Abstract
An effective HIV-1 vaccine should elicit sufficient breadth of immune recognition to protect against the genetically diverse forms of the circulating virus. Evaluation of the breadth and magnitude of cellular immune responses to epitope variants is important for HIV-1 vaccine assessment. We compared HIV-1 Gag-specific T-lymphocyte responses in 20 HIV-1-infected individuals representing two different HIV-1 subtypes, B and C. By assessing T lymphocyte responses with peptides based on natural HIV-1 variants, we found evidence for limited cross-reactivity and significantly enhanced within-clade responses among clade B-infected subjects, and not among clade C-infected subjects.
INTRODUCTION
The extraordinary genetic diversity of HIV-1 poses a major obstacle in the effort for vaccine development against the virus. In order to be protective against multiple strains, an HIV-1 vaccine must elicit cellular immune responses with robust magnitude and breadth. Therefore, to design a successful T lymphocyte-based HIV-1 vaccine it is extremely important to characterize the cross-reactive potential of the T lymphocyte responses in the setting of a natural HIV-1 infection. Whether T lymphocytes from an individual infected with one clade of HIV-1 are capable of recognizing epitope variants from other clades of the virus would help in vaccine design. It has been shown that Gag-specific T-lymphocytes from an individual infected with one clade respond preferentially to peptides related to the infecting clade [1]. Previously, we have shown that in rhesus monkeys vaccinated with a clade B immunogen, the breadth of vaccine-elicited cellular immune responses (number of epitopes recognized by peptides derived from natural strains) was significantly higher than responses to other clades [2]. In this study we have tested whether such within-clade higher reactivity is evident in 20 HIV-infected subjects, 10 infected with clade B, and 10 with clade C viruses.
MATERIALS AND METHODS
Ethical Statement
CHAVI Protocol 001 (Pro00006579) is an Acute HIV-1 Infection Prospective Cohort Study to study the early-transmitted HIV-1 virus, and to evaluate the host response and the genetic factors that determine HIV transmission and the viral set point. This protocol was approved by the Duke Institutional Review board at a full board committee. The Duke University Health System Institutional Review Board for Clinical Investigations (DUHS IRB) is duly constituted, fulfilling all requirements for diversity, and has written procedures for initial and continuing review of human research protocols. The DUHS IRB complies with the Guidelines of the International Conference on Harmonization to the extent required by the U. S. Food and Drug Administration.
The research was conducted according to the principles expressed in the Declaration of Helsinki. Written informed consents were obtained from all subjects.
Human subjects
Cryopreserved PBMC from10 clade B-infected and 10 clade C-infected subjects from CHAVI001 cohort were used in the study. All 20 subjects had CD4 counts >600 and were not on antiretroviral therapy. Plasma viral loads of these subjects ranged from 2000 copies/ml and 98,000 copies/ml.
HIV-1 Gag peptide sets and design of peptide matrices
We used 4 sets of HIV-1 Gag peptides (15-mer peptides overlapping by 11, spanning the entire protein), one protein each from clades A, B, C and G. The 4 natural strains of HIV-1 Gag that were used in this study were a subset of a larger set of Gag peptides that was designed based on 10 natural strains, that we have used in previous studies to assess the cross-reactivity of vaccine responses to natural variants. Four Gag peptide sets that were representative of the diversity were selected, as cryopreserved PBMC were limiting and the full set of 10 Gag proteins could not be tested [3]. We selected one clade A sequence 1152NG from Cameroon, one clade B sequence PCM013 from Columbia, one clade C sequence TRA3011 from Uruguay, and one clade G sequence 4049HAN from Cameroon; GenBank accession numbers AY371163, AY561237, AY563169, and AY371121 respectively [3]. Each Gag peptide set consisted of 120 overlapping peptides, which were used to make the peptidematrices. These peptides included up to 4 variants for every 15-mer and gave extensive global coverage of 9-mer length fragments in the Los Alamos HIV database. But unlike consensus [4] or Potential T cell Epitope (PTE) peptides [5], they also capture some strain-specific natural variants. All natural strains have some rare and distinctive amino acids, thus using these diverse natural forms of Gag as a basis for peptide design gives a more realistic view of cross-reactivity than studies using peptides based on common epitope variants. The PeptGen tool at the Los Alamos HIV database (http://www.hiv.lanl.gov/content/hivdb/PEPTGEN/PeptGenSubmitForm.html) was used for peptide design. For the matrix mapping 22 pools, each consisting of 15 peptides, were made so that each peptide was represented in two different pools for identification by positive ELISpot responses. A representative design of the matrix for one set of Gag peptides is included in Supplemental online material.
