Abstract
Free radical-induced oxidation of phospholipids contributes significantly to pathologies associated with inflammation and oxidative stress. Detection of covalent interaction between oxidized phospholipids (oxPL) and proteins by LC-MS/MS could provide valuable information about the molecular mechanisms of oxPL effects. However, such studies are very limited because of significant challenges in detection of the comparatively low levels of oxPL–protein adducts in complex biological systems. Current approaches have several limitations, most important of which is the inability to detect protein modifications by naturally occurring oxPL. We now report, for the first time, an enrichment method that can be applied to the global analysis of protein adducts with various naturally occurring oxPL in relevant biological systems. This method exploits intrinsic properties of peptides modified by oxPL, allowing highly efficient enrichment of oxPL-modified peptides from biological samples. Very low levels of oxPL–protein adducts (<2 ppm) were detected using this enrichment method in combination with LC-MS/MS. We applied the method to several model systems, including oxidation of high density lipoprotein (HDL) and interaction of human platelets with a specific oxPL, and demonstrated its extremely high efficiency and productivity. We report multiple new modifications of apolipoproteins in HDL and proteins in human platelets.
Phospholipids are a major component of biological membranes of cells, intracellular organelles, and lipoproteins. The polyunsaturated fatty acid chain at the sn-2 position of phospholipids is a major oxidative target under oxidative stress. Oxidized phospholipids (oxPL) were detected in apoptotic cells, plasma, and various tissues including aorta, small intestine, dermal fibroblast, brain, retina, and others.1 OxPL can act as markers of “modified self” and induce the recognition by soluble and cell-associated receptors of innate immunity,1 including C-reactive protein, natural antibodies, and scavenger receptors,2 thus promoting the removal of apoptotic cells or oxidized lipoproteins. OxPL can also modulate intracellular signaling by activating signal-transducing receptors,1 such as scavenger receptors,3 PAF receptors,4 prostaglandin receptors,5 VEGF receptors,6 toll-like receptors,7 and others. OxPL can regulate second messengers, including Ca2+i8 and cAMP,9 activate protein kinases,10 phosphatases,8 and small GTPases,11 and regulate expression of genes related to inflammation, lipid metabolism, cellular stress, proliferation, and differentiation.12 OxPL contribute significantly to various diseases associated with oxidative stress, including atherosclerosis, Alzheimer’s disease, Parkinson’s disease, diabetes, lung injury, leprosy, ischemia, rheumatoid arthritis, and others.1,13
Most of the oxPL have electrophilic functional groups at the sn-2 position, which can form covalent adducts with lysine, histidine, and cysteine residues of proteins. The studies on the covalent interaction between oxPL and proteins in a relevant biological system using LC-MS/MS analysis could provide detailed information about the oxPL–proteins adducts, including (a) the identity of proteins preferentially modified by oxPL in a relevant biological system (such as plasma or cells), (b) specific function of protein affected through revealing specific sites on proteins that are covalently modified, and (c) species of oxPL modifying proteins and types of adducts formed. Such studies could provide important information about the molecular mechanisms by which oxPL exert their effects. However, the reports of LC-MS/MS analysis of proteins covalently modified by oxPL are very limited.14,15 One major reason for this is the relatively low levels of oxPL–protein adducts compared to a highly diverse pool of proteins in a complex biological systems. Approaches providing a means of high affinity capture of the adducted proteins, or adducted peptides derived from enzymatic digestion of the adducted proteins, are indispensable for the analysis of protein targets of oxPL.
We now report an enrichment method that can be applied to the global analysis of protein adducts with various naturally occurring oxPL in relevant biological systems. Using the enrichment method in combination with LC-MS/MS analysis, we were able to detect very low levels of oxPL–protein adducts (at least <2 ppm) in cell lysate. It allowed us to detect various covalent adducts on multiple sites of major and minor apolipoproteins of oxidized HDL formed by a variety of naturally occurring oxPL. Furthermore, the method demonstrated high efficiency in detection of oxPL–protein adducts in such a complex biological system as a whole cell. The enrichment method described here is highly promising for the study on the covalent interactions between naturally occurring oxPL and proteins in various biological systems.
EXPERIMENTAL SECTION
Isolation and Oxidation of HDL
HDL was isolated from fresh human plasma as described elsewhere,16 extensively dialyzed against PBS at 4 °C, and adjusted to 0.8 mg HDL protein/mL using PBS. HDL was oxidized by dialysis against PBS containing 10 µM copper sulfate for 6 h at 37 °C, followed by reduction using 100 mM NaBH4 to stabilize the reversible adducts formed between oxPL and HDL proteins. The sample volume was reduced to 1/10 of the original volume by centrifugation at 4000 × g for 15 min using centrifugal filter (10K cutoff, Millipore). Then the volume was restored using 100 mM Tris buffer (pH 7.8), and the procedure was repeated twice. A 12% gel SDS-PAGE was ran to monitor the oxidation of HDL (Figure S1, Supporting Information).
Generation of KODA-PC Modified HDL
KODA-PC (300 µg) (abbreviations are defined in Table 2) and lysoPC (2.1 mg) were mixed in chloroform, dried under nitrogen flow, and dissolved in 0.5 mL of phosphate buffer (100 mM, pH 7.4). To the resulting solution, 0.68 mg of HDL in 0.5 mL of PBS was added, followed by incubation at room temperature for 1 h. Then 100 mM NaBH4 was added to reduce the imine group in the Schiff base adduct and carbonyl group in the Michael adduct formed between KODA-PC and HDL proteins, thus stabilizing these reversible adducts. The volume was reduced to 1/10 of the original volume by centrifugation at 4000g for 15 min using a centrifugal filter (10K cutoff, Millipore). Then the volume was restored using 100 mM Tris buffer (pH 7.8), and the procedure was repeated twice. A 12% gel SDS-PAGE was ran to monitor the modification of HDL by KODA-PC (Figure S1, Supporting Information).
