Abstract
Ribosome biogenesis drives cell growth and proliferation but mechanisms that modulate this process within specific lineages remain poorly understood. Here we identify a Drosophila RNA polymerase I (Pol I) regulatory complex composed of Under-developed (Udd), TAF1B, and a TAF1C-like factor. Disruption of udd or TAF1B results in reduced ovarian germline stem cell (GSC) proliferation. Female GSCs display high levels of rRNA transcription, and Udd becomes enriched in GSCs relative to their differentiating daughters. Increasing Pol I transcription delays differentiation whereas reducing rRNA production induces both morphological changes that accompany multicellular cyst formation and specific decreased expression of the BMP pathway component Mad. These findings demonstrate that modulating rRNA synthesis fosters changes in the cell fate, growth and proliferation of female Drosophila GSCs and their daughters.
Lineage-specific stem cell populations help to maintain tissues that experience high rates of cell turnover (1). Self-renewal and differentiation must be finely tuned to replace cells lost under normal physiological conditions and to rapidly compensate for acute cell loss. Although external cues from niches influence stem cell-based homeostasis (2–4), the intrinsic mechanisms that regulate differential growth and proliferation within stem cell lineages remain poorly understood.
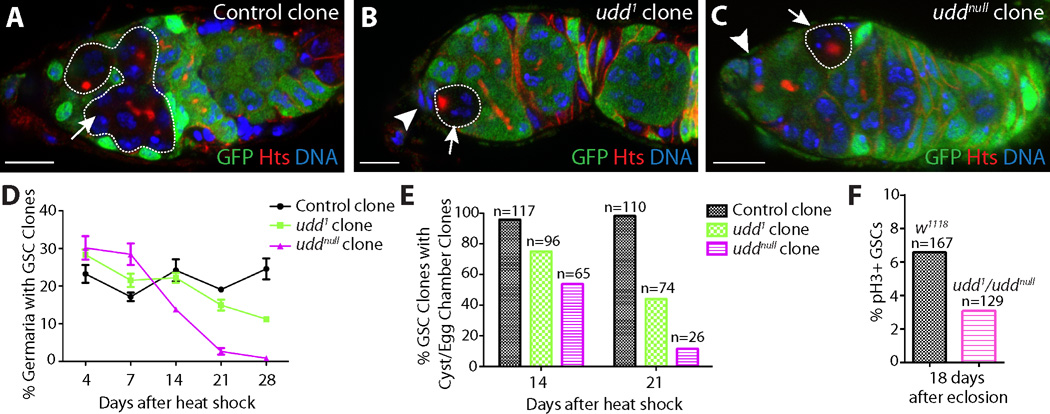
We isolated the Drosophila recessive mutation under-developed1 (udd1) based on its sterile phenotype. Staining for the germline markers Vasa and Hts (5, 6) revealed that udd1 mutants exhibit germ cell loss in ovaries and testes (figs. S1 and S2). Non-complementation tests, RT-PCR and cDNA rescue experiments indicated that udd1 disrupts the expression of a divergent gene CG18316, referred to as udd hereafter (figs. S2 and S3). uddnull homozygotes exhibited embryonic lethality, which was rescued by expression of the udd ORF (fig. S3). Mosaic analysis revealed udd1 and uddnull homozygous clones displayed egg chamber degeneration similar to udd1/udd1 and udd1/uddnull mutants (fig. S1). Over time, udd1 and uddnull mutant GSCs became less proliferative and were eventually lost from the cap cell niche (Fig. 1).
Fig. 1.
Disruption of udd results in reduced GSC proliferation and loss. Negatively marked (A) control, (B) udd1 and (C) uddnull clones (white dotted lines) dissected 21 days after clone induction stained for GFP (green), Hts (red) and DNA (blue). Arrowheads mark cap cells. (D) Percentage of germaria with GSCs clones over time. (E) Percentage of GSC clones that produce a differentiating cyst over time. (F) Percentage of GSCs positive for the mitotic marker phospho-histone H3 (pH3). Scale bars represent 10 µm.