IFN-γ ELISpot Assay
IFN-γ ELISpot assays were performed using cryopreserved peripheral blood mononuclear cells (PBMC) from infected subjects as previously described [2]. Briefly, cryopreserved PBMC were thawed and allowed to rest for 6 hours at 37°C in a 5% CO2 environment. Multiscreen ninety-six well plates were coated overnight with 100µl per well of 5 µg/ml anti-human interferon-γ (IFN-γ) (B27; BD Pharmingen) in endotoxin-free Dulbecco’s-PBS (D-PBS). The plates were then washed three times with D-PBS containing 0.1% Tween-20, blocked for 2 h with RPMI containing 10% FBS to remove the Tween-20, and incubated with peptide pools and 2 × 105 PBMCs in duplicate in 100 µl reaction volumes. Each peptide pool was comprised of 15 amino acid peptides overlapping by 11 amino acids. Each peptide in a pool was present at a 1 µg/ml concentration. Phytohemagglutinin-M (PHAM) was used as positive control. Following an 18-h incubation at 37°C, the plates were washed nine times with D-PBS containing 0.1% Tween-20 and once with distilled water. The plates were then incubated with 2 µg/ml biotinylated rabbit anti-human IFN-γ (U-Cytech, The Netherlands) for 2 h at room temperature, washed six times with D-PBS containing 0.1% Tween-20, and incubated for 1.5h with a 1:500 dilution of streptavidin-AP (Southern Biotechnology, Birmingham, AL). After five washes with D-PBS containing 0.1% Tween-20 and three washes with D-PBS alone, the plates were developed with bromochloroindolyl phosphate–nitro blue tetra-zolium (BCIP-NBT) chromogen (Pierce), stopped by washing with tap water, air dried, and read with an ELISpot reader (Cellular Technology, Ltd) using ImmunoSpot Analyzer software (Cellular Technology, Ltd, Ohio).
PBMC obtained from each of the 20 subjects were evaluated for their recognition of discrete epitopes of each of the 4 indicator Gag proteins. Epitope enumeration was accomplished by matrix mapping. A peptide was scored as positive if it stimulated more than 4 times the background response and a minimum of 55 spot-forming cells (SFC) per million PBMC.
Statistical methods
We analyzed the breadth and magnitude of the responses (as estimated above) using two strategies. The first was a non-parametric, rank-based permutation statistic [6], designed to test whether the observed fraction of responses above the median to within-clade proteins was higher than across clades. By randomizing the assignments of “within-clade” and “between-clade”, 10,000 times, this test accounts for the fact that we had subjects from two different clades (B and C), and that the study subjects had different response levels,. The number of occurrences that were higher than the actual values in the 10,000 randomized data sets was used to estimate the probability of observing the difference between the intra- and inter-clade responses in the actual data by chance alone. We next applied a mixed-effects generalized linear model (GLM) to the data to estimate the impact of and relationships between different parameters implemented using glmer in R (www.r-project.org; lme4.r-forge.r-project.org). We compared nested models simplifying to models with fewer variables in a step-wise manner, using the Akaike Information Criterion (AIC) implemented using the ANOVA function [7], keeping only variables whose significance was below 0.05. This allowed us to test for possible interactions, as well as model the impact of different response levels in different subjects and different epitopes nested within subjects. We used a Poisson distribution to model the minimum numbers of detected epitopes, due to the discrete nature of the data (counts). For magnitude comparisons, a Gaussian distribution gave the best fit.