Table 2.
oxPL-Modified Peptides Detected in Oxidized HDL by LC-MS/MSa
| proteins | oxPL-modified peptides | OxPL (adduct types) |
|---|---|---|
| apoA-I | ATEH(199)LSTLSEK | KOHA-PL or HOHA-PL (MAb); KOOA-PL or HOOA-PL (MAb); KODA-PL or HODA-PL (MAb) |
| LAEYHAK(195)ATEHLSTLSEK | OV-PL (SBb); HODA-PL (CH) or KODA-PL (KAb); KOHA-PL or HOHA-PL (MAb); KOOA-PL or HOOA-PL (MAb); KODA-PL or HODA-PL (MAb) | |
| LAEYH(193)AKATEHLSTLSEK | KOOA-PL or HOOA-PL (MAb); KODA-PL or HODA-PL (MAb); HOOA-PL, HODA-PL (CH) | |
| LEALK(182)ENGGAR | KOHA-PL, KOOA-PL (SBb); HOHA-PL (CH) or KOHA-PL (KAb); HOOA-PL (CH) or KOOA-PL (KAb); HOOA-PL (HF) | |
| AHVDALRTH(162)LAPYSDELR | KOHA-PL or HOHA-PL (MAb); KOOA-PL or HOOA-PL (MAb); KODA-PL or HODA-PL (MAb); KOOA-PL, KODAPL (furan); HOHA-PL, HOOA-PL, HODA-PL (CH) | |
| AH(155)VDALR | KOHA-PL or HOHA-PL (MAb); KOOA-PL or HOOA-PL (MAb); KODA-PL or HODA-PL (MAb); KOOA-PL, KODAPL (furan); HOHA-PL (CH) | |
| LHELQEK(140)LSPLGEEMR | OV-PL, ON-PL (SBb) | |
| QK(133)LHELQEK | HOOA-PL (HF) | |
| QK(118)VEPLRAELQEGAR | HOOA-PL (HF) | |
| VKDLATVYVDVLK(23)DSGR | ON-PL (SBb); KOOA-PL or HOOA-PL (MAb) | |
| VK(12)DLATVYVDVLK | ON-PL, KODA-PL (SBb); KOOA-PL or HOOA-PL (MAb); KODA-PL or HODA-PL (MAb) | |
| apoA-II | SK(46)EQLTPLIK | OV-PL, KOOA-PL, KOHA-PL (SBb); KOOA-PL or HOOA-PL (MAb); HOHA-PL (CH) or KOHA-PL (KAb) |
| SYFEK(44)SKEQLTPLIK | OV-PL (SBb) | |
| SPELQAEAK(39)SYFEK | OV-PL, ON-PL, KOOA-PL, KODA-PL (SBb); KOOA-PL or HOOA-PL (MAb); HODA-PL (CH) or KODA-PL (KAb) | |
| DLMEKVK(30)SPELQAEAK | KODA-PL or HODA-PL (MAb); ON-PL (SBb) | |
| DLMEK(28)VKSPELQAEAK | KOOA-PL or HOOA-PL (MAb); | |
| apoC-III | TAK(44) DALSSVQESQVAQQAR | ON-PL (SBb) |
The abbreviations used are as follows: MA, Michael adduct; SB, Schiff base adduct; KA, ketoamide adduct; HF, 2,3-dihydrofuran adduct; CH, cyclic hemiacetal adduct; HODA-PL, the 9-hydroxy-12-oxo-10-dodecenoic acid esters of lysophospholipid; HOOA-PL, the 5-hydroxy-8-oxo-6-octenoic acid esters of lysophospholipid; HOHA-PL, the 4-hydroxy-7-oxo-5-heptenoic acid esters of lysophospholipid; KODA-PC, the 9-keto-12-oxo-10-dodecenoic acid esters of lysophospholipid; KOOA-PC, the 5-keto-8-oxo-6-octenoic acid esters of lysophospholipid; KOHA-PC, the 4-keto-7-oxo-5-heptenoic acid esters of lysophospholipid; OV-PC, the 5-oxovaleric acid esters of lysophospholipid; ON-PC, the 9-oxononanoic acid esters of lysophospholipid.
Adducts reduced by NaBH4.
Reaction between KODA-PC and Human Platelets
Human platelets were isolated as described elsewhere.16 KODA-PC (110 µg) and POPC (3 mg) were mixed in chloroform and dried under a stream of nitrogen. The resulting residue was resuspended in 2 mL of modified Tyrode’s buffer (137 mM NaCl, 2.8 mM KCl, 12 mM NaHCO3, 1 mM CaCl2, 1 mM MgC12, and 20 mM HEPES, pH 7.4) and passed through a 30 nm polycarbonate filter 11 times using an Avanti Mini-Extruder Set (Avanti Polar Lipids, Inc., Alabaster, AL) to make 30 nm vesicles. The obtained KODA-PC vesicles were added to human platelets (2 × 108 platelets/mL) at a final concentration of 20 µM in modified Tyrode’s buffer, followed by incubation at room temperature with gentle shaking for 30 min. Then 50 mM NaBH4 was added, followed by 30 min incubation at room temperature. Then platelets were pelleted by centrifugation at 4000 rpm for 15 min at 22 °C. The resulting platelet pellet was then lysed using 8 M urea in 50 mM Tris (pH 8.0), followed by dithiothreitol reduction (10 mM) and iodoacetamide alkylation (40 mM). Then more dithiothreitol was added (20 mM) to consume the excessive iodoacetamide. The resulting solution was diluted using Tris buffer (50 mM, pH 7.6) to reduce the concentration of urea to 2M, followed by tryptic digestion (trypsin:protein = 1:100) at 37 °C for 24 h.
Preliminary Size Separation Procedure
The tryptic digest of platelet lysate was diluted 1 time using 100 mM ammonium bicarbonate buffer (pH 7.8) and lysoPC was added to a final concentration of 500 µM. The resulting mixture was transferred to 15 mL centrifugal filter unit (Millipore, 30K cutoff) and centrifuged at 4000g for 10 min to reduce the volume to 1/10 of the original volume. The volume was restored by adding 100 mM ammonium bicarbonate buffer (pH 7.8) and centrifuged again using the same conditions. The procedure was repeated one more time, and samples were used directly in the C18 enrichment procedure.
C18 Enrichment Procedure
The C18 solid-phase extraction (SPE) column (500 mg bed weight, Discovery DSC-18, Supelco) was first activated by 15 bed volume of methanol, followed by elution with 5 bed volume of 50% methanol, 5% methanol with 0.1% formic acid and water sequentially. Then the peptide mixture obtained by a size separation procedure was loaded to the column, followed by stepwise elution using 10 bed volumes of water, 5%, 30%, 50%, and 70% methanol with 0.1% formic acid. After that, the column was equilibrated using 5 bed volumes of 5% methanol and water sequentially before eluting the column using 5 bed volume of 30% ammonium hydroxide or 40% methylamine. Then the column was filled with 30% ammonium hydroxide or 40% methylamine and sealed at both ends. The aminolysis was carried out at room temperature for 40 h before eluting the column using 70% methanol with 0.1% formic acid to collect the modified peptides (PLEAs). The eluent was blown with nitrogen to remove the ammonia gas or methylamine and then dried using a Speedvac (Savant Speedvac SC110), followed by suspension in 1 mL of water. The homogenate was sonicated for 10 min using an FS30 sonicator (Fisher scientific) and then filtered using a 0.22 µm filter (Corning Inc.). The filtrate was loaded onto a C18 SPE column (100 mg bed weight, Discovery DSC-18, SUPELCO) and eluted using 10 bed volumes of water and 5% methanol with 0.1% formic acid sequentially. Then the modified peptides were eluted using 15 bed volumes of 50% methanol with 0.1% formic acid. The eluant was dried using a Speedvac and resuspended in 50 µL of 1% acetic acid for LC-MS-MS analysis.
