Keski-Rahkonen A, Lopponen M, Muhonen M, Kajander J, Allonen T, et al. Brain serotonin 1A receptor binding in bulimia nervosa. Biol Psychiatry. 2004;55:871–873. doi: 10.1016/j.biopsych.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 34.Bailer UF, Frank GK, Henry SE, Price JC, Meltzer CC, Mathis CA, et al. Exaggerated 5-HT1A but normal 5-HT2A receptor activity in individuals ill with anorexia nervosa. Biol Psychiatry. 2007;61:1090–1099. doi: 10.1016/j.biopsych.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 35.Hsiung SC, Adlersberg M, Arango V, Mann JJ, Tamir H, Liu KP. Attenuated 5-HT1A receptor signaling in brains of suicide victims: involvement of adenylyl cyclase, phosphatidylinositol 3-kinase, Akt and mitogen-activated protein kinase. J Neurochem. 2003;87:182–194. doi: 10.1046/j.1471-4159.2003.01987.x. [DOI] [PubMed] [Google Scholar]
- 36.Tsetsenis T, Ma XH, Lo Iacono L, Beck SG, Gross C. Suppression of conditioning to ambiguous cues by pharmacogenetic inhibition of the dentate gyrus. Nat Neurosci. 2007;10:896–902. doi: 10.1038/nn1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holick KA, Lee DC, Hen R, Dulawa SC. Behavioral Effects of Chronic Fluoxetine in BALB/cJ Mice Do Not Require Adult Hippocampal Neurogenesis or the Serotonin 1A Receptor. Neuropsychopharmacology. 2008;33:406–417. doi: 10.1038/sj.npp.1301399. [DOI] [PubMed] [Google Scholar]
- 38.Rajkowska G. Depression: what we can learn from postmortem studies. Neuroscientist. 2003;9:273–284. doi: 10.1177/1073858403252773. [DOI] [PubMed] [Google Scholar]
- 39.Rajkowska G, Halaris A, Selemon LD. Reductions in neuronal and glial density characterize the dorsolateral prefrontal cortex in bipolar disorder. Biol Psychiatry. 2001;49:741–752. doi: 10.1016/s0006-3223(01)01080-0. [DOI] [PubMed] [Google Scholar]
- 40.Hamidi M, Drevets WC, Price JL. Glial reduction in amygdala in major depressive disorder is due to oligodendrocytes. Biol Psychiatry. 2004;55:563–569. doi: 10.1016/j.biopsych.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Choudary PV, Molnar M, Evans SJ, Tomita H, Li JZ, Vawter MP, et al. Altered cortical glutamatergic and GABAergic signal transmission with glial involvement in depression. Proc Natl Acad Sci U S A. 2005;102:15653–15658. doi: 10.1073/pnas.0507901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wennstrom M, Hellsten J, Ekstrand J, Lindgren H, Tingstrom A. Corticosterone-induced inhibition of gliogenesis in rat hippocampus is counteracted by electroconvulsive seizures. Biol Psychiatry. 2006;59:178–186. doi: 10.1016/j.biopsych.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 43.Basel-Vanagaite L, Attia R, Yahav M, Ferland RJ, Anteki L, Walsh CA, et al. The CC2D1A, a member of a new gene family with C2 domains, is involved in autosomal recessive non-syndromic mental retardation. J Med Genet. 2006;43:203–210. doi: 10.1136/jmg.2005.035709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogaeva A, Galaraga K, Albert PR. The Freud-1/CC2D1A family: Transcriptional regulators implicated in mental retardation. J Neurosci Res. 2007;85:2833–2888. doi: 10.1002/jnr.21277. [DOI] [PubMed] [Google Scholar]
- 45.Rogaeva A, Ou XM, Jafar-Nejad H, Lemonde S, Albert PR. Differential repression by Freud-1/CC2D1A at a polymorphic site in the dopamine-D2 receptor gene. J Biol Chem. 2007;282:20897–20905. doi: 10.1074/jbc.M610038200. [DOI] [PubMed] [Google Scholar]
- 46.Marchler-Bauer A, Anderson JB, DeWeese-Scott C, Fedorova ND, Geer LY, He S, et al. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 2003;31:383–387. doi: 10.1093/nar/gkg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