RESULTS
Epitope enumeration
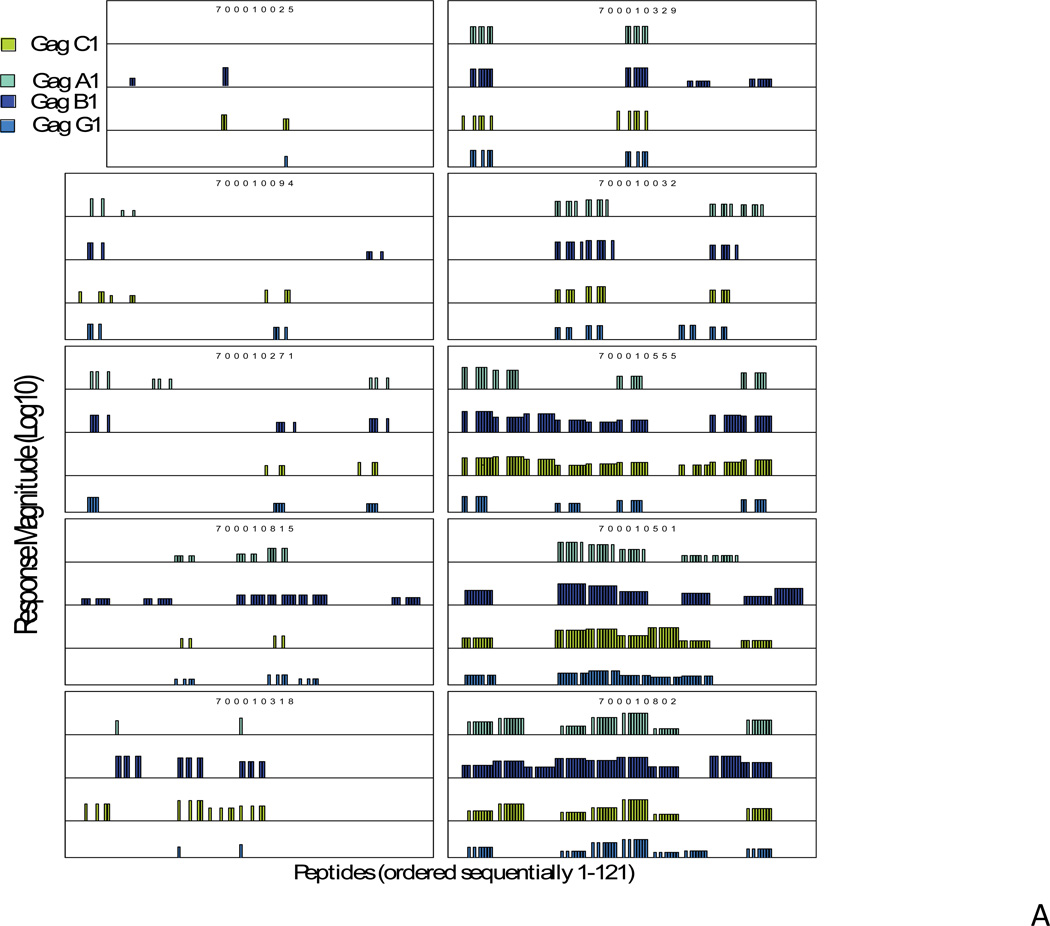
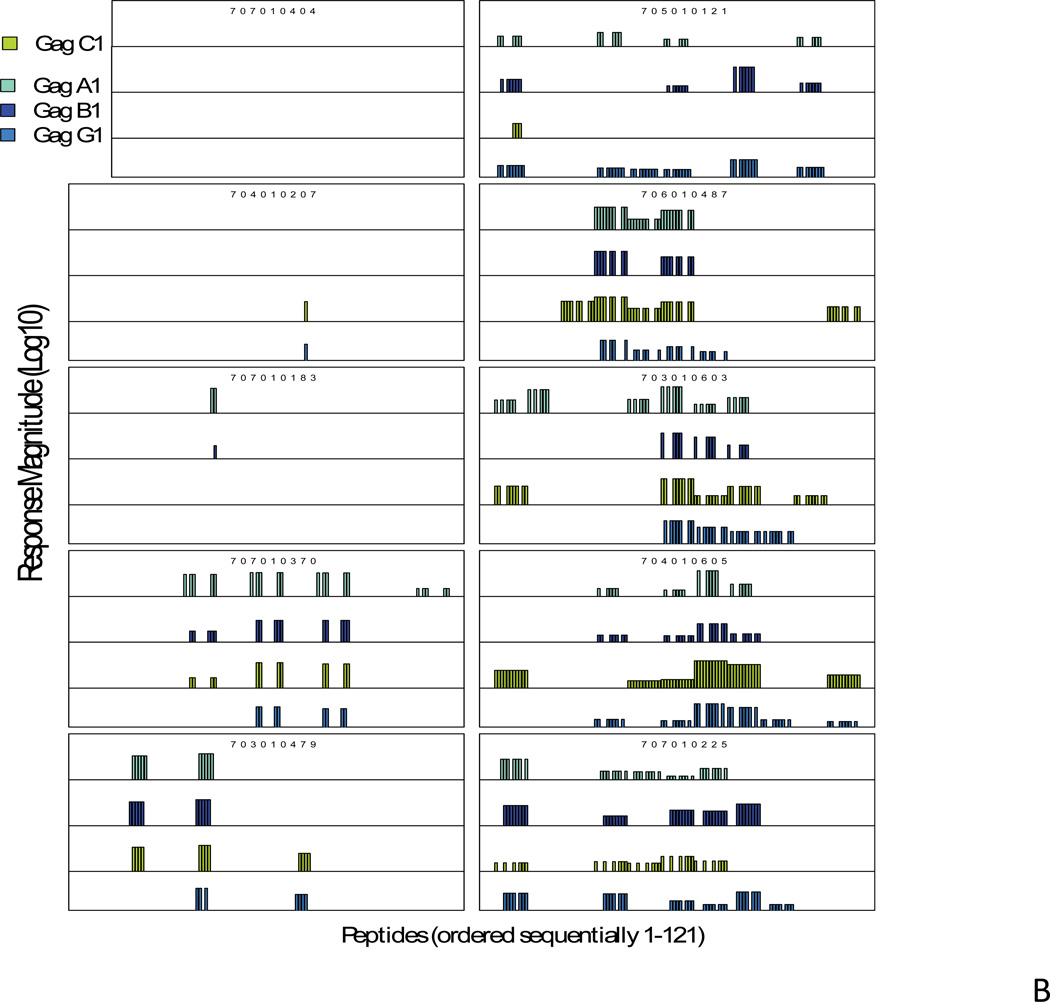
All subjects in this study had CD4 counts >600 and were not on antiretroviral therapy. These two criteria were chosen for the study in order to get subjects with relatively healthy immune system (CD4 count >600) and with enough viral antigen load (off ART therapy) so that cross-clade breadth of T lymphocyte responses in these HIV-1 infected subjects can be measured. Table 1 shows the CD4 counts and viral load of the subjects used in this study. There were three clade B-infected (700010318, 700010555, 700010815), and one clade C-infected subjects (706010487) who had controlled viremia well (viral load <400). However, inclusion of these subjects was based on the criteria that were applied for other subjects. IFN-γ ELISpot assays were performed using cryopreserved peripheral blood mononuclear cells (PBMC) from infected subjects as previously described [2]. Frozen PBMC were evaluated for their recognition of peptide pools derived from the 4 Gag proteins, one each from clades A, B, C and G (Figure 1). Parallel matrices were made for each protein to enable direct comparisons of cross-reactivity.
Table 1.
Viral load and CD4 count of the study subjects
| Infecting Clade |
Subject | Viral load (copies/ml) |
CD4 count/mm3 of blood |
|---|---|---|---|
| B | 700010025 | 86,474 | 546 |
| B | 700010032 | 98,829 | 773 |
| B | 700010094 | 50,889 | 712 |
| B | 700010271 | 26,703 | 620 |
| B | 700010318 | 434 | 1036 |
| B | 700010329 | 6,957 | 726 |
| B | 700010501 | 25,285 | 514 |
| B | 7000010555 | <400 | 905 |
| B | 700010802 | 2029 | 756 |
| B | 700010815 | <400 | 995 |
| C | 703010479 | 37,585 | 612 |
| C | 703010603 | 38,504 | 846 |
| C | 704010605 | 4,030 | 519 |
| C | 705010121 | 7,736 | 796 |
| C | 707010183 | 2,985 | 918 |
| C | 707010225 | 1,672 | 719 |
| C | 707010370 | 7,776 | 796 |
| C | 707010404 | 18,354 | 778 |
| C | 706010487 | <400 | 785 |
| C | 704010207 | 92,800 | 714 |
Figure 1.