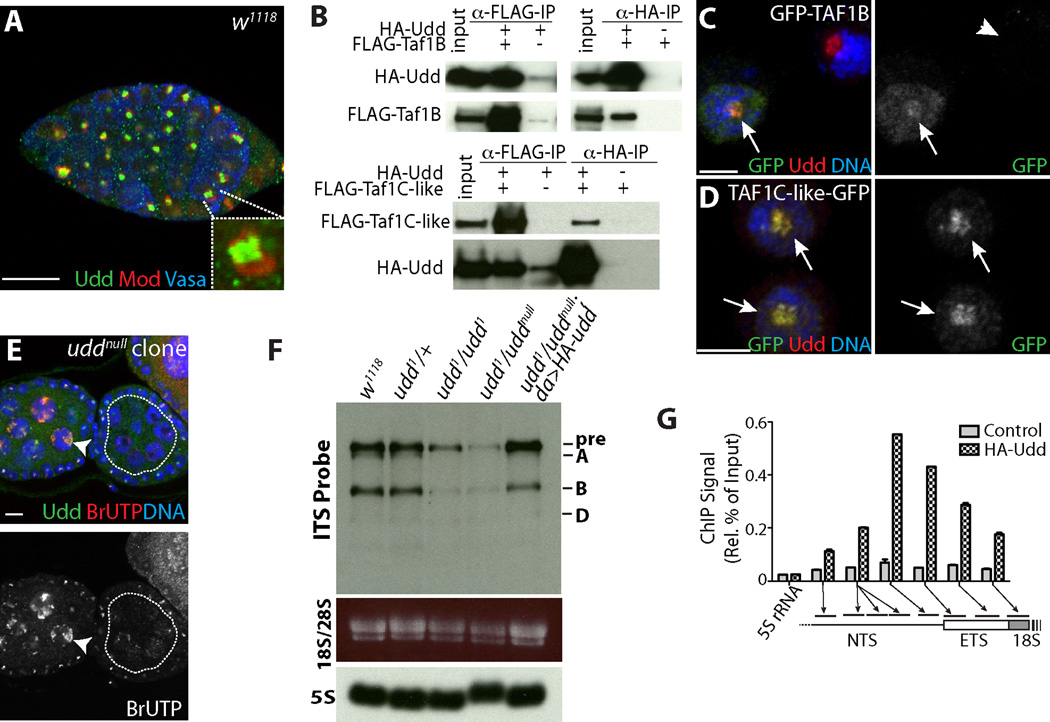
Co-staining with Modulo (Mod) (7) revealed that Udd protein exhibits ubiquitous expression and localizes to the nucleoli of non-dividing cells (n>100 cells) (Fig. 2A; fig. S3). Tandem purification and mass spectrometry (fig. S4), followed by co-immunoprecipitation (Fig. 2B; fig. S4) revealed that Udd associates with two proteins, CG6241 and CG10496. CG6241 shares homology with human TAF1B and yeast Rrn7 (8, 9)(fig. S5), whereas CG10496 resembles human TAF1C based on sequence and secondary structure analyses (fig. S6). CG6241 and CG10496 will be referred to as TAF1B and TAF1C-like, respectively. Human TAF1B and TAF1C are components of the Selectivity Factor 1 (SL1) complex, which promotes Pol I transcription (10–12). Drosophila TAF1B and the TAF1C-like factor localized to nucleoli (Fig. 2C and 2D; fig. S4) and TAF1B was required for the localization and stability of Udd (fig. S7). Udd and TAF1B associated with the Pol I specific subunit RpI135 (13) (fig. S4) and knock-down of TAF1B in the germline resulted in phenotypes similar to udd1 (fig. S7).
Fig. 2.
Characterization of a Drosophila SL1-like complex. (A) w1118 germarium stained for endogenous Udd (green), Mod (red) and Vasa (blue). (B) Co-immunoprecipitation of FLAG-tagged TAF1B and TAF1C-like, and HA-tagged Udd from transfected S2 cells. (C) GFP-TAF1B and (D) TAF1C-like-GFP co-localize with endogenous Udd in S2 cells. Arrows mark transfected cells and arrowheads mark non-transfected cells. (E) uddnull mosaics stained for Udd (green), BrUTP (red) and DNA (blue). Heterozygous nurse cells (arrowhead) exhibit Udd expression and BrUTP incorporation, whereas uddnull mutant cells (white-dotted line) show little BrUTP labeling. (F) Northern blot of total ovarian RNA isolated from indicated genotypes probed using a fragment of Internal Transcribed Spacer (ITS). Ethidium Bromide stained mature 28S and 18S rRNA. 5S rRNA was used as a loading control. (G) ChIP-qPCR analysis of da>HA-udd ovaries reveals Udd associates with specific sites within the rRNA promoter and External Transcribed Spacer (ETS), as indicated by arrows and bars. Control represents anti-HA IP from the da-gal4 background. (C, D) Scale bars represent 5 µm. Scale bars represent 10 µm in other panels.