Mass Spectrometry and Data Processing
Chromatographic separation of the peptide samples was performed by a UltiMate 3000 RSLCnano LC system equipped with a reversed-phase capillary chromatography column (Dionex- Acclaim Pepmap C18, 15 cm × 75 µm i.d., 2 µm, 100 Å). An elution gradient was used by mixing mobile phase A (0.1% formic acid in water) with solvent B (0.1% formic acid in acetonitrile) as follows: isocratic elution with 2% B from 0 to 5 min; increase to 40% B from 5 to 76 min; increase to 70% B from 76 to 78 min; isocratic elution with 70% B from 78 to 85 min; decrease to 2% B from 85 to 86 min; isocratic elution with 2% B from 86 to 100 min. ESI mass spectrometry was performed with a Thermo Scientific LTQ-Orbitrap-Elite MS in the positive ion mode. Five microliters of the samples was injected. The peptides eluted from the column at a flow rate of 0.25 µL/min were introduced into the source of the mass spectrometer online. The nanoelectrospray ion source was operated at 2.5 kV. The inlet capillary temperature was maintained at 200 °C. The samples were analyzed by either a data-dependent acquisition (DDA) mode, in which each full MS scan was acquired in the Orbitrap at a resolution of 60 000 and followed by 15 MS/MS scans, or a selective reaction monitoring (SRM) mode, in which specific peptides were targeted over the entire course of the LC experiments. For the DDA mode, the dynamic exclusion option was enabled after three repeat acquisitions within 20 s duration, and the exclusion duration was set at 90 s. The MS/MS collision energy was set to 35%. The MS/MS spectra obtained were searched against a human protein database. Common modifications in the form of various types of adducts formed between proteins and fragmented oxPL, including Schiff base adduct, Michael adduct, ketoamide adduct, cyclic hemiacetal adduct, and other previously reported adducts,17 were used for the database search. The corresponding mass shifts were shown in Table S1, Supporting Information. The search parameters used were two missed cleavage sites, a mass tolerance of 10 ppm for the parent ion, and 1.2 Da for the fragment ion.
RESULTS
Strategy for Enrichment of Peptides Covalently Modified by oxPL
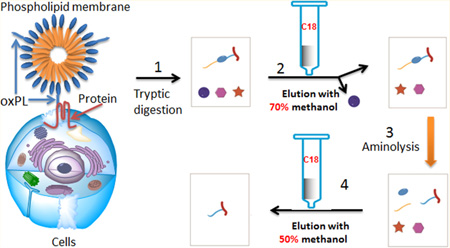
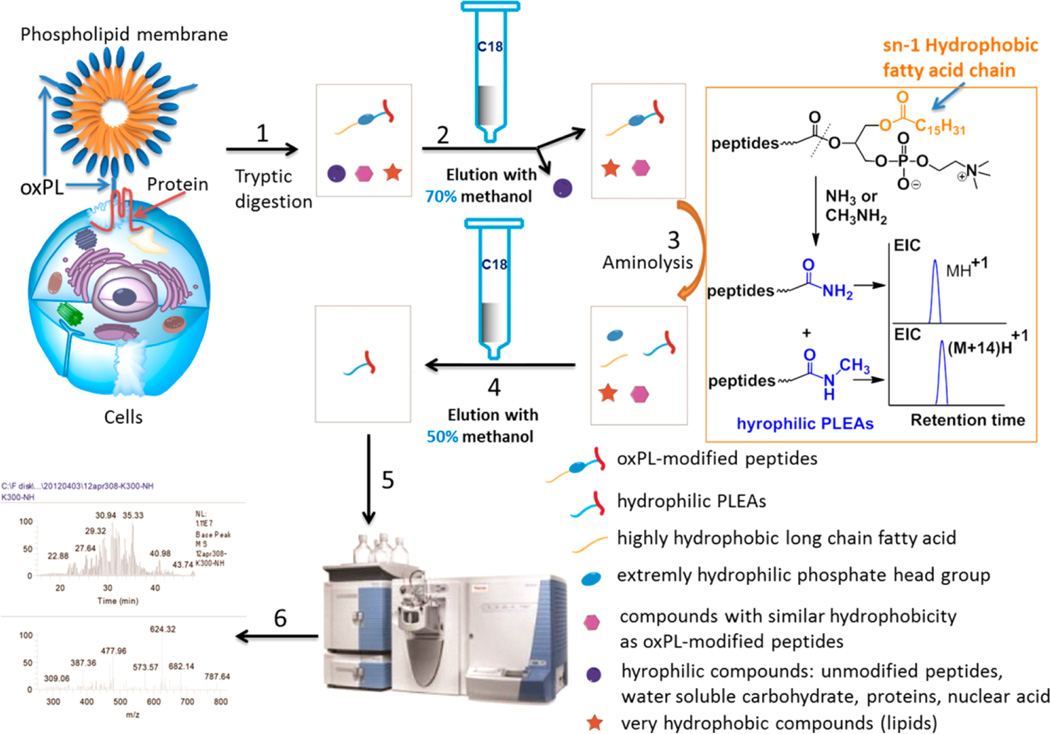
Upon covalent interaction with oxPL, modified peptides acquire significant hydrophobicity due to the presence of the highly hydrophobic long chain fatty acid at the sn-1 position of oxPL, which reacted with proteins as shown in Figure 1. The dramatic difference in hydrophobicity can be exploited to separate oxPL-modified peptides from more hydrophilic unmodified peptides and other interfering compounds using hydrophobic interaction chromatography.
Figure 1.
Strategy for the enrichment of oxPL-modified peptides using C18 columns. (1) Proteolytic digestion of oxPL–protein adducts to generate oxPL-modified peptides, (2) enrichment using a C18 column to remove hydrophilic unmodified peptides and other interfering compounds, (3) aminolysis to transform oxPL-modified peptides into hydrophilic PLEAs, (4) enrichment using a C18 column to separate PLEAs from other interfering hydrophobic unmodified peptides and compounds, (5) peptide analysis by LC-MS/MS, and (6) peptide identification.