Magnitude of pooled peptide responses in each of the clade B-infected (A) and clade C-infected subjects (B). For each subject the magnitude of IFN-γ ELISpot responses are shown on a logarithmic scale for all the peptides in each of the four sets of HIV-1 Gag peptides (range 30–4605 SFC responses/million PBMC, 1.5–3.7 on a logarithmic scale). The peptide numbers that elicited responses are shown on the “X” axis. The responses for clade A peptides are shown in aqua, clade B in dark blue, clade C in green and clade G in blue. One of the clade C-infected subjects (707010404) had no responses against any peptides but had responses against positive control, PHAM.
Due to limited PBMC availability, we were not able to determine the precise epitope reactivity. Therefore, for breadth assessment, we estimated the minimum number of responses by counting the number of positive responses in each row and column of the matrix. If two overlapping peptide pools elicited a response, those responses were counted as a single epitope. Three or 4 overlapping responses were counted as 2 epitopes, 5 or 6 as 3. We multiplied the minimum number of epitope responses thus estimated in each column by the number of reactive rows. To estimate magnitude, for each protein, we summed all responses across all rows in each matrix, we summed all responses across all columns in the table, and, finally, we took the maximum between the two.
Specificity of pooled responses
Among all 20 subjects tested, one of the clade C-infected subjects had no positive IFN-γ ELISpot responses to any Gag peptides, whereas, in all other subjects we observed responses, with a minimum of 1 and a maximum of 97 positive peptide responses, including responses across all four Gag variants. In this summary count, a response may be counted multiple times. Of all reactive pools, 43% reacted to one Gag variant only, 22% reacted to 2, 14% to 3, and 21% to all 4 peptides (Fig. 1). In a previously reported nonhuman primate vaccination study, we observed a similar specificity in the vaccine-elicited T-lymphocyte responses, where the majority of responses were directed against one of the variants [8].
