Abstract
Three plant growth-promoting bacteria (PGPB; Bacillus pumilus ES4, B. pumilus RIZO1, and Azospirillum brasilense Cd) were tested for their ability to enhance plant growth and development of the native Sonoran Desert shrub quailbush (Atriplex lentiformis) and for their effect on the native bacterial community in moderately acidic, high-metal content (AHMT) and in neutral, low metal content natural tailings (NLMT) in controlled greenhouse experiments. Inoculation of quailbush with all three PGPB significantly enhanced plant growth parameters, such as germination, root length, dry weight of shoots and roots, and root/shoot ratio in both types of tailings. The effect of inoculation on the indigenous bacterial community by the most successful PGPB Bacillus pumilus ES4 was evaluated by denaturating gradient gel electrophoresis (PCR-DGGE) fingerprinting and root colonization was followed by specific fluorescent in situ hybridization (FISH). Inoculation with this strain significantly changed the bacterial community over a period of 60 days. FISH analysis showed that the preferred site of colonization was the root tips and root elongation area. This study shows that inoculation of native perennial plants with PGPB can be used for developing technologies for phytostabilizing mine tailings.
Keywords: Bacillus, DGGE, FISH, phytostabilization, mine tailings
1. Introduction
In the deserts of the southwestern United States and northern Mexico, large mounds of mine tailings, the main waste product of mineral ore processing of abandoned and productive mines, are potentially a health hazard to nearby urban populations in the form of wind-blown dust and ground water sources. This happens because tailings, lacking plant cover and soil structure, easily degrade by wind and rain action and serve as a continuous source of metal pollution (Pilon-Smits, 2005). Phytostabilization, using native plants has been proposed as an economic strategy to reduce these hazards (McCutcheon and Schnoor, 2003). This phytoremediation approach involves creation of a plant cover on the tailings that is sufficient to prevent erosion. A major challenge is that some of these tailings cannot serve, in their natural state, as growth substrate for most plant species because of metal toxicity, low pH, lack of essential minerals, lack of clay and organic matter to retain water, lack of soil structure, lack of a seed bank of nearby native plants, or some combination of these environmental parameters (Mendez and Maier, 2008). Thus, tailings can remain devoid of plants for decades or have only a slight plant cover (Gonzalez-Chavez et al., 2008). A partial solution and an upgrade of this inhospitable substrate to a “soil-like” status can be accomplished by adding large quantities of compost, biosolids, and irrigation (Ye et al., 2001; Chiu et al., 2006). The drawback of the compost-water strategy is that applying large quantities of compost is often not economically feasible because of the extensive area of tailings and the remoteness of the sites. Furthermore, water and irrigation facilities are largely absent in deserts, especially at long-abandoned mines.
Inoculation with plant growth-promoting bacteria (PGPB; Bashan and Holguin, 1998) has been proposed to aid in establishment of plants on tailings at reduced compost concentrations (Petrisor et al., 2004; Zhuang et al., 2007; Mendez and Maier, 2008). Successful attempts have been made to isolate and use potential PGPB from tailings (Grandlic et al., 2008). The premise for this approach is that PGPB isolated from tailings may aid in plant survival and growth through several mechanisms including: increasing nutrient availability to plants, increasing plant resistance to metal toxicity, or decreasing toxic metal bioavailability in the rhizosphere (Burd et al., 1998, 2000; Pishchik et al., 2002; Glick, 2003; Belimov et al., 2004; Reed and Glick, 2005; Reed et al., 2005; Vivas et al., 2006; Wu et al., 2006a; Li et al., 2007; Rajkumar and Freitas, 2008 a,b).
Alternatively, there are many well-studied PGPB used in other applications that could be useful in helping plant establishment in mine tailings. For example, strains from the genus Bacillus have been shown to enhance the growth of agricultural crops, wild plants, trees, microalgae, and model plants, through different mechanisms of plant growth-promotion (Bashan et al., 2000; Enebak et al., 1998; Hernandez et al., 2009; Kloepper et al., 2004 a,b; Ryu et al., 2005; Vessey, 2003). Bacillus is also a genus commonly found in various types of mine tailings (Natarajan, 1998; Vijayalakshmi and Raichur, 2003; Wu et al., 2006b; Tsuruta, 2007; Zhang et al., 2007). A second genus, Azospirillum, is also commonly used for its PGP activities. Azospirillum is perhaps the best studied of the PGPB except for rhizobia (Bashan et al., 2004).
The hypothesis of this study is that inoculation with classical agricultural PGPB can support establishment and growth of the common, native Sonoran Desert shrub, quailbush (Atriplex lentiformis) in mine tailings at reduced compost concentrations. The specific objectives were to: (1) Evaluate the growth of quailbush in two types of tailings: an acidic, high-metal content tailing (AHMT) and neutral low-metal content tailing (NLMT) supplemented with a sub-optimal level of compost and inoculated with one of three PGPB (Bacillus pumilus ES4, B. pumilus RIZO1, and A. brasilenese Cd), comparing the performance of the three PGPB on plant growth; (2) Monitor root colonization capacity of the most successful PGPB using the technique of fluorescent in situ hybridization (FISH) and confocal laser microscopy; and (3) Assess whether there is an effect in structure on the tailings bacterial community by PGPB inoculation using PCR-DGGE fingerprint analysis.
2. Material and methods
2.1. Organisms, bacterial growth conditions and inoculant preparation
The plant growth-promoting bacteria Azospirillum brasilense Cd (DSM 1843, the type strain of A. brasilense; Braunschweig, Germany), Bacillus pumilus ES4, and Bacillus pumilus RIZO1 (Puente et al., 2004) and the evergreen shrub quailbush (Atriplex lentiformis) native to southwestern North America, served as model organisms. The two B. pumilus strains were identified by sequencing their entire 16S rRNA gene (100% similarity to deposited B. pumilus) and their sequences were deposited in the GenBank (B. pumilus RIZO1 accession number is FJ032016 and B. pumilus ES4 is FJ032017). Both B. pumilus strains are diazotrophs, where strain ES4 is also a phosphate solubilizer. Both strains can degrade various types of rocks, were isolated from the rhizoplane of a cactus, and promoted growth of the giant cardon cactus. Strain ES4 also promoted the growth of the microalgae Chlorella vulgaris by supplying it with fixed atmospheric nitrogen (Hernandez et al. 2009; Puente et al. 2004). Nomenclature of PGPB in the latter reference differs from the current nomenclature as B. pumilus.
The PGPB were cultivated on nutrient broth (Sigma) for 24 h at 30 °C and stirred at 120 rpm. The resulting bacteria cultures were formulated into a dry, microbead inoculant made of alginate, as described by Bashan et al. (2002), using specialized equipment (http://www.bashanfoundation.org/bead.html, accessed 8 April, 2010). Alginate inoculants, during numerous greenhouse and field experiments, proved to be an inert and reliable carrier for inoculation of PGPB for crops (Bashan 1986; Bashan et al. 2002) and various desert plants (Bashan et al. 2009, a,b; Grandlic et al. 2009). Bacterial microbead inoculant was mixed with wild quailbush seeds in the same planting hole in the greenhouse experiment at a level of 1.2 × 106 CFU g−1 soil, as described below.
2.2. Tailings characterization
Two types of substrate were used: Acidic, high-metal content tailings (AHMT) and neutral, low-metal content tailings (NLMT). The physiochemical properties of AHMT were previously characterized (Grandlic et al., 2008). A single composite sample of NLMT tailings was oven-dried at 105 °C, sieved through a 2-mm mesh, and analyzed for texture (Soil Survey Laboratory Methods Manual, 2004). Total organic carbon (TOC) and total nitrogen (TN) were measured in the solid phase using a Shimadzu high temperature combustion TOC/TN analyzer (organic and inorganic nitrogen). Electrical conductivity (EC), pH, dissolved organic carbon (DOC), and dissolved nitrogen (DN, organic plus inorganic) were measured in a saturated paste extract (1:1) in deionized water. Samples were prepared by microwave acid digestion (EPA method 3051; EPA, 2004) for total element (As, Cd, Cu, Fe, K, Mn, Na, Pb, Zn) analysis by ICP-MS. Analysis was done as a service by a laboratory in the Department of Soil, Water, and Environmental Science University of Arizona, Tucson, AZ. Total heterotrophic bacterial counts were done by suspending 1 g of tailing material in sterile 0.85% saline solution with shaking at 120 rpm for 1 h; serial dilution were done in 0.85% saline solution and plated on nutrient agar; the plates were incubated at 37 °C and developed colonies were counted. Enumerated bacteria were expressed as CFU g−1 soil.