The general strategy for the enrichment of oxPL-modified peptides is shown in Figure 1. A protein sample containing oxPL-modified proteins was digested with trypsin. The resulting digest was then loaded onto a reverse phase (C18) column. Hydrophobic components of the digests, such as oxPL-modified peptides, possess high binding affinity to the C18 stationary phase, allowing removal of hydrophilic compounds, including unmodified hydrophilic peptides, by elution with the appropriate mobile phase (methanol/water). Next, to separate the hydrophobic oxPL-modified peptides from hydrophobic unmodified peptides and unrelated hydrophobic compounds also retained on the C18 stationary phase, we exploited specific structural characteristics of oxPL-modified peptides. To dramatically and specifically reduce the hydrophobicity of oxPL-modified peptides, we used aminolysis, a reaction that will break the bond between the phosphoglycerol backbone and the residues at the sn-2 position of oxPL, thus separating the hydrophobic fatty acid at the sn-1 position from the modified peptide. The resulting peptide–lipid electrophile adducts (PLEAs) are much more hydrophilic than their precursors (Figure 1). Consequently, the PLEAs can be eluted from the C18 column using a polar solvent, allowing separation from other interfering hydrophobic compounds that will be retained on the C18 stationary phase. Thus, this strategy permits removal of both hydrophilic and hydrophobic unmodified peptides, as well as other interfering compounds, from the sample in two simple steps.
For the detection of oxPL–proteins adducts of very low abundance or in a complex biological system with relatively low levels of oxPL–protein adducts, large amounts of protein digest have to be loaded on a C18 column. In this situation, we found that to prevent the overloading of the column, we need additional preliminary enrichment of the samples containing oxPL–peptide adducts. To this end, we used a property of lysophosphatidylcholine (lysoPC) to form mixed micelles incorporating oxPL–peptide adducts. According to our experiments, most of the unmodified peptides under these conditions are not incorporated in vesicles and can be easily separated from vesicles using a size separation procedure. Experiments with several peptides demonstrated about 80–100 fold enrichment of oxPL-modified peptides over unmodified peptides using this procedure (see SI methods and Figure S2 in Supporting Information). The association of oxPL-modified peptides with lysoPC vesicles is most probably a reflection of amphiphilic properties and long hydrophobic fatty acid chain that both possess.
Hydrophobic Interaction Chromatography Allows Effective Separation of Unmodified Peptides from oxPL-Modified Peptides
To test whether the separation of oxPL-modified peptides from unmodified peptides can be achieved using hydrophobic interaction chromatography, we first examined elution conditions of unmodified peptides on a C18 column. To find universal elution conditions for unmodified peptides, we generated various peptides by tryptic digestion of a combination of several proteins, including human serum albumin, β-lactoglobulin, α-lactalbumin, protein kinase A, and myelin basic protein. The digest was loaded onto a C18 column and eluted stepwise by aqueous solution of increasing concentrations of methanol (see SI methods in Supporting Information). We found that most of the peptides were eluted using 10 bed volumes of 50% and 70% methanol successively (Figure S3a, Supporting Information).
We next examined the elution condition for oxPL-modified peptides. We chose apoA-I, a main component of HDL, for experiments because it is a major target of biotinylated oxPL in plasma,14 and because the oxPL modification may impair its major function in reverse cholesterol transport, leading to clinical consequences.18 As a model oxPL, we used KODA-PC, a biologically active oxPL known to be present in significant amounts in vivo in plasma and at sites of oxidative stress.19 ApoA-I was incubated with KODA-PC at room temperature for 1 h, followed by NaBH4 reduction and tryptic digestion. The digest, containing various KODA-PC-modified peptides, was loaded onto a C18 column and eluted stepwise by aqueous solution with increasing concentrations of methanol. Eluent fractions were collected and subjected to aminolysis, thus removing the phospholipid moiety which could interfere with the LC-MS/MS detection of oxPL-modified peptides by suppressing ionization and competing with peptide fragmentation. The resulting samples were further purified using a second C18 column and analyzed using LC-MS/MS. The MS/MS spectra were searched against an apoA-I sequence database using mass spectrometry data analysis program SEQUEST. Ten KODA-PC-modified peptides were identified in the 100% methanol eluent fraction (Table S2, Supporting Information). Of these, four KODA-PC-modified peptides were also detected in the 90% methanol eluent fraction, but no modified peptides were found in the 70% and 80% methanol eluent fractions. These results indicated that a high concentration of methanol (90–100%) is required to elute oxPL-modified peptides from a C18 column. The above experiments demonstrated that while most of the unmodified peptides can be eluted off the C18 column using 50–70% methanol, oxPL-modified peptides will be retained on the C18 column under this elution condition, allowing the separation of oxPL-modified peptides from hydrophilic unmodified peptides and other hydrophilic compounds.
We reasoned that, in addition to oxPL-modified peptides, hydrophobic unmodified peptides and nonprotein hydrophobic compounds could also be retained on the C18 column, interfering with the subsequent analysis. To separate the oxPL-modified peptides from other hydrophobic interfering compounds, we used aminolysis to transform the hydrophobic oxPL-modified peptides into more hydrophilic PLEAs (Figure 1). Studies of the elution conditions using KODA-PC-modified apoA-I demonstrated that the hydrophilic PLEAs can be eluted off the C18 column using 50% methanol (Table S3, Supporting Information). Under this mild elution condition, the interfering hydrophobic unmodified peptides and other compounds will be retained on the C18 column. Thus, by a combination of approaches described above, both hydrophilic and hydrophobic unmodified peptides as well as other interfering compounds can be separated from oxPL-modified peptides.
Optimization of the Enrichment Procedure and Enrichment Efficiency Study
Next, we optimized the elution gradients and aminolysis conditions (Table S4, Supporting Information) to increase the enrichment efficiency. We then studied the enrichment efficiency of the procedure using apoA-I modified by KODA-PC. In one set of samples, the KODA-PC-modified peptides were enriched using the C18 enrichment procedure (see Experimental Section). In the control set of samples, the tryptic digest was directly subjected to aminolysis without the C18 enrichment procedure. Both samples were subsequently desalted using a second C18 column and subjected to LC-MS/MS analysis using a synthetic peptide (LVNEVTEFAK) as an internal standard. Selected reaction monitoring (SRM) mode was used to monitor five unmodified peptides and five KODA-PC-modified peptides of various length and polarity. The peak area of each peptide in the enriched samples was normalized to that in the control set of samples. The analysis demonstrated that after the C18 enrichment procedure, about 40–60% KODA-PC-modified peptides were recovered, while less than 0.5% of unmodified peptides were not removed (Figure S3b, Supporting Information), demonstrating about 100-fold relative enrichment and good recovery of modified peptides.