Breadth of responses
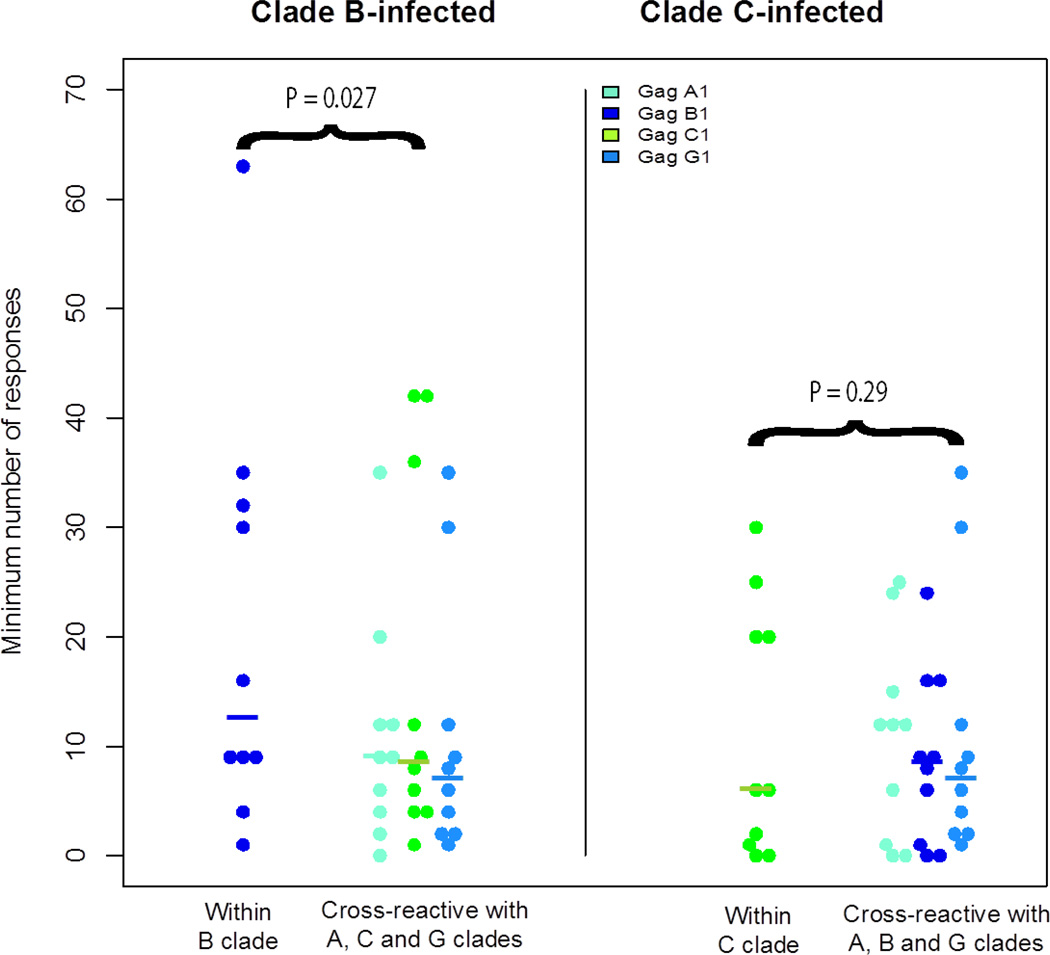
We fitted a mixed-effects Poisson model with random effects for subject and for different clade responses nested within the inter-clade variable. We chose the Poisson family due to the discrete nature of the data (counts). We found a significant interaction between clade and the inter-clade variable (P=0.008), so we analyzed the two clades separately. We found statistical support for within B-clade advantage (P=0.027 by permutation test, P=3.1e-08 by GLM) showing greater number of responses (Figure 2), but not for within C-clade (P=0.36 by permutation test, P=0.29 by GLM). We also asked the question whether there were any correlation between T lymphocyte responses and viral load of the tested subjects by performing a Spearman’s rank correlation test. However, there were no correlation between minimum number of responses and viral load of the subjects (p=0.29).
Figure 2.
Minimum number of epitope responses. Each dot represents the estimated number of epitope responses for each subject across the four Gag proteins. B clade subjects are represented on the left panel, and C-clade subjects on the right panel. Medians are denoted with horizontal bars. The number of within-clade epitope responses was higher in the B-clade subjects (p=0.027 by permutation test) but not the C-clade subjects (p=0.29 by permutation test).