2.3. Greenhouse cultivation
Three treatments were tested in this experiment. Treatment 1 evaluated the effect of three individual PGPB on the growth of quailbush in composted tailings. Treatment 2 evaluated the growth of quailbush in composted tailings in the absence of PGPB (the uninoculated control). Treatment 3 was an unplanted control consisting of pots of composted tailings that did not receive either plants or PGPB inoculation. Mature compost with relatively low levels of resident bacteria (de-Bashan et al., unpublished data) was applied. The physico-chemical compositions of the compost were previously described (Mendez et al. 2007).
In this experiment, quailbush was grown from seeds (Carter Seeds, Vista, CA, USA) in black plastic commercial pots (15 × 10 cm dia.) with a drainage hole, each containing a thoroughly mixed substrate of 530 g made of 9:1 (non-sieved tailings: dairy compost, w/w). As discussed by Grandlic et al. (2008), this is a suboptimal compost concentration for these tailings (the optimal tailings:compost ratio is 17:3). Initially, 20 seeds per pot were planted at ~0.5 cm with a sterile tweezers and were checked for germination five days later (day 0). Then, each pot was thinned to 10 seedlings. At day 5, one seedling was removed from each pot. This procedure was repeated at day 15, 30, and 60.
At the end of the experiment, 6 plants per pots were left and were used for measuring growth parameters and for FISH analysis of their roots, as explained below. A total of 50 pots were used (10 per treatment in a randomized layout). All plants were grown in a greenhouse on elevated metal net beds at controlled temperature (20–30 °C, n/d) and drip irrigated with tap water every 3 h with 93.4 ml pot−1. These are optimal irrigation conditions for this species. The experiment lasted 60 days, after which plant growth parameters were measured.
2.4. Extraction of DNA from soil and PCR amplification
“Rhizosphere tailing soil” was defined according to the original definition of rhizosphere soil by Hiltner (Hartmann et al. 2008), namely, tailing particles adhere to the roots after the plant was removed from the tailing substrate. Rhizosphere samples from each treatment (inoculated plants, non inoculated plants, soil amended with compost without plant, and compost) were collected in sterile 1.5-ml snap cap tubes and stored at –80 °C until analysis. DNA was extracted using a kit (Fast DNA SPIN for soils, MP Biomedicals, Solon, OH), following the manufacturer’s protocol. In samples containing humic acids, the binding matrix-DNA complex was further rinsed with saturated 5.5 M guanidine thiocyanate (Flucka Sigma-Aldrich, Buchs, Switzerland) until the supernatant lost its brown tint (Rosario et al., 2007). Samples (0.4 g soil) were used for each extraction and three replicates for each treatment were analyzed.
Once extracted, DNA was quantified in a mini-fluorometer (TBS 380, Turner BioSystems, Sunnyvale, CA), using the protocol for Quant-iT PicoGreen dsDNA (Molecular Probes, Eugene, OR). About 100 pg µl−1 of each sample was used for PCR reaction.
2.4.1. Polymerase chain reaction (PCR)
The V9 variable region of the 16S rRNA gene was PCR amplified using the bacteria primers 1070F (5′-ATG GCT GTC GTC AGC T-3′) and 1406R (5′-ACG GGC GGT GTG TAC-3′) with a 40-bp GC clamp (Ferris et al., 1996). PCR for DGGE was done using a modification of the method of Colores et al. (2000). Briefly, 25 µl of the reaction mix contained 2.5 µl of 10× buffer with 15 mM MgCl2 (Qiagen Sciences, Germantown, MD), 200 µM dNTP, 0.4 µM of each primer, 5% dimethyl sulfoxide (Sigma, St. Louis, MO), 0.4 g l−1 unacetylated bovine serum albumin (Sigma), 0.5 units µl−1 HotStarTaq DNA polymerase (Qiagen Sciences), and 100 pg of community DNA extract. Reaction was run on an automated PCR thermoblock (Peltier thermal cycler 200, MJ Research, Waltham, MA) for 95 °C for 15 min and 30 cycles (94 °C for 45 s, 55 °C for 45 s. 72 °C for 30 s). PCR products were viewed after electrophoresis by running a 2% agarose gel (GenePure LE ISC BioExpress, Kaysville, UT), followed by ethidium bromide (EMD Chemicals, Darmstat, Germany) staining.
2.5. Denaturing gradient gel electrophoresis (DGGE) analysis
DGGE of the 16s rRNA gene products was performed using a D-Code Universal Mutation Detection System (Bio-Rad Laboratories, Hercules, CA). Acrylamide gels (7%) were prepared with a 45 to 70 % urea-formamide denaturing gradient, according to the manufacturer’s protocol. Lanes were loaded with 10–15 µl of PCR product.
Electrophoresis was run at a constant 50 V for 16.5 h at 60 °C. Gels were stained with SYBR Green I nucleic acid gel stain (Lonza, Rockland, ME) and gel images were recorded with an AlphaImager HP (Alpha Innotech, San Leandro, CA) gel documentation system. DGGE profiles were analyzed using Quantity One 4.5.2 software (Bio-Rad Laboratories).
2.6. Probes for fluorescent in situ hybridization (FISH)
Two probes were used: EUB338 (Amman et al., 1990), which is specific for the domain Bacteria and the specific probe that targets the complementary region of 16S rRNA in Bacillus (BAC07; Probe Base accession number: pB00403; Liu et al., 2001). BAC07 was checked with the Probe Match tool of the Ribosomal Database Project and with Greengenes (http://greengenes.ibl.gov). Oligonucleotides probes were labeled at the 5′ end with the fluorescent dyes Cy3 (specific probe) and Cy5 (universal probe) (Integrated DNA Technologies, Coralville, IA).
2.6.1. Preparation of roots for fluorescent in situ hybridization
Root samples were taken at 15, 30, and 60 d. Roots were carefully separated from the soil, washed with 1× PBS (200 mM NaH2PO4, 200 mM Na2HPO4), and fixed with 4% paraformaldehyde (Acros Organics, Geel, Belgium) for 2 h at −4 °C. After fixation, roots were washed with 1 × PBS and stored in a mix of 1× PBS/96% Ethanol (1:1 v/v) at −20 °C until hybridization.
2.6.2. In situ hybridization
FISH analysis was performed as described by Iverson and Maier (2009). Briefly, slices of the fixed root were thawed and placed on a gelatin (0.1% w/v, 0.01% w/v chromium potassium sulfate)-coated microscope slides and fixed to the slide by adding 1 drop of warm, low-melt agarose solution (0.2 % w/v; Agarose LMP, Promega, Madison, WI) and dried at 37 °C for 45 min. Samples were dehydrated by successive 50, 80, and 96% ethanol washes (3 min each), then air dried. Prior to hybridization, dehydrated roots were treated with 10 mg ml−1 lysozyme (Sigma) for 15 min to enhance permeation of the probe into the Gram-positive Bacillus (Mogge et al., 2000).
Hybridization was performed at 15% stringency at 46 °C for 2 h (Daims et al. 2005). The final concentration of the probe was 3 ng µl −1. A careful washing was performed at 48 °C for 15 min with 50 ml warm washing buffer. The slides were then rinsed for a few seconds with ice-cold deionized water and then air dried. Slides not visualized immediately were stored at −20 °C in the dark.