Preliminary Enrichment Using a Size Separation Procedure Can Further Improve the Limit of Peptide Detection
We then tested the efficiency of the preliminary enrichment method exploiting lysoPC capacity to incorporate oxPL-modified peptides, but not unmodified peptides, into lysoPC vesicles, allowing subsequent separation by a size separation procedure. We analyzed a tryptic digest using either the C18 enrichment procedure or using a combination of two procedures. To further increase the complexity of the system, instead of using apoA-I, in these experiments we used human HDL modified by KODA-PC. The HDL was modified by KODA-PC, digested with trypsin, and mixed with a tryptic digest of RAW cell lysate in various mass ratios (1/25000, 1/125000, 1/625000) (Table 1). Samples were analyzed by LC-MS/MS using a data-dependent acquisition (DDA) mode, as well as a SRM mode. The LC-MS/MS analysis (Table 1) demonstrated a significant advantage of the combination of two enrichment procedures under conditions of a large excess of unmodified proteins. Encouragingly, even at a mass ratio of 1/625 000, one modified peptide was still detected by SRM and even by DDA analysis in the experiment using both enrichment procedures. These results demonstrated that this preliminary enrichment procedure further enhances the enrichment efficiency and increases the detection limit in the subsequent LC-MS/MS analysis. The proposed method allows detection of very low levels of oxPL–protein adducts (at least <2 ppm) in complex protein mixtures by LC-MS/MS.
Table 1.
KODA-PC-Modified Peptides Detected by LC-MS/MS in the Detection Limit Studya
| KODA-PC-modified peptides detected by LC-MS/ MS |
|||
|---|---|---|---|
| mass ratio of digestion mixture (HDL–KODA-PC adduct/cell lysate) |
enrichment methods |
SRM mode | DDA mode |
| 1/25000 | SSP + C18 | P1, P2, P3, P4, P5, P6, P7 | P1, P2, P3, P4, P6, P7 |
| 1/125000 | SSP + C18 | P1, P2, P3, P4, P7 | P2, P3, P7 |
| 1/625000 | SSP + C18 | P2 | P2 |
| 1/25000 | C18 | P2 | ND |
| 1/125000 | C18 | ND | ND |
SSP, size separation procedure; C18, C18 enrichment procedure;
ND, no detection; P1, AH*VDALR; P2, TH*LAPYSDELR; P3, LAEYH*AK; P4, ATEH*LSTLSEK; P5, LH*ELQEK; P6, LEALK* ENGGAR; P7, TAK*DALSSVQESQVAQQAR.
Aminolysis Using Ammonium Hydroxide and Methylamine, Respectively, Generates Doublet Peaks in LC-MS/MS Analysis
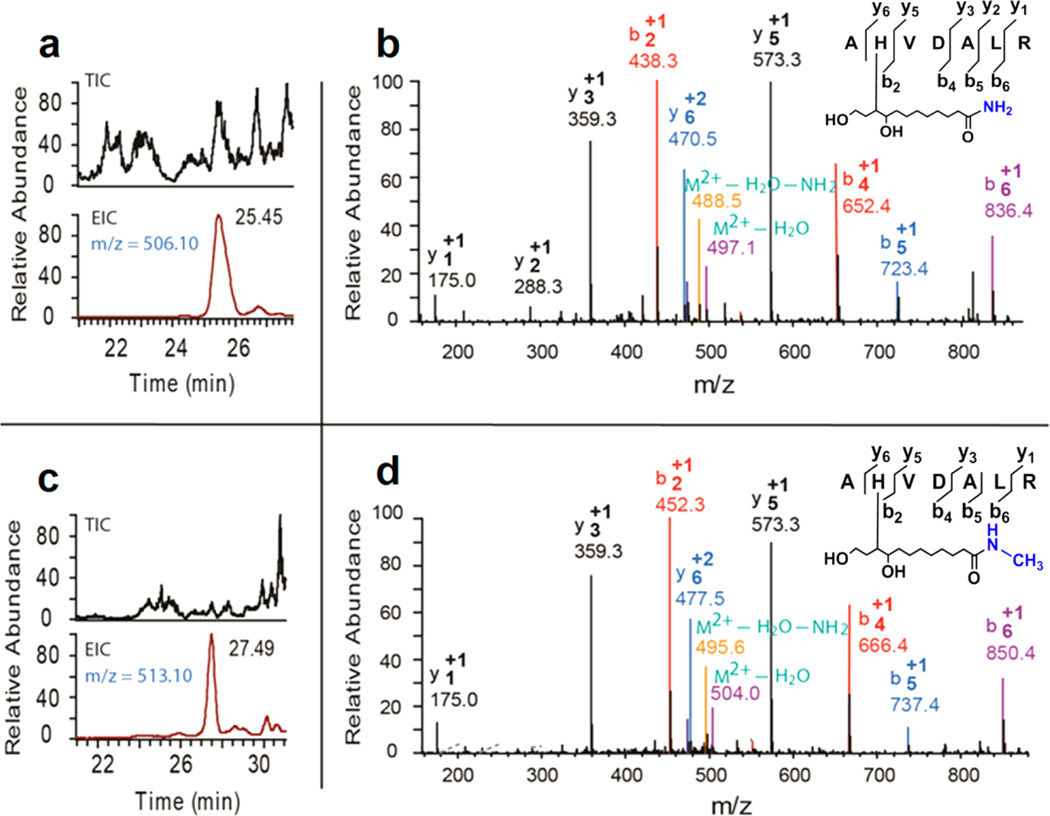
Another technique that was designed to facilitate the identification of oxPL-modified peptides is a use of either ammonium hydroxide or methylamine in parallel experiments for aminolysis, generating amide and N-methylamide accordingly (Figure 1). The molecular weight difference between the amide and N-methylamide is 14 Da. Thus, in the subsequent LC-MS/MS analysis, two sets of peaks are detected, with a mass-to-charge ratio (m/z) difference of 14 Da for singly charged ions and 7 Da for doubly charged ions. The extracted ion chromatogram and MS/MS spectra of a pair of PLEAs, generated by aminolysis of a KODA-PC-modified peptide (AH*VDALR) using either ammonium hydroxide or methylamine (see Experimental Section), is shown in Figure 2. The m/z difference between the two doubly charged precursor ions is 7 Da (Figure 2a and 2c, m/z 506.1 VS 513.10). In addition, the MS/MS spectra demonstrated a m/z difference of 14 Da between the two sets of singly charged b ions (b2, b4, b5, and b6), and 7 Da between two sets of doubly charged y ions (y6). The detection of two sets of MS/MS peaks with a m/z difference of 14 Da (singly charged ions) or 7 Da (doubly charged ions) confirms that the peaks belong to oxPL-modified peptides, thus facilitating the identification of oxPL-modified peptides. This approach is especially helpful for identification of peptides with low matching scores, especially when the peptide has poor fragmentations and the resulting MS/MS spectra cannot provide enough information for peptide identification.
Figure 2.