Magnitude of responses
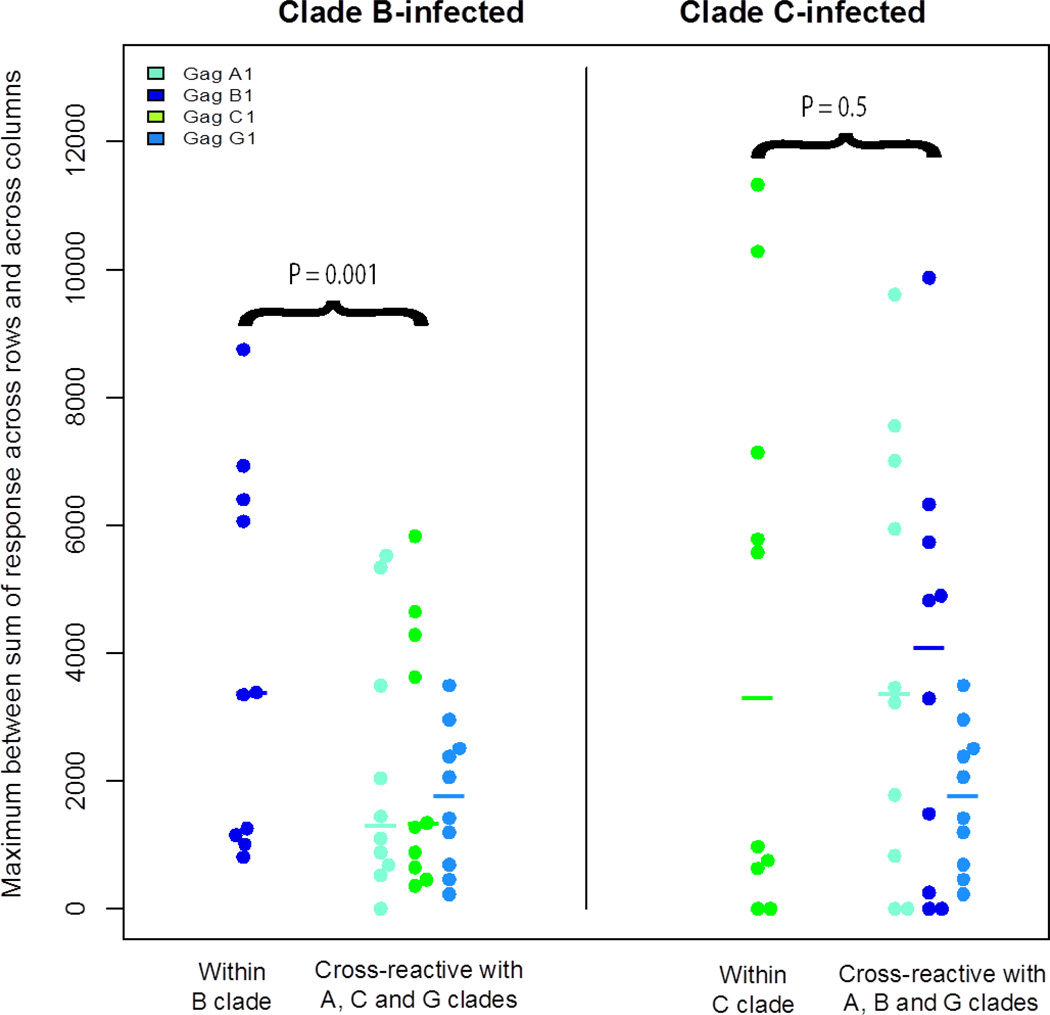
In 9 of the 10 clade B-infected individuals, the highest magnitude of response was to clade B peptides, and in 4 of the 9 clade C-infected individuals the highest magnitude of response was to clade C peptides (Figure 3). We fitted a mixed-effects model with random effects for subjects and for different clade responses nested within the inter-clade variable similar to that done for minimum epitope enumeration. We assumed a Gaussian distribution for the model in this case, as the data was consistent with a Gaussian by a Shapiro-Wilkoxon test. We found the interaction between clade and the inter-clade variable to be marginally significant (P=0.054). Therefore we analyzed the two clades separately. We found statistical significance for within B clade advantage (P=0.001 by permutation test, P=0.0001 by GLM) for higher magnitude of responses (Figure 2), but not for within C clade (P=0.5 by permutation test, P=0.654 by GLM).
Figure 3.
Magnitude of epitope responses. Each dot represents the maximum magnitude of epitope responses for each subject across the four Gag proteins). B clade subjects are represented on the left panel, and C-clade subjects on the right panel. Medians are denoted with vertical bars. The magnitude of within-clade epitope responses was higher in the B-clade subjects (p=0.001 by permutation test) but not the C-clade subjects (p=0.5 by permutation test).