2.6.3. Visualization of root colonization by confocal laser scanning microscopy
Before visualization, the slides were mounted in AF1 anti-fading reagent (Citifluor, Electron Microscopy Sciences Hatfield, PA). Microscopy was performed with a confocal scanning laser microscope (LMS 510, Carl Zeiss MicroImaging, Thornwood, NY). B. pumilus and other bacteria were viewed simultaneously by using two helium neon lasers for excitation of the dyes Cy3 at 543 nm and Cy5 at 633 nm. The pinhole was set for both channels at 1.2 Airy units. Samples were observed with a 63× water immersion objective (Carl Zeiss MicroImaging). For visualization, fluorescence from Cy3 and Cy5-labeled oligonucleotide probes were assigned green and red colors, respectively. B. pumilus cells were identified when the two single-color images were merged into one picture that produced a yellow color. Other bacterial colonies appeared red in the photomicrographs.
The images were combined and analyzed with a software (LSM 510, version 4.2 (Carl Zeiss International, Oberkochen, Germany). Images were processed with Adobe Photoshop, version 8.0 (Adobe Systems, Mountain View, CA).
2.7. Measurement of plant parameters
Plant growth parameters measured at day 60 of cultivation were: shoot and root length, number of developing leaves, and dry weight of shoot and roots. Once harvested, roots and shoots were separated and rinsed gently under running water to remove all substrate particles. Plant roots and shoots were then placed in individual aluminum foil packets, dried for 48 h in a 60 °C oven, and immediately weighed (Bashan and de-Bashan, 2005b).
2.8. Experimental design and statistical analysis
Greenhouse experiments were conducted in a completely randomized design with six plants per pot (60 plants per treatment and 300 plants per experiment). The experiment was analyzed statistically and repeated. The presented data is from one experiment. After normalization of the data, results of all experiments were analyzed by one-way ANOVA and then by Tukey’s HSD post-hoc analysis. Significance was set at P < 0.05. Data in percentage was converted to arcsine before analysis. These statistical measures used Statistica, ver. 6.0 (StatSoft, Tulsa, OK).
The profiles obtained from DGGE gels were analyzed for similarity using the Dice coefficient. A dendrogram was formed from the unweighted pair group matching band average (UPGMA). Similarity varies from 0 to 1, where 1 indicates 100% similarity. Additionally, similarities among profiles for all treatments at each time frame were evaluated by the Kruskal’s isotonic multi-dimensional scaling analysis (KNMDS) (Venables and Ripley, 2002) using the statistical software R (R Foundation for Statistical Computing, Vienna, Austria), as described for mine tailings by Rosario et al. (2007) at the significance level of P < 0.05. A stress factor was calculated to reflect goodness-of-fit of the models. Values <0.1 are considered a good fit.
3. Results
3.1. Mine tailing analysis
The NLMT tailings have the following physiochemical properties: pH 7.0; 0.2% total carbon; 0.02% total organic carbon; <0.01% total nitrogen; 450 mg kg−1 total phosphorus; 1.4 mg kg−1 organic phosphorus; 7400 mg kg−1 total sulfur; 1550 mg kg−1 SO4–2; 2.3 mmhos cm−1 electrical conductivity; 3.89 meq 100g−1 CEC; texture, 76.8% sand, 16.6% silt, 6.6% clay. Elemental analysis (mg kg−1): Fe (12600); Ca (9600); Mg (3550); K (3040); Na (783); Cu (423); Mn (190); Zn (39); Ni (11); Cr (8); Pb (3.4); As (1.4); Se (0.73); and Cd (0.37). The tailings lack a defined soil structure and have a culturable neutrophilic heterotrophic bacterial count of 8 ± 0.12 × 103 CFU g−1 dry tailings.
Analysis of the AHMT tailings has been presented previously (Grandlic et al. 2008). Here we summarize selected properties for comparison to the NLMT. The AHMT have a pH of 4.54; 0.004% total organic carbon; < 0.01% total nitrogen; texture, 51.9% sand, 26.4% silt, 21.7% clay. The major metals in the tailings are (mg kg−1): Fe (26,600); Pb (4620); Mn (2810); Zn (1400); Cu (653); As (91); Cd (2.4). The tailings lacked a defined soil structure and had a culturable neutrophilic heterotrophic bacterial count of 2.95 ± 0.97 × 103 CFU g−1 dry tailings (de-Bashan et al., unpublished data).
3.2. Effect of inoculation with three PGPB on growth parameters of quailbush
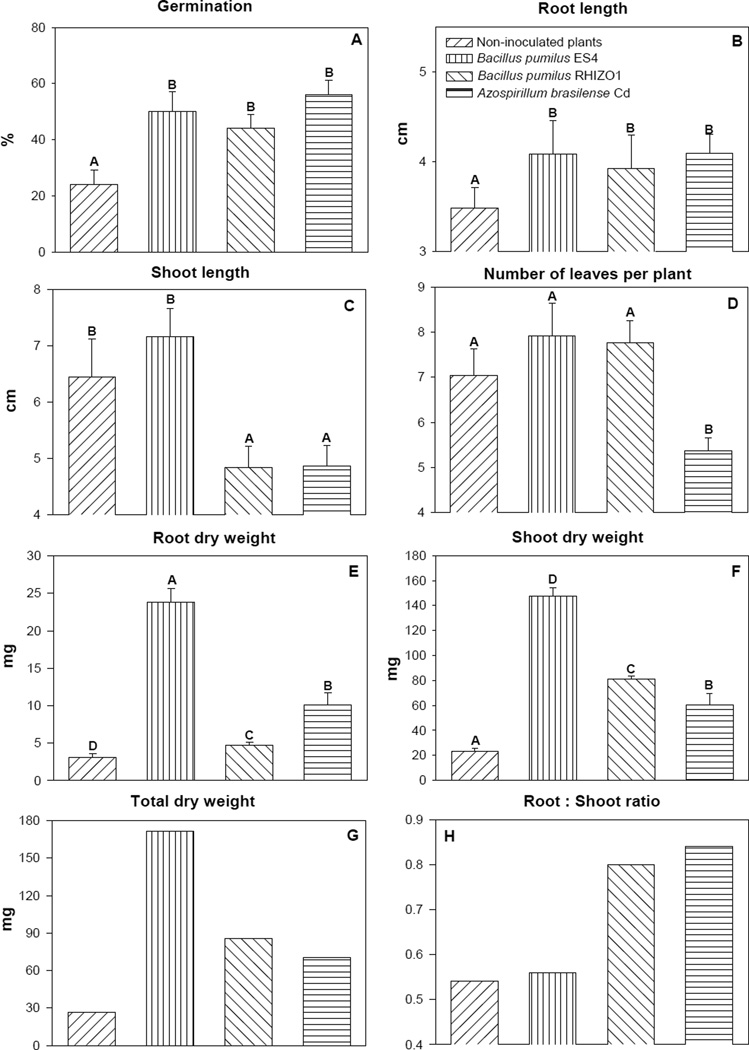
Promotion of growth of quailbush after inoculation with PGPB was significant, and variable in the two tailings. In the AHMT, the three PGPB increased seed germination (Fig. 1A) and root length (Fig. 1B). None of the PGPB significantly increased shoot length (Fig. 1C) or the number of leaves per plant (Fig. 1D). All PGPB strains enhanced root, shoot, and total dry weight of the plants (Fig. 1E, 1F, 1G); however, B. pumilus ES4 had the greatest effect in these parameters. The root:shoot ratio was enhanced only by B. pumilus RIZO1 and A. brasilense Cd.
Fig. 1.
Growth parameters of quailbush uninoculated or inoculated with one of three PGPB and grown in acidic, high-metal content mine tailings. Column denoted by a different letter in each subfigure, differ significantly by one-way ANOVA at P < 0.05. Bars represent standard error.