Aminolysis of oxPL-modified peptides using ammonium hydroxide and methylamine respectively generates doublet peaks in LC-MS/MS analysis. LC-MS extracted ion chromatogram of KODA-PC modified peptide (AHVDALR) aminolyzed using ammonium hydroxide (a), and methylamine (c), and the corresponding MS/MS spectra b and d.
Detection of Various oxPL–Protein Adducts in Oxidized HDL (oxHDL)
We applied the method described above to study the oxPL–protein adducts generated in the process of oxidation of HDL. HDL (0.8 mg/mL) was subjected to a strong oxidation by dialysis against 10 µM copper sulfate as described,2 followed by NaBH4 reduction to stabilize the reversible oxPL–protein adducts (see Experimental Section and Figure S1, Supporting Information). After the tryptic digestion of oxHDL, oxPL-modified peptides were enriched using the double enrichment method and subjected to LC-MS/ MS analysis. The MS/MS spectra were searched against a human protein sequence database using SEQUEST software. The LC-MS/MS analysis (Table 2) demonstrated that eleven amino acid residues on apoA-I were modified by a variety of oxPL in the form of several adduct types. We also detected modifications of five amino acid residues in less abundant apoA-II, and one residue in minor HDL apolipoprotein apoC-III. This study demonstrated the versatility of the present enrichment method to enrich a wide range of oxPL–peptide adducts formed between peptides of different lengths and oxPL with a variety of reactive functional groups at their sn-2 position.
Detection of oxPL–Protein Adducts in Human Platelets Exposed to KODA-PC
We have recently shown that specific oxidized phospholipids (oxPCCD36) bind to platelets via scavenger receptor CD36 and, at pathophysiological levels, promote platelet activation via CD36.19 In the present study, we tested whether the proposed enrichment method can be applied to study the covalent modifications of proteins in whole cells, using human platelets. Isolated human platelets were incubated with 20 µM KODA-PC, subsequently treated with NaBH4, lysed with 8 M urea, and digested with trypsin. OxPL-modified peptides were enriched using the double enrichment method and subjected to LC-MS/MS analysis. The MS/MS spectra were searched against a human protein database using SEQUEST software. We detected six proteins modified by KODA-PC in platelets (Table 3). Lysine 40 on CD36 was found to form a Schiff base adduct with KODA-PC (Figure S4a, Supporting Information, MS/MS spectra), which is consistent with the previous report that CD36 (AA 28–93) contains a binding domain for oxPL.20 Talin, an adaptor protein required for platelet integrin activation and for platelet function in hemostasis and thrombosis,21 was found to be modified by KODA-PC at Cys1023. In addition, two low abundance proteins in platelet (<50 ppm), myosin-13 and helicase, were demonstrated to form ketoamide and furan adducts (Figure S4b, Supporting Information, MS/MS spectra) with KODA-PC at Lys1835 and Lys1035, respectively.
Table 3.
KODA-PC–Protein Adducts Identified in KODA-PC-Treated Human Platelets
| SEQUEST score | ||||||
|---|---|---|---|---|---|---|
| protein targets of KODA-PC in platelets |
protein location |
molecule No. (abundance in platelet, ppm)a |
sequences modified | position (adduct type) |
XCorr | ΔM (ppm) |
| CD36 | membrane | 16700 (726) | K*QVVLEEGTIAFK | K40 (SB) | 3.86 | 0.58 |
| talin-1 | cytoplasm | 116000 (5043) | ASVPTIQDQASAMQLSQC*AK | C1023 (MA) | 3.90 | −0.23 |
| helicase SKI2W | cytoplasm | 1000 (43) | ERMQIQK*EMER | K1035 (furan) | 2.94 | −0.89 |
| myosin-13 | cytoplasm | <500 (<22) | ELENELDVEQK*R | K1835 (KA) | 2.12 | 1.08 |
| protein SCAF11 | cytoplasm | NA | SC*NEQIEESEK | C450 (furan) | 2.13 | −5.09 |
| dynein heavy chain 12, axonemal isoform 1 | cytoplasm | NA | C*IIFIDDMNMPALEK | C1955 (MA) | 2.58 | −4.07 |
The ppm value was calculated as follows: molecule number of each protein divided by the molecule number of total proteins in one platelet.
DISCUSSION
Oxidative truncation of the polyunsaturated fatty acid chain at the sn-2 position of phospholipids generates a variety of phospholipids with various truncated sn-2 fatty acid moieties and their short-chain aldehyde counterparts such as 4-hydroxy-2-nonenal (HNE) and 4-oxo-2-nonenal (ONE).1,17 The bioactivity and signaling functions of these phospholipid oxidation products have drawn widespread interest because of their contribution to chronic inflammatory pathologies.1,17 Characterization of the covalent interaction of phospholipid oxidation products with proteins by LC-MS/MS could provide critical information about the molecular mechanisms by which phospholipid oxidation products exert their effects. Over the past two decades, covalent interactions of proteins with HNE and ONE, two diffusible short chain aldehydes, have been extensively studied.17 In the case of HNE, several enrichment methods for HNE–protein adducts have been developed.22 These methods have dramatically accelerated the discovery of HNE–protein adducts in a relevant biological systems.
Compared with the studies on HNE–protein adducts, the studies on oxPL–protein adducts are lagging because of the lack of appropriate enrichment methods. Unlike HNE, oxPL represent a wide variety of species with varying functional groups at both sn-2 and sn-3 positions. The previously reported enrichment methods14,15 using biotinylated phospholipid probes to detect protein modifications produced significant results but have several limitations. While protein adducts with biotinylated oxPL can be easily enriched using streptavidin resin, one major disadvantage of these methods is the inability to enrich protein adducts with naturally occurring oxPL generated either in vitro or in vivo. In addition, the interaction between proteins and biotinylated oxPL, to some extent, may not reflect the actual interaction between proteins and naturally occurring oxPL because of the structural difference between biotinylated oxPL and naturally occurring oxPL.
In the present study, we describe the development of a high efficiency enrichment method for oxPL-modified peptides and its application for analysis of complex biological samples. This method takes advantage of the highly hydrophobic long chain fatty acid of oxPL moieties in oxPL-modified peptides. Most importantly, this feature engenders a significant difference in hydrophobicity between unmodified and oxPL-modified peptides, allowing a very effective separation by hydrophobic interaction chromatography. Furthermore, the “in column” aminolysis allows us to selectively reduce the hydrophobicity of oxPL-modified peptides and separate them from hydrophobic unmodified peptides and other interfering compounds by eluting from C18 column with polar solvents. Also, the hydrophobic fatty acid chain is likely to enable the oxPL-modified peptides to be incorporated into lysoPC micelles and facilitate their preliminary enrichment by a size separation procedure (we used centrifugal filtration for simplicity and speed), allowing removal of the small-size unmodified peptides and various other compounds which are not incorporated into lysoPC micelles.