DISCUSSION
We assessed cross-clade T-lymphocyte responses against 4 natural sequences of HIV-1 Gag proteins in 10 clade B-infected and 10 clade C-infected subjects. Our reason for using Gag is that it is the focus of the most frequent and intense immune responses at the population level in natural infection in people. Gag-specific responses have been shown to be correlated with delayed disease progression. Moreover, in a recent study Riou et al. described the kinetics of Gag-specific CD4+ and CD8+ T lymphocyte responses during acute HIV-1 infection in 12 different subjects from the same CHAVI 001 cohort, where highly activated, Gag-responsive IFN-γ+CD8+ T lymphocytes were detectable throughout the course of infection in the peripheral blood (9). Enhanced responses may have been detected in subjects receiving supervised treatment interruption from therapy (10), however we had detectable Gag responses in all of our subjects, adequate for enabling the direct comparison of cross reactivity of natural T lymphocyte responses the issue we set out to investigate. The majority of the T lymphocyte responses in these subjects recognized peptides from only 1 of the 4 proteins tested, and few were cross-reactive with all 4 proteins. In clade B-infected subjects, the number and magnitude of within-clade responses were significantly higher, whereas in clade C-infected subjects using a clade-matched peptide set did not impact the number and magnitude of detected responses. Some previous studies of epitope cross-reactivity in HIV-1 have found evidence for preferential within-clade reactivity [11–13], while others have observed high levels of cross-clade reactivity [14, 15]. Here we extend this earlier work by using a set of full-length natural Gag proteins as basis for our peptide design, rather than focusing on particular epitopes or using consensus proteins for peptide design. While this provides a realistic measure of the cross-reactive potential of a response, still a single protein was used to represent each clade, thus detection of within-clade preferential cross-reactivity in B clade but not in C clade may be a consequence of the specific protein used. Furthermore, the detection of CTL activity using exogenous synthetic peptides in ELISpot assays can give an overly optimistic portrait of T-lymphocyte cross-reactivity, and epitopes expressed from within cells can be more sensitive to the impact of mutations [11, 16]. Despite this, we found strong evidence for limited T-lymphocyte cross-reactivity, as 43% of T-lymphocyte responses recognize only one of the 4 Gag variants, and T-lymphocyte response detection for B-clade infections showed within-clade preferential reactivity.
The development of immune models to determine HIV-1 vaccine efficacy is a major challenge. Our strategy of using sets of peptides designed from circulating natural strains of HIV-1 for assessing T lymphocyte responses requires many more peptides, includes more variants. But this assessment provides us a very realistic picture of how many cross-reactive responses are generated to actual representative circulating virus strains and in more detail than can be obtained with a conventional approach. With the conventional approach the lack of a response to an overlapping peptide may be either be a consequence of the peptide boundaries, or the amino acid variation. With the natural strain peptides, all variants start and end at the same position so “no response” has to be a consequence of variation, not boundaries. Second, natural strains very often have a less common amino acid or combination of amino acids that are not represented in the peptide sets used in the conventional approach. Thus, using sets of natural proteins as basis for peptide reagent design may have important applications in developing realistic predictive models and in monitoring immunity in HIV-1 vaccine trials.
Supplementary Material
Highlights.
Cross-clade breadth of Gag-specific T lymphocyte responses were evaluated in HIV-1 clade B and clade C-infected subjects.
Subjects infected with clade B but not clade C, had limited cross-clade and greater within-clade responses
Use of sets of natural proteins sequences for peptide reagent design has important applications in HIV-1 vaccine trials.