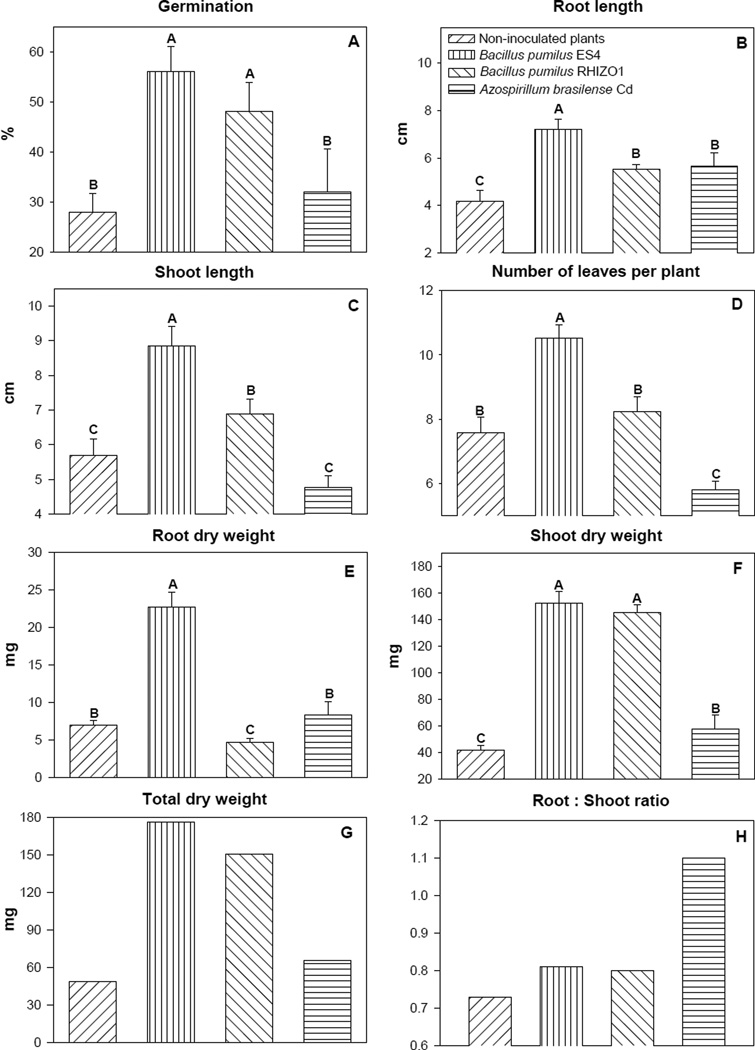
In the NLMT, seed germination, shoot length, and the number of leaves per plant were enhanced by the two B. pumilus strains but not by A. brasilense Cd (Fig 2A, 2C, 2D). Root length was enhanced by all PGPB strains (Fig. 2B). Root dry weight was enhanced only by B. pumilus ES4 (Fig. 2E). Shoot dry weight and total dry weight of plants were enhanced by the three PGPB (Fig. 2G). The root:shoot ratio was enhanced only by A. brasilense Cd (Fig. 2H).
Fig. 2.
Growth parameters of quailbush uninoculated or inoculated with one of three PGPB and grown in neutral, low-metal content mine tailings. Column denoted by a different letter in each subfigure, differ significantly by one-way ANOVA at P < 0.05. Bars represent standard error.
3.3. Effect of B. pumilus ES4 inoculation on tailings bacterial community structure
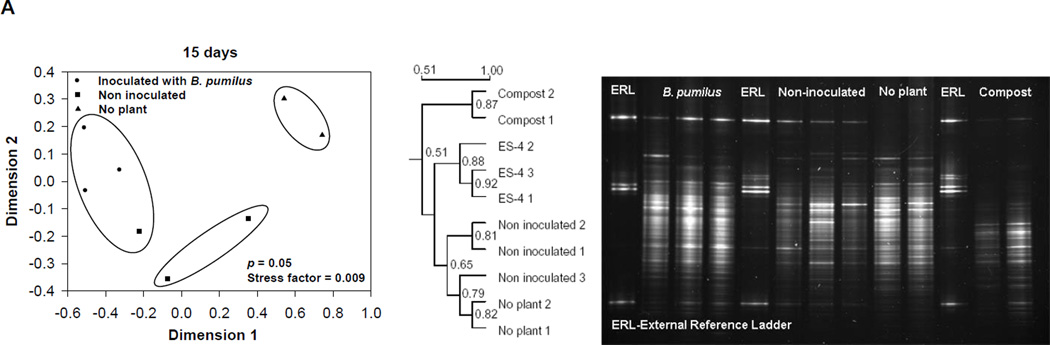
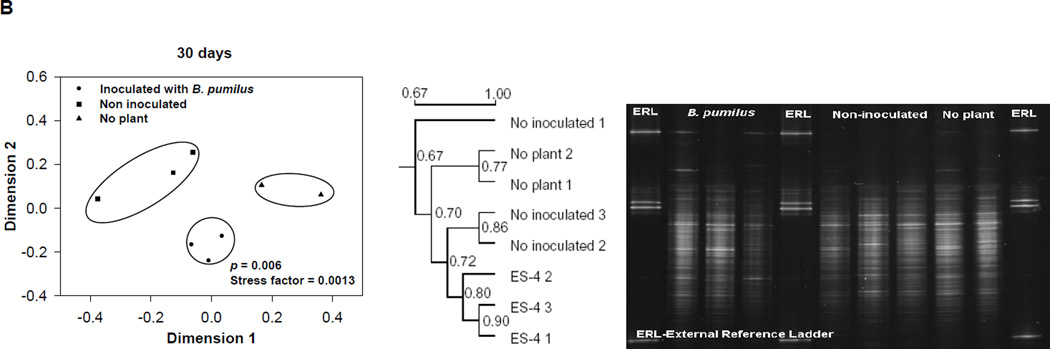
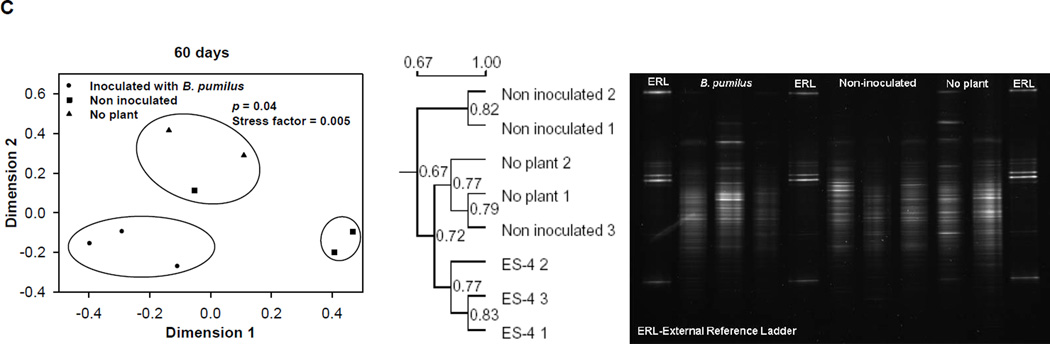
PCR-DGGE gels revealed that the rhizosphere bacterial community appeared to be different among the treatments, composted mine tailings inoculated with B. pumilus ES4 and planted, uninoculated planted controls, and unplanted controls at each time point examined (15, 30, and 60 d) (Fig. 3). Multidimensional scaling analysis was used to evaluate the relationship among the replicate sample profiles from each treatment. At 15 days, the sample profiles grouped into three clusters that were significantly different from each other (P < 0.05, Fig. 3A). One cluster contained the two unplanted control samples, the second cluster contained two of the three uninoculated control samples, and the third contained all of the inoculated samples as well as one uninoculated control sample. At 30 days, there were three clusters (P = 0.006, Fig. 3B). The first contained the two unplanted control samples, the second cluster contained the three uninoculated control samples, and the third contained the three inoculated samples. Finally, at 60 days there were three clusters (P = 0.04, Fig. 3C). The first contained the two unplanted control samples and one uninoculated control sample. The second contained the remaining two uninoculated control samples. The third contained all three inoculated samples.
Fig. 3.