The previous study using the biotinylated phospholipid probe reported that apoA-I in HDL was one of the most highly modified proteins in plasma.14 We applied the new method to study the formation of protein adducts with various naturally occurring oxPL during free radical-induced oxidation of HDL. These experiments demonstrated the versatility of the current method which allows enrichment of a wide range of oxPL–peptide adducts formed between peptides of various lengths and oxPL with a number of reactive functional groups at sn-2 position. A wide variety of adducts formed between various oxPL and the major as well as minor apolipoproteins in HDL were detected using LC-MS/MS (Table 2). All the histidine and lysine residues in the region (155–199) of apoA-I were intensively modified by various oxPL. Among them, His155, His162, and Lys182 are located in a region (144–186) that is critical for promoting ABCA1-mediated lipid efflux,23 activation of lecithin–cholesterol acyltransferase, and subsequent HDL remodeling and maturation.24 In addition, His193, Lys195, and His199 are located in the C-terminal region of apoA-I, which is critical for cell surface binding and lipid efflux.25 These amino acids are also inside the binding domain for myeloperoxidase which forms a ternary complex with apoA-I and PON1 and is involved in apoA-I modification.26 ApoA-II, the second most abundant proteins of the HDL, involved in visceral fat accumulation and metabolism of triglyceride rich particles,27 was intensively modified at Lys 39 and Lys 46 by various oxPL. Lys 39 has been reported to play a role in the secondary structure of APO-AII.28 ApoC-III, a minor HDL apolipoprotein involved in triglyceride metabolism,29 was modified at Lys 44 by ON-PL. The oxPL modifications on HDL proteins reported here are likely to impair HDL functions in lipid metabolism including its role in reverse cholesterol transport.
Investigation of the interaction between oxPL and proteins targets in cells could provide important information on the molecular mechanism by which oxPL impact cell function. Gugiu and co-workers have recently studied covalent modification of proteins by biotinylated oxPL in endothelial cells using the biotinylated phospholipid probe.15 This approach allowed to successfully identify 20 proteins interacting with the biotinylated oxPL. However, no modification sites on target proteins and adduct types were reported in this study. In the current study, the application of the new method allowed us to identify six proteins in human platelet that covalently interact with KODA-PC, forming various adduct types, and to locate the exact oxPL modification sites on the target proteins. Our previous study demonstrated that oxPL are recognized by and activate platelets via scavenger receptor CD36.19 However, no covalent modification of CD36 has been reported previously. In the present study, we detected a Schiff base adduct formed between KODA-PC and CD36 and identified one oxPL modification site (Lys 40) on CD36. Surprisingly, other target proteins of KODA-PC are located not on the platelet membrane but in the cytoplasm (Table 3). Of particular interest is modification of talin, an adaptor protein controlling platelet integrin αIIbβ3 activation and platelet function in hemostasis and thrombosis.21 Talin is located in cytoplasm in resting platelets. Its translocation to the membrane during platelet activation is critical for integrin activation. While the effect of oxPL modification of Cys1023, located in the actin binding domain of talin, on talin function is unknown, it is likely to induce constitutive association of talin with membrane. Such constitutive association with the membrane can lead to enhanced function of talin and may play a role in platelet activation by specific oxPL.19 Two cytoplasmic proteins with a low abundance in platelets, helicase SKI2W (about 43 ppm) and myosin-13 (less than 22 ppm),30 were also modified by KODA-PC at Lys1035 and Lys1835, respectively. This last result highlights the most surprising result of this experiment, namely a clear demonstration of highly selective modification of proteins with oxPL. This result strongly suggests that the secondary and tertiary structure of the protein and intracellular localization play a role in the highly selective modification of cellular proteins by oxPL. In the case of platelets, there is a possibility that KODA-PC is translocated into the platelet via CD36, providing some specificity for interaction with intracellular proteins. Taken together, these results demonstrate the high efficiency of the proposed method for the enrichment and subsequent detection of oxPL–peptide adducts derived from low abundance proteins in a relevant biological system.
CONCLUSION
In the current study, we successfully developed a highly efficient enrichment method for oxPL–peptide adducts. To our knowledge, this is the first enrichment method that can be applied to global analysis of protein adducts with various naturally occurring oxPL in a relevant biological system. This method has enabled us to detect various adducts between proteins and a wide variety of naturally occurring oxPL, to detect them at very low level (the lower boundary of sensitivity is <2 ppm) and to identify target proteins and sites in proteins selectively modified by oxPL. The enrichment method is highly promising for the study on the covalent interactions between naturally occurring oxPL and proteins in various biological systems, which can provide mechanistic insights into oxidative damage in pathologies associated with inflammation and oxidative stress.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Alejandro Zimman for helpful discussions and Dr. Ling Li for technical assistance on LC-MS/MS analysis. This work is supported in part by National Institute of Health grants HL077213, 5P01HL073311-06, and 1S10RR031537-01.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Additional information as noted in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Bochkov VN, Oskolkova OV, Birukov KG, Levonen AL, Binder CJ, Stockl J. Antioxid. Redox Signal. 2010;12:1009–1059. doi: 10.1089/ars.2009.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Podrez EA, Poliakov E, Shen Z, Zhang R, Deng Y, Sun M, Finton PJ, Shan L, Gugiu B, Fox PL, Hoff HF, Salomon RG, Hazen SL. J. Biol. Chem. 2002;277:38503–38516. doi: 10.1074/jbc.M203318200. [DOI] [PubMed] [Google Scholar]