ACKNOWLEDGEMENTS
This work was supported by grant from the NIH, NIAID U19-AI067854-07 (the Center for HIV/AIDS Vaccine Immunology). We thank Jennifer Kirchherr of Duke Human Vaccine Institute for PBMC sample coordination and also the CHAVI-001 protocol participants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Geldmacher C, Currier JR, Gerhardt M, Haule A, Maboko L, Birx D, Gray C, Meyerhans A, Cox J, Hoelscher M. In a mixed subtype epidemic, the HIV-1 Gag-specific T-lymphocyte response is biased towards the infecting subtype. AIDS. 2007;21:135–143. doi: 10.1097/01.aids.0000247589.77061.f7. [DOI] [PubMed] [Google Scholar]
- 2.Santra S, Korber BT, Muldoon M, Barouch DH, Nabel GJ, Gao F, Hahn BH, Haynes BF, Letvin NL. A centralized gene-based HIV-1 vaccine elicits broad cross-clade cellular immune responses in rhesus monkeys. Proc. Natl. Acad. Sci. USA. 2008;105:10489–10494. doi: 10.1073/pnas.0803352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santra S, Liao HX, Zhang R, Muldoon M, Watson S, Fischer W, Theiler J, Szinger J, Balachandran H, Buzby A, Quinn D, Parks RJ, Tsao CY, Carville A, Mansfield KG, Pavlakis GN, Felber BK, Haynes BF, Korber BT, Letvin NL. Mosaic vaccines elicit CD8+ T lymphocyte responses that confer enhanced immune coverage of diverse HIV strains in monkeys. Nat Med. 2010;16:324–328. doi: 10.1038/nm.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, Novitsky V, Haynes B, Hahn BH, Bhattacharya T, Korber B. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 5.Li F, Malhotra U, Gilbert PB, Hawkins NR, Duerr AC, McElrath JM, Corey L, Self SG. Peptide selection for human immunodeficiency virus type 1 CTL-based vaccine evaluation. Vaccine. 2006;24:6893–6904. doi: 10.1016/j.vaccine.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP, Jr, Bjorkman PJ, Wilen CB, Doms RW, O'Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH. Phenotypic properties of transmitted founder HIV-1. Proc. Natl. Acad. Sci. USA. 2013;110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers JM, Hastie TJ. Statistical Models in S. Pacific Grove, CA: Wadsworth & Brooks/Cole; 1992. [Google Scholar]
- 8.Santra S, Muldoon M, Watson S, Buzby A, Balachandran H, Carlson KR, Mach L, Kong WP, McKee K, Yang ZY, Rao SS, Mascola JR, Nabel GJ, Korber BT, Letvin NL. Breadth of cellular and humoral immune responses elicited in rhesus monkeys by multi-valent mosaic and consensus immunogens. Virology. 2012;428:121–127. doi: 10.1016/j.virol.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riou C, Ganusov VV, Campion S, Mlotshwa M, Liu MKP, Whale VE, Goonetilleke N, Borrow P, Ferrari G, Betts MR, Haynes BF, McMichael AJ, Gray CM. Distinct kinetics of Gag-specific CD4+ and CD8+ T cell responses during acute HIV-1 infection. J Immunol. 2012;188:2198–2206. doi: 10.4049/jimmunol.1102813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altfeld M, Lunzen JV, Frahm N, Yu XG, Schneider C, Eldridge RL, Feeney ME, Meyer-Olson D, Stellbrink H-J, Walker BD. Expansion of pre-existing, lymph node-localized CD8+ T cells during supervised treatment interruptions in chronic HIV infection. J Clin Invest. 2002;109:837–843. doi: 10.1172/JCI14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett MS, Ng HL, Ali A, Yang OO. Cross-clade detection of HIV-1-specific cytotoxic T lymphocytes does not reflect cross-clade antiviral activity. J Infect Dis. 2008;197:390–397. doi: 10.1086/525281. [DOI] [PubMed] [Google Scholar]
- 12.Altfeld M, Addo MM, Shankarappa R, Lee PK, Allen TM, Yu XG, Rathod A, Harlow J, O'Sullivan K, Johnston MN, Goulder PJ, Mullins JI, Rosenberg ES, Brander C, Korber B, Walker BD. Enhanced detection of human immunodeficiency virus type 1-specific T-lymphocyte responses to highly variable regions by using peptides based on autologous virus sequences. J Virol. 2003;77:7330–7340. doi: 10.1128/JVI.77.13.7330-7340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malhotra U, Nolin J, Mullins JI, McElrath MJ. Comprehensive epitope analysis of cross-clade Gag-specific T-lymphocyte responses in individuals with early HIV-1 infection in the US epidemic. Vaccine. 2007;25:381–390. doi: 10.1016/j.vaccine.2006.07.045. [DOI] [PubMed] [Google Scholar]
- 14.Barugahare B, Baker C, K'Aluoch O, Donovan R, Elrefaei M, Eggena M, Jones N, Mutalya S, Kityo C, Mugyenyi P, Cao H. Human immunodeficiency virus-specific responses in adult Ugandans: patterns of cross-clade recognition. J. Virol. 2005;79:4132–4139. doi: 10.1128/JVI.79.7.4132-4139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao H, Kanki P, Sankalé JL, Dieng-Sarr A, Mazzara GP, Kalams SA, Korber B, Mboup S, Walker BD. Cytotoxic T-lymphocyte crossreactivity among different human immunodeficiency virus type 1 clades: implications for vaccine development. J. Virol. 1997;71:8615–8623. doi: 10.1128/jvi.71.11.8615-8623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valentine LE, Piaskowski SM, Rakasz EG, Henry NL, Wilson NA, Watkins DI. Recognition of escape variants in ELISPOT does not always predict CD8+ T-lymphocyte recognition of simian immunodeficiency virus-infected cells expressing the same variant sequences. J. Virol. 2008;82:575–581. doi: 10.1128/JVI.00275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.