A comparison of the bacterial community structure from the three treatments studied (composted tailings planted and inoculated with Bacillus pumilus ES4, uninoculated planted composted tailings, unplanted uninoculated composted tailings at 15 (A), 30 (B), and 60 (C) days. The tailings used for this analysis was the neutral, low-metal content mine tailings. Each subfigure contains the original DGGE gel and a dendrogram and Kruskal’s isotonic multidimentional scaling analysis derived from the data of that gel. Data in circles are grouped together and differ from data from other groups at the indicated level of significance.
3.4. Root colonization of quailbush plants by B. pumilus ES4
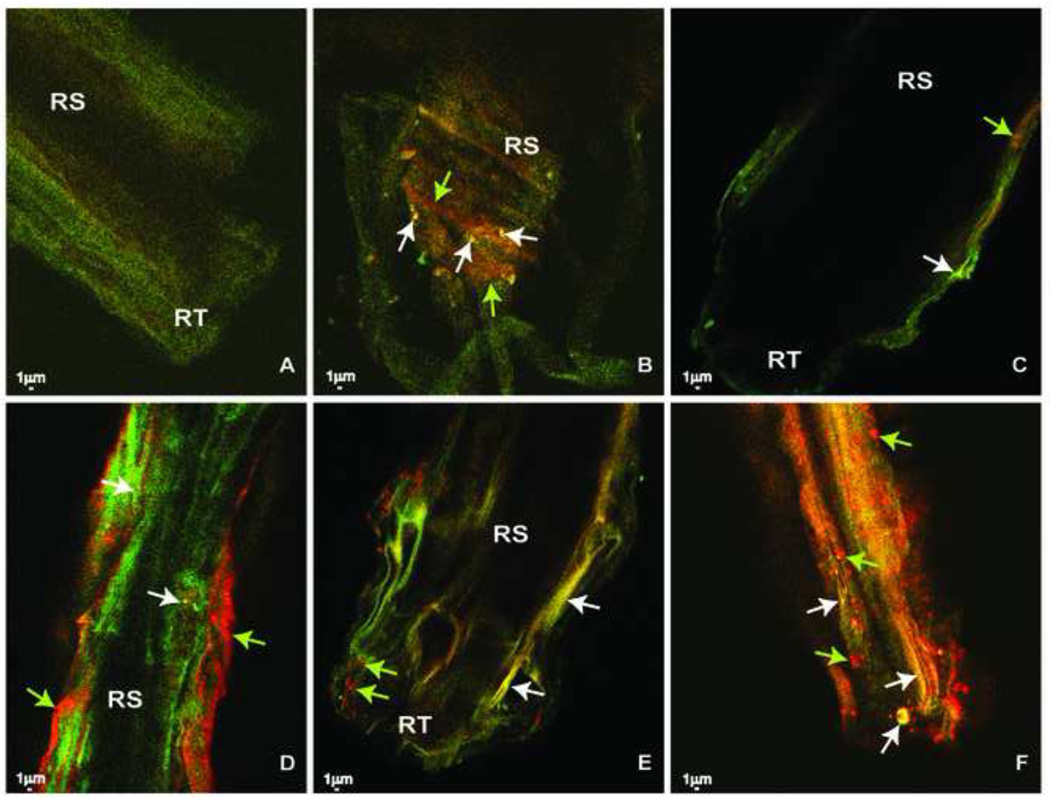
Bacterial colonization of same-age quailbush roots was examined using two sets of FISH primers, one specific for B. pumilus ES4 and one for the domain Bacteria. Originally, tailings had very limited populations of unidentified bacteria and most root surfaces were devoid of bacteria. This continued for the 60 days of the experiment (Fig. 4A). Inoculation with B. pumilus ES4 significantly and consistently changed the visual rhizosphere profile of these roots. Specific detection of microcolonies of B. pumilus ES4 after day 15 was observed exclusively in the root tip and elongation zone area of the root (Fig. 4B, C) and this was observed up to day 60 of the experiment (Fig. 4D, E). With time, the population of bacteria of other species also increased and became dominant over the root surfaces (Fig. 4D, E, F).
Fig. 4.
Colonization of roots of quailbush by Bacillus pumilus ES4 detected by fluorescent in situ hybridization (FISH) using double hybridization with the specific probe BAC07 and the universal probe EUB338 and viewed by confocal laser microscopy. (A) non-inoculated roots; (B,C) root tip and elongation zone 15 days after inoculation, respectively, (D) root elongation zone 60 days after inoculation (E) root tip of the same root 60 days after inoculation, and (F) root tip and elongation zone 60 days after inoculation. Yellow colonies and yellow patches are labeled cells of Bacillus pumilus ES4 (marked with white arrows). Red colonies and red patches are labeled cells of unidentified bacteria on the roots (marked with green arrows). Because all non-inoculated roots showed the same bacterial population, only one control microphotograph is presented. RS = Root surface; RT = Root tip.
4. Discussion
Phytostabilization of mine tailings is a type of phytoremediation that uses plants capable of surviving on these stressed substrates, with the objective of eliminating dispersal of contaminant and particulates from the site. In general, this has been done with large quantities of organic matter coupled with irrigation. Alternatively, the use of PGPB in phytostabilization of mine tailings is proposed (Petrisor et al., 2004; Zhuang et al., 2007 Mendez and Maier, 2008). There are two possibilities to consider in the search for PGPB compatible with mine tailings. One is to isolate native, potential PGPB from tailings or from plants already growing in the tailings, propagate them, and use them as inoculants. This is a labor intensive procedure that has been successfully tried (Grandlic et al., 2008, 2009). The other approach, described in this study, is to examine isolates from the large number of available and proven PGPB strains, mainly used in agriculture (Bashan and de-Bashan, 2005).
All three PGPB used in this study proved to be growth promoters of quailbush in the two types of mine tailings tested. Although the two types of tailings were very different, the same small amount of compost was added to give the same baseline for comparison of the performance of the PGPB. Addition of compost is essential for normal development of native plants in the desert on toxic and even less toxic mine tailings (Iverson and Maier 2009; Mendez and Maier 2008; Grandlic C., pers. comm.). Inoculation with PGPB can reduce the amount of compost from 15% to 10% for raising quailbush but in buffalo grass PGPB enhanced growth in the absence of compost for a period of time (Grandlic et al. 2008). Yet, for practical revegetation of mine tailings, and for most native plant species tested, complete elimination of compost was impossible (Grandlic C., pers. comm.); a significant difficulty for this technology, so far. Of these isolates, B. pumilus ES4 performed the best, as measured by plant biomass produced. Increasing plant mass at the whole plant level is a common effect of PGPB (Bashan and Dubrovsky, 1996). Consequently, it was chosen as the candidate PGPB for detailed studies of root colonization and its effect on the native substrate community of bacteria. It is notable that the PGPB stimulated plant growth in both types of tailings even though the pH and metal content were very different.
Inoculation with B. pumilus ES4 significantly changed the tailings bacterial community structure as early as 15 days into the experiment. The triplicate B. pumilus ES4 inoculated treatment samples were completely distinguishable from the unplanted control by day 15 and from the uninoculated control by day 30. These results are in contrast to three studies using the PGPB Azospirillum brasilense that examined maize grown in pots in a controlled greenhouse. Analysis of the structure of the bacterial community in these experiment, using two fingerprinting methods, DGGE and automated ribosomal intergenic spacer analysis (ARISA), showed that inoculation with A. brasilense strains Cd and Sp 245 had no effect or very marginal effect, at best, on the size or the structure of the bacterial communities and that plant age was far more significant in affecting bacterial communities (Herschkovitz et al., 2005, a,b and Lerner et al., 2006). Another study employing tomatoes and Azospirillum brasilense and B. subtilis in a peat-based substrate and evaluated by DGGE showed similar trends (Felici et al., 2008).