- 3.Podrez EA, Poliakov E, Shen Z, Zhang R, Deng Y, Sun M, Finton PJ, Shan L, Febbraio M, Hajjar DP, Silverstein RL, Hoff HF, Salomon RG, Hazen SL. J. Biol. Chem. 2002;277:38517–38523. doi: 10.1074/jbc.M205924200. [DOI] [PubMed] [Google Scholar]
- 4.Androulakis N, Durand H, Ninio E, Tsoukatos DC. J. Lipid Res. 2005;46:1923–1932. doi: 10.1194/jlr.M500074-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Li R, Mouillesseaux KP, Montoya D, Cruz D, Gharavi N, Dun M, Koroniak L, Berliner JA. Circ. Res. 2006;98:642–650. doi: 10.1161/01.RES.0000207394.39249.fc. [DOI] [PubMed] [Google Scholar]
- 6.Zimman A, Mouillesseaux KP, Le T, Gharavi NM, Ryvkin A, Graeber TG, Chen TT, Watson AD, Berliner JA. Arterioscler., Thromb., Vasc. Biol. 2007;27:332–338. doi: 10.1161/01.ATV.0000252842.57585.df. [DOI] [PubMed] [Google Scholar]
- 7.Imai Y, Kuba K, Neely GG, Yaghubian-Malhami R, Perkmann T, van Loo G, Ermolaeva M, Veldhuizen R, Leung YH, Wang H, Liu H, Sun Y, Pasparakis M, Kopf M, Mech C, Bavari S, Peiris JS, Slutsky AS, Akira S, Hultqvist M, Holmdahl R, Nicholls J, Jiang C, Binder CJ, Penninger JM. Cell. 2008;133:235–249. doi: 10.1016/j.cell.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bochkov VN, Mechtcheriakova D, Lucerna M, Huber J, Malli R, Graier WF, Hofer E, Binder BR, Leitinger N. Blood. 2002;99:199–206. doi: 10.1182/blood.v99.1.199. [DOI] [PubMed] [Google Scholar]
- 9.Cole AL, Subbanagounder G, Mukhopadhyay S, Berliner JA, Vora DK. Arterioscler., Thromb., Vasc. Biol. 2003;23:1384–1390. doi: 10.1161/01.ATV.0000081215.45714.71. [DOI] [PubMed] [Google Scholar]
- 10.Kronke G, Bochkov VN, Huber J, Gruber F, Bluml S, Furnkranz A, Kadl A, Binder BR, Leitinger N. J. Biol. Chem. 2003;278:51006–51014. doi: 10.1074/jbc.M304103200. [DOI] [PubMed] [Google Scholar]
- 11.Birukov KG, Bochkov VN, Birukova AA, Kawkitinarong K, Rios A, Leitner A, Verin AD, Bokoch GM, Leitinger N, Garcia JG. Circ. Res. 2004;95:892–901. doi: 10.1161/01.RES.0000147310.18962.06. [DOI] [PubMed] [Google Scholar]
- 12.Yeh M, Gharavi NM, Choi J, Hsieh X, Reed E, Mouillesseaux KP, Cole AL, Reddy ST, Berliner JA. J. Biol. Chem. 2004;279:30175–30181. doi: 10.1074/jbc.M312198200. [DOI] [PubMed] [Google Scholar]; Yeh M, Cole AL, Choi J, Liu Y, Tulchinsky D, Qiao JH, Fishbein MC, Dooley AN, Hovnanian T, Mouilleseaux K, Vora DK, Yang WP, Gargalovic P, Kirchgessner T, Shyy JY, Berliner JA. Circ. Res. 2004;95:780–788. doi: 10.1161/01.RES.0000146030.53089.18. [DOI] [PubMed] [Google Scholar]
- 13.Greig FH, Kennedy S, Spickett CM. Free Radical Biol. Med. 2012;52:266–280. doi: 10.1016/j.freeradbiomed.2011.10.481. [DOI] [PubMed] [Google Scholar]
- 14.Szapacs ME, Kim HY, Porter NA, Liebler DC. J. Proteome Res. 2008;7:4237–4246. doi: 10.1021/pr8001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gugiu BG, Mouillesseaux K, Duong V, Herzog T, Hekimian A, Koroniak L, Vondriska TM, Watson AD. J. Lipid Res. 2008;49:510–520. doi: 10.1194/jlr.M700264-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Valiyaveettil M, Kar N, Ashraf MZ, Byzova TV, Febbraio M, Podrez EA. Blood. 2008;111:1962–1971. doi: 10.1182/blood-2007-08-107813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Drug Metab. Rev. 2006;38:651–675. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- 18.Podrez EA. Clin. Exp. Pharmacol. Physiol. 2010;37:719–725. doi: 10.1111/j.1440-1681.2010.05380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Podrez EA, Byzova TV, Febbraio M, Salomon RG, Ma Y, Valiyaveettil M, Poliakov E, Sun M, Finton PJ, Curtis BR, Chen J, Zhang R, Silverstein RL, Hazen SL. Nat. Med. (N. Y., NY, U. S.) 2007;13:1086–1095. doi: 10.1038/nm1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearce SF, Roy P, Nicholson AC, Hajjar DP, Febbraio M, Silverstein RL. J. Biol. Chem. 1998;273:34875–34881. doi: 10.1074/jbc.273.52.34875. [DOI] [PubMed] [Google Scholar]
- 21.Petrich BG, Marchese P, Ruggeri ZM, Spiess S, Weichert RAM, Ye F, Tiedt R, Skoda RC, Monkley SJ, Critchley DR, Ginsberg MH. J. Exp. Med. 2007;204:3103–3111. doi: 10.1084/jem.20071800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ullery JC, Marnett LJ. Biochim. Biophys. Acta, Biomembr. 2012;1818:2424–2435. doi: 10.1016/j.bbamem.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chroni A. J. Biol. Chem. 2002;278:6719–6730. doi: 10.1074/jbc.M205232200. [DOI] [PubMed] [Google Scholar]
- 24.Frank PG, Marcel YL. J. Lipid Res. 2000;41:853–872. [PubMed] [Google Scholar]; Roosbeek S, Vanloo B, Duverger N, Caster H, Breyne J, De Beun I, Patel H, Vandekerckhove J, Shoulders C, Rosseneu M, Peelman F. J. Lipid Res. 2001;42:31–40. [PubMed] [Google Scholar]
- 25.Burgess JW, Frank PG, Franklin V, Liang P, McManus DC, Desforges M, Rassart E, Marcel YL. Biochemistry. 1999;38:14524–14533. doi: 10.1021/bi990930z. [DOI] [PubMed] [Google Scholar]
- 26.Huang Y, Wu Z, Riwanto M, Gao S, Levison BS, Gu X, Fu X, Wagner MA, Besler C, Gerstenecker G, Zhang R, Li X-M, DiDonato AJ, Gogonea V, Tang WHW, Smith JD, Plow EF, Fox PL, Shih DM, Lusis AJ, Fisher EA, DiDonato JA, Landmesser U, Hazen SL. J. Clin. Invest. 2013;123:3815–3828. doi: 10.1172/JCI67478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scanu AM, Edelstein C. FASEB J. 2008;22:4044–4054. doi: 10.1096/fj.08-117150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar MS, Carson M, Hussain MM, Murthy HM. Biochemistry. 2002;41:11681–11691. doi: 10.1021/bi026069w. [DOI] [PubMed] [Google Scholar]
- 29.Jong MC, Hofker MH, Havekes LM. Arterioscler., Thromb., Vasc. Biol. 1999;19:472–484. doi: 10.1161/01.atv.19.3.472. [DOI] [PubMed] [Google Scholar]
- 30.Burkhart JM, Vaudel M, Gambaryan S, Radau S, Walter U, Martens L, Geiger J, Sickmann A, Zahedi RP. Blood. 2012;120:e73–e82. doi: 10.1182/blood-2012-04-416594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.