The two tailings used in this study have an extremely low population of heterotrophic bacteria (Mendez et al., 2007, this study), comparable in size to other tailings from the same area (Iverson and Maier 2009). Even with the addition of 10% compost, the total population of heterotrophic bacteria in these tailings is relatively low, depending on the type of tailings (103–105 CFU g−1, Iverson and Maier 2009); 104–106 CFU g−1, Mendez et al. 2007). The low initial heterotrophic numbers is a possible and plausible explanation for the changes that B. pumilus ES4 induced in the bacterial community of the tailings. In comparison to previous studies on changing of bacterial community structure, maize ,in those studies, is a large and fast-growing plant that produces a large amount of roots in the pots and consequently transformed the entire soil of the pot to a rhizosphere-like mini-ecosystem where a large population of bacteria exists, normally 108–109 CFU culturable bacteria g−1 soil. Similarly, this occurs with microbial populations in the tomato rhizosphere (Meyer and Linderman, 1986; de Brito Alvarez et al., 1995). On this elevated native bacterial background, inoculation with Azospirillum, normally done at the level of 106 CFU g−1 or about 0.1–1% of the background population (Bashan, 1986), has only marginal, if any, effect on the general population observed in the above studies (Felici et al., 2008; Herschkovitz et al., 2005, a,b; and Lerner et al., 2006) and is below the limits of the molecular techniques used. In complex microflora, PCR-DGGE does not detect microorganisms present at a level lower than 1% of the total microbial population (Felske et al., 1998). However, our experiments were done with quailbush, a relatively slow growing small plant with roots that did not completely colonize the substrate even after 60 days; this might have had a small effect on the relatively low initial bacterial community. For relatively low community size of bacteria, as occurs in the tailings (Iverson and Maier 2009; Mendez et al. 2007), inoculation of B. pumilus ES4 (106 CFU g−1) created a large shift in the populations, allowing the molecular techniques to detect it.
To verify that the effects on plant growth resulted from inoculation, it is essential to demonstrate that seeds inoculated with bacterial strains yield colonized roots. Numerous methods for identifying root colonization by PGPB have been developed. They include traditional microbiology, immunology, molecular insertion of specific marker genes (Jacoud et al., 1998), insertion of the gfp green fluorescent protein (Ramos et al., 2002; Bacilio et al., 2004), lacZ markers (Arsene et al., 1994), and FISH (Kirchhof et al., 1997). Because the tailings were non-sterile, we employed a modified FISH technique to follow root colonization by the PGPB. FISH monitored with confocal laser microscopy is a powerful tool to detect PGPB in soils and rhizosphere samples (Assmus et al., 1995). B. pumilus cells were specifically detected by the FISH technique on the colonized root tips and the elongation zone areas at day 15 and 60. Furthermore, the PGPB changed the bacterial colonization of the tailings by inducing proliferation of other rhizosphere bacteria on the root surfaces, as detected by DGGE and FISH analyses. Proliferation of other bacteria may have been the result of an increase in exudation from the roots induced by the PGPB on the roots. Enhanced proton and organic acid extrusion are one of the mechanisms proposed for PGPB (Bashan and Levanony 1991; Carrillo et al. 2002; Bashan and de-Bashan 2005a). Taken together, this suggests that this PGPB has sufficient rhizosphere fitness to serve as an effective PGPB. Rhizosphere fitness is a major condition for the success of any PGPB (Lugtenberg and Dekkers, 1999; Bashan et al., 2004).
In summary, this study identified an efficient PBPB strain to promote growth of a native plant in two distinct types of mine tailings and its capacity to modify the community of bacteria of the tailings.
Acknowledgements
We thank Julia Neilsen, Antje Legatzki, Sadie Iverson, and Fernando Solis-Dominguez from our research group at the University of Arizona for advice and discussion, Anton Hartmann and Michael Schmid of the Hemholtz Zentrum, München, Germany for advice on FISH development, and Christopher Grandlic at Synthetic Genomics, California, for advice on compost application in tailings. This research was supported by the National Institute of Environmental Health Sciences Superfund Basic Research Program, NIH, USA (grant 2 P42 ES04940-11) and by Consejo Nacional de Ciencia y Tecnologia of Mexico (CONACYT basic research grant 50052-Z).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amman RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsene F, Katupitiya S, Kennedy IR, Elmerich C. Use of LacZ fusions to study the expression of nif genes of Azospirillum brasilense in association with plants. Mol. Plant-Microbe Int. 1994;7:748–757. [Google Scholar]
- Assmus B, Hutzler P, Kirchhof G, Amann R, Lawrence JR, Hartmann A. In situ localization of Azospirillum brasilense in the rhizosphere of wheat with fluorescently labeled rRNA-targeted oligonucleotide probes and scanning confocal laser microscopy. Appl. Environ. Microbiol. 1995;61:1013–1019. doi: 10.1128/aem.61.3.1013-1019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacilio M, Rodriguez H, Moreno M, Hernandez J-P, Bashan Y. Mitigation of salt stress in wheat seedlings by a gfp-tagged Azospirillum lipoferum . Biol. Fertil. Soils. 2004;40:188–193. [Google Scholar]
- Bashan Y. Significance of timing and level of inoculation with rhizosphere bacteria on wheat plants. Soil Biol. Biochem. 1986;18:297–301. [Google Scholar]
- Bashan Y. Alginate beads as synthetic inoculant carriers for the slow release of bacteria that affect plant growth. Appl. Environ. Microbiol. 1986;51:1089–1098. doi: 10.1128/aem.51.5.1089-1098.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashan Y, de-Bashan LE. Bacteria / Plant growth-promotion. In: Hillel D, editor. Encyclopedia of soils in the environment. Vol. 1. Oxford, U.K: Elsevier; 2005a. pp. 103–115. [Google Scholar]
- Bashan Y, de-Bashan LE. Fresh-weight measurements of roots provide inaccurate estimates of the effects of plant growth-promoting bacteria on root growth: a critical examination. Soil Biol. Biochem. 2005b;37:1795–1804. [Google Scholar]
- Bashan Y, Dubrovsky JG. Azospirillum spp. participation in dry matter partitioning in grasses at the whole plant level. Biol. Fertil. Soils. 1996;23:435–440. [Google Scholar]
- Bashan Y, Hernandez J-P, Leyva LA, Bacilio M. Alginate microbeads as inoculant carrier for plant growth-promoting bacteria. Biol. Fertil. Soils. 2002;35:359–368. [Google Scholar]
- Bashan Y, Holguin G. Proposal for the division of plant growth-promoting rhizobacteria into two classifications: biocontrol-PGPB (Plant Growth-Promoting Bacteria) and PGPB. Soil Biol. Biochem. 1998;30:1225–1228. [Google Scholar]
- Bashan Y, Holguin G, de-Bashan LE. Azospirillum-plant relationships: physiological, molecular, agricultural, and environmental advances (1997–2003) Can. J. Microbiol. 2004;50:521–577. doi: 10.1139/w04-035. [DOI] [PubMed] [Google Scholar]
- Bashan Y, Levanony H. Alterations in membrane potential and in proton efflux in plant roots induced by Azospirillum brasilense . Plant Soil. 1991;137:99–103. [Google Scholar]
- Bashan Y, Moreno M, Troyo E. Growth promotion of the oilseed halophyte Salicornia bigelovii in seawater inoculated with mangrove rhizosphere bacteria and Azospirillum . Biol. Fertil. Soils. 2000;32:265–272. [Google Scholar]
- Bashan Y, Salazar B, Puente ME. Responses of native legume desert trees used for reforestation in the Sonoran Desert to plant growth-promoting microorganisms in screen house. Biol. Fertil. Soils. 2009;45:655–662. [Google Scholar]
- Bashan Y, Salazar B, Puente ME, Bacilio M, Linderman RG. Enhanced establishment and growth of giant cardon cactus in an eroded field in the Sonoran Desert using native legume trees as nurse plants aided by plant growth-promoting microorganisms and compost. Biol. Fertil. Soils. 2009;45:585–594. [Google Scholar]
- Belimov AA, Kunakova AM, Safronova VI, Stepanok VV, Yudkin LV, Alekseev YV, Kozhemyakov AP. Employment of rhizobacteria for the inoculation of barley plants cultivated in soil contaminated with lead and cadmium. Microbiol. 2004;73:99–106. [PubMed] [Google Scholar]
- Burd GI, Dixon DG, Glick BR. A plant growth-promoting bacterium that decreases nickel toxicity in seedlings. Appl. Environ. Microbiol. 1998;64:3663–3668. doi: 10.1128/aem.64.10.3663-3668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd GI, Dixon DG, Glick BR. Plant growth-promoting bacteria that decrease heavy metal toxicity in plants. Can. J. Microbiol. 2000;46:237–245. doi: 10.1139/w99-143. [DOI] [PubMed] [Google Scholar]
- Carrillo AE, Li CY, Bashan Y. Increased acidification in the rhizosphere of cactus seedlings induced by Azospirillum brasilense . Naturwissenschaften. 2002;89:428–432. doi: 10.1007/s00114-002-0347-6. [DOI] [PubMed] [Google Scholar]
- Chiu KK, Ye ZH, Wong MH. Growth of Vetiveria zizanioides and Phragmities australis on Pb/Zn and Cu mine tailings amended with manure compost and sewage sludge: A greenhouse study. Bioresource Technol. 2006;97:158–170. doi: 10.1016/j.biortech.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Colores GM, Macur RE, Ward DM, Inskeep WP. Molecular analysis of surfactant-driven microbial population shifts in hydrocarbon-contaminated soil. Appl. Environ. Microbiol. 2000;66:2959–2964. doi: 10.1128/aem.66.7.2959-2964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H, Stoecker K, Wagner M. Fluorescence in situ hybridization for the detection of prokaryotes. In: Osborn AM, Smith CJ, editors. Molecular Microbial Ecology. New York: Taylor and Francis Group; 2005. [Google Scholar]
- de Brito-Alvarez MA, Gagne S, Antoun H. Effect of compost on rhizosphere microflora of the tomato and on the incidence of plant growth-promoting rhizobacteria. Appl. Environ. Microbiol. 1995;61:194–199. doi: 10.1128/aem.61.1.194-199.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPA. Test Methods for Evaluating Solid Waste. EPA SW-846. Washington, DC: US Environmental Protection Agency; 2004. [Google Scholar]
- Enebak SA, Wei G, Kloepper JW. Effects of plant growth-promoting rhizobacteria on loblolly and slash pine seedlings. For. Sci. 1998;44:139–144. [Google Scholar]
- Felske A, Akkermans ADL, de Vos WM. Quantification of 16S rRNA in complex bacterial communities by multiple competitive reverse transcription-PCR in temperate gradient gel electrophoresis fingerprintings. Appl. Environ. Microbiol. 1998;64:4581–4587. doi: 10.1128/aem.64.11.4581-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris MJ, Muyzer G, Ward DM. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BR. Phytoremediation: synergistic use of plants and bacteria to clean up the environment. Biotechnol. Adv. 2003;21:383–393. doi: 10.1016/s0734-9750(03)00055-7. [DOI] [PubMed] [Google Scholar]
- Gomez NCM, Heuer H, Schonfeld J, Costa R, Hagler-Mendonca L, Smalla K. Bacterial diversity of the rhizosphere of maize (Zea mays) grown in tropical soil studied by temperature gradient gel electrophoresis. Plant Soil. 2001;232:167–180. [Google Scholar]
- Gonzalez-Chavez MC, Carrillo-Gonzalez R, Gutierrez-Castorena MC. Natural attenuation in a slag heap contaminated with cadmium: The role of plants and arbuscular mycorrhizal fungi. J. Hazard Mater. 2009;161:1288–1298. doi: 10.1016/j.jhazmat.2008.04.110. [DOI] [PubMed] [Google Scholar]
- Grandlic CJ, Mendez MO, Chorover J, Machado B, Maier RM. Plant growth-promoting bacteria for phytostabilization of mine tailings. Environ. Sci. Technol. 2008;42:2079–2084. doi: 10.1021/es072013j. [DOI] [PubMed] [Google Scholar]
- Grandlic CJ, Palmer MW, Maier RM. Optimization of plant growth-promoting bacteria-assisted phytostablization of mine tailings. Soil Biol. Biochem. 2009;41:1734–1740. doi: 10.1016/j.soilbio.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann A, Rothballer M, Schmid M. Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil. 2008;312:7–14. [Google Scholar]
- Hernandez J-P, de-Bashan LE, Rodriguez DJ, Rodriguez Y, Bashan Y. Growth promotion of the freshwater microalga Chlorella vulgaris by the nitrogen-fixing, plant growth-promoting bacterium Bacillus pumilus from arid zone soils. Eur. J. Soil Biol. 2009;45:88–93. [Google Scholar]
- Herschkovitz Y, Lerner A, Davidov Y, Okon Y, Jurkevitch E. Azospirillum brasilense does not affect population structure of specific rhizobacterial communities of inoculated maize (Zea mays) Environ. Microbiol. 2005a;7:1847–1852. doi: 10.1111/j.1462-2920.2005.00926.x. [DOI] [PubMed] [Google Scholar]
- Herschkovitz Y, Lerner A, Davidov Y, Rothballer M, Hartmann A, Okon Y, Jurkevitch E. Inoculation with the plant growth promoting rhizobacterium Azospirillum brasilense causes little disturbance in the rhizosphere and rhizoplane of maize (Zea mays) Microbial Ecol. 2005b;50:277–288. doi: 10.1007/s00248-004-0148-x. [DOI] [PubMed] [Google Scholar]
- Iverson SL, Maier RM. Effects of compost on colonization of roots of plants grown in metalliferous mine tailings, as examined by fluorescence in situ hybridization. Appl Environ Microbiol. 2009;75:842–847. doi: 10.1128/AEM.01434-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoud C, Faure D, Wadoux P, Bally R. Development of a strain-specific probe to follow inoculated Azospirillum lipoferum CRT1 under field conditions and enhancement of maize root development by inoculation. FEMS Microbiol. Ecol. 1998;27:43–51. [Google Scholar]
- Kirchhof G, Schloter M, Assmus B, Hartmann A. Molecular microbial ecology approaches applied to diazotrophs associated with non-legumes. Soil Biol. Biochem. 1997;29:853–862. [Google Scholar]
- Kloepper JW, Reddy MS, Rodríguez-Kabana R, Kenney DS, Kokalis-Burelle N, Martinez-Ochoa N, Vavrina CS. Application for rhizobacteria in transplant production and yield enhancement. Acta Horticulturae. 2004a;631:217–229. [Google Scholar]
- Kloepper JW, Ryu C-M, Zhang S. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology. 2004b;94:1259–1266. doi: 10.1094/PHYTO.2004.94.11.1259. [DOI] [PubMed] [Google Scholar]
- Ledin M, Pedersen K. The environmental impact of mine wastes – Roles of microorganisms and their significance in treatment of mine wastes. Earth-Science Reviews. 1996;41:67–108. [Google Scholar]
- Lerner A, Herschkovitz Y, Baudoin E, Nazaret S, Moenne-Loccoz Y, Okon Y, Jurkevitch E. Effect of Azospirillum brasilense inoculation on rhizobacterial communities analyzed by denaturing gradient gel electrophoresis and automated ribosomal intergenic spacer analysis. Soil Biol. Biochem. 2006;38:1212–1218. [Google Scholar]
- Li WC, Ye ZH, Wong MH. Effects of bacteria on enhanced metal uptake of the Cd/Zn-hyperaccumulating plant, Sedum alfredii . J. Exper. Bot. 2007;58:4173–4182. doi: 10.1093/jxb/erm274. [DOI] [PubMed] [Google Scholar]
- Liu W-T, Mirzaberok AD, Stahl DA. Optimization of an oligonucleotide microchip for microbial identification studies: a non-equilibrium dissociation approach. Environ. Microbiol. 2001;3:619–629. doi: 10.1046/j.1462-2920.2001.00233.x. [DOI] [PubMed] [Google Scholar]
- Lugtenberg BJJ, Dekkers LC. What makes Pseudomonas bacteria rhizosphere competent. Environ. Microbiol. 1999;1:9–13. doi: 10.1046/j.1462-2920.1999.00005.x. [DOI] [PubMed] [Google Scholar]
- McCutcheon SC, Schnoor JL. Phytoremediation: Transformation and control of contaminants. Hoboken, NJ: Wiley-Interscience; 2003. [Google Scholar]
- Mendez MO, Glenn EP, Maier RM. Phytostabilization potential of quailbush for mine tailings: Growth, metal accumulation, and microbial community changes. J Environ. Qual. 2007;36:245–253. doi: 10.2134/jeq2006.0197. [DOI] [PubMed] [Google Scholar]
- Mendez MO, Maier RM. Phytostabilization of mine tailings in arid and semiarid environments–an emerging remediation technology. Environ. Health Perspec. 2008;116:278–283. doi: 10.1289/ehp.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JR, Linderman RG. Selective influence on populations of rhizosphere or rhizoplane bacteria and actinomycetes by mycorrhizas formed by Glomus fasciculatum . Soil Biol. Biochem. 1986;18:191–196. [Google Scholar]
- Moter A, Göbel UB. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods. 2000;4:85–112. doi: 10.1016/s0167-7012(00)00152-4. [DOI] [PubMed] [Google Scholar]
- Mogge B, Loferer C, Agerer R, Hutzler P, Hartmann A. Bacterial community structure and colonization patterns of Fagus sylvatica L. ectomycorrhizospheres as determined by fluorescence in situ hybridization and confocal laser scanning microscopy. Mycorrhiza. 2000;9:271–278. [Google Scholar]
- Natarajan KA. An integrated biotechnological approach to gold processing-An Indian experience. Mineral Processing and Extractive Metallurgy Review. 1998;19:235–251. [Google Scholar]
- Petrisor I, Dobrota S, Komnitsas K, Lazar I, Kuperberg JM, Serban M. Artificial inoculation–perspectives in tailings phytostabilization. Int. J. Phytoremediation. 2004;6:1–15. doi: 10.1080/16226510490439918. [DOI] [PubMed] [Google Scholar]
- Pilon-Smits E. Phytoremediation. Annu. Rev. Plant Biol. 2005;56:15–39. doi: 10.1146/annurev.arplant.56.032604.144214. [DOI] [PubMed] [Google Scholar]
- Pishchik VN, Vorobyev NI, Chernyaeva II, Timofeeva SV, Kozhemyakov AP, Alexeev YV, Lukin SM. Experimental and mathematical simulation of plant growth promoting rhizobacteria and plant interaction under cadmium stress. Plant Soil. 2002;243:173–186. [Google Scholar]
- Puente ME, Bashan Y, Li CY, Lebsky VK. Microbial populations and activities in the rhizoplane of rock-weathering desert plants I. Root colonization and weathering of igneous rocks. Plant Biol. 2004;6:629–642. doi: 10.1055/s-2004-821100. [DOI] [PubMed] [Google Scholar]
- Ramos HJO, Roncato-Maccari LDB, Souza EM, Soares-Ramos JRL, Hungria M, Pedrosa FO. Monitoring Azospirillum-wheat interactions using the gfp and gusA genes constitutively expressed from a new broad-host range vector. J. Biotechnol. 2002;97:243–252. doi: 10.1016/s0168-1656(02)00108-6. [DOI] [PubMed] [Google Scholar]
- Reed MLE, Glick BR. Growth of canola (Brassica napus) in the presence of plant growth–promoting bacteria and either copper or polycyclic aromatic hydrocarbons. Can. J. Microbiol. 2005;51:1061–1069. doi: 10.1139/w05-094. [DOI] [PubMed] [Google Scholar]
- Reed MLE, Warner BG, Glick BR. Plant growth-promoting bacteria facilitate the growth of the common reed Phragmites australis in the presence of copper or polycyclic aromatic hydrocarbons. Current Microbiol. 2005;51:425–429. doi: 10.1007/s00284-005-4584-8. [DOI] [PubMed] [Google Scholar]
- Rajkumar M, Freitas H. Effects of inoculation of plant growth-promoting bacteria on Ni uptake by Indian mustard. Bioresource Technol. 2008a;99:3491–3498. doi: 10.1016/j.biortech.2007.07.046. [DOI] [PubMed] [Google Scholar]
- Rajkumar M, Freitas H. Influence of metal-resistant plant growth-promoting bacteria on the growth of Ricinus communis in soil contaminated with heavy metals. Chemosphere. 2008b;71:834–842. doi: 10.1016/j.chemosphere.2007.11.038. [DOI] [PubMed] [Google Scholar]
- Rosario K, Iverson SL, Henderson DA, Chartrand S, McKeon C, Glenn EP, Maier RM. Bacterial community changes during plant establishment at the San Pedro River mine tailings site. J. Environ. Qual. 2007;3:1249–1259. doi: 10.2134/jeq2006.0315. [DOI] [PubMed] [Google Scholar]
- Ryu C-M, Hu C-H, Locy RD, Kloepper JW. Study of mechanisms for plant growth promotion elicited by rhizobacteria in Arabidopsis thaliana . Plant Soil. 2005;268:285–292. [Google Scholar]
- Burt R, editor. Laboratory Methods Manual. Soil Survey Laboratory Investigations Report No. 42, Version 4.0. Lincoln, NE: National Resources Conservation Service; 2004. [Google Scholar]
- Tsuruta T. Removal and recovery of uranium using microorganisms isolated from North American uranium deposits. Amer. J Environ. Sci. 2007;3:60–66. [Google Scholar]
- Venables WN, Ripley BD. Modern applied statistics with S. 4th ed. New York: Springer; 2002. pp. 385–389. [Google Scholar]
- Vessey JK. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil. 2003;255:571–586. [Google Scholar]
- Vijayalakshmi SP, Raichur AM. The utility of Bacillus subtilis as a bioflocculant for fine coal. Colloids and Surfaces B: Biointerfaces. 2003;29:265–275. [Google Scholar]
- Vivas A, Biro B, Ruiz-Lozano JM, Barea JM, Azcon BR. Two bacterial strains isolated from a Zn-polluted soil enhance plant growth and mycorrhizal efficiency under Zn-toxicity. Chemosphere. 2006;62:1523–1533. doi: 10.1016/j.chemosphere.2005.06.053. [DOI] [PubMed] [Google Scholar]
- Wu SC, Cheung KC, Luo YM, Wong HM. Effects of inoculation of plant growth-promoting rhizobacteria on metal uptake by Brassica juncea . Environ. Poll. 2006a;140:124–135. doi: 10.1016/j.envpol.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Wu SC, Luo YM, Cheung KC, Wong MH. Influence of bacteria on Pb and Zn speciation, mobility and bioavailability in soil: A laboratory study. Environ. Poll. 2006b;144:765–773. doi: 10.1016/j.envpol.2006.02.022. [DOI] [PubMed] [Google Scholar]
- Ye ZH, Yang ZY, Chan GYS, Wong MH. Growth response of Sesbania rostrata and S. cannabina to sludge-amended lead/zinc mine tailings. A greenhouse study. Environ. Int. 2001;26:449–455. doi: 10.1016/s0160-4120(01)00026-5. [DOI] [PubMed] [Google Scholar]
- Zhang H-B, Yang M-X, Shi W, Zheng Y, Sha T, Zhao Z-W. Bacterial diversity in mine tailings compared by cultivation and cultivation-independent methods and their resistance to lead and cadmium. Microbial Ecol. 2007;54:705–712. doi: 10.1007/s00248-007-9229-y. [DOI] [PubMed] [Google Scholar]
- Zhuang X, Chen J, Shim H, Bai Z. New advances in plant growth-promoting rhizobacteria for bioremediation. Environ. Int. 2007;33:406–413. doi: 10.1016/j.envint.2006.12.005. [DOI] [PubMed] [Google Scholar]