To test whether the Udd, TAF1B and TAF1C-like complex promotes rRNA generation, we performed BrUTP in situ run-on transcription assays to label nascent rRNA in uddnull clonal ovaries. BrUTP pulse-labeling revealed co-localization between nascent rRNA and Udd protein in control cells, but little BrUTP incorporation in homozygous uddnull mutant cells (Fig. 2E; figs. S8 and S9). RNAi knockdown of TAF1B also reduced the synthesis of rRNA (fig. S7). Northern blot analysis (14) showed udd mutants displayed a reduction of both pre-rRNA and processed rRNA intermediates (Fig. 2F; fig. S9). Chromatin immunoprecipitation (ChIP) experiments revealed that Udd associates with the rDNA promoter (Fig. 2G; fig. S9). Together these data indicate the Udd, TAF1B and TAF1C-like complex likely functions in a manner analogous to the human SL1 complex to promote Pol I transcription (fig. S7). As expected, disruption of Pol I transcription impeded ribosome production based on the nuclear accumulation of GFP-tagged RpS2 in udd1/uddnull mutant cells (fig. S9).
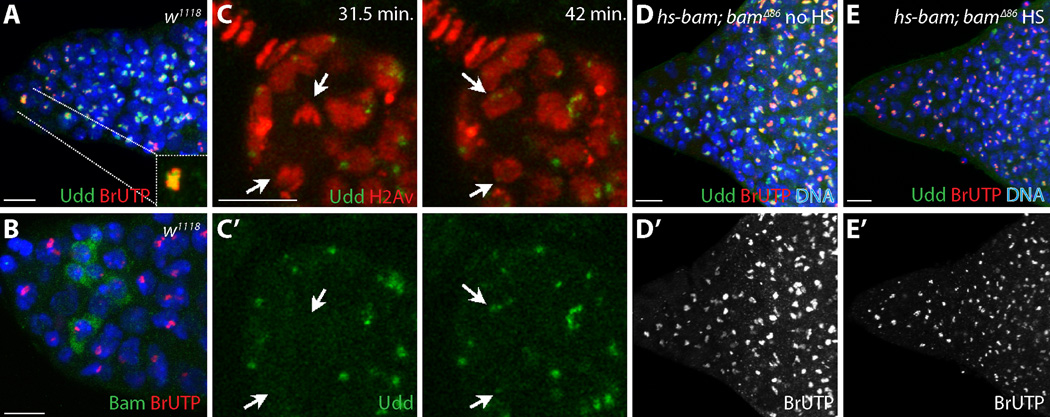
GSCs exhibited higher levels of rRNA synthesis and nucleolar Udd relative to their immediate progeny (Fig. 3A; fig. S10). These differences correlated with the expression of Bam, a key differentiation factor (15–17) (Fig. 3B; fig. S10). Wicked, a component of the rRNA processing U3 snoRNP complex, becomes enriched in cytoplasmic particles that asymmetrically segregate to presumptive GSCs during mitosis (18). To determine whether Udd also becomes asymmetrically enriched within GSCs, we performed immunofluorescence analysis of endogenous Udd and time-lapse microscopy using a rescuing GFP-tagged Udd genomic transgene (Fig. 3C; figs. S11 and S12; Movies S1–S8). Live imaging showed discrete Udd-GFP localization during prophase. Udd-GFP dispersed during metaphase and anaphase, but a small amount of endogenous Udd remained associated with chromosomes through most of mitosis (fig. S11). At the end of telophase, GFP-tagged and endogenous Udd re-coalesce within the nucleoli of newly formed GSCs more quickly and at higher levels relative to their sibling cells oriented away from the cap cells (Fig. 3C; fig. S11). In contrast, Udd appeared evenly distributed in multicellular cyst nucleoli immediately after mitosis (fig. S12).
Fig. 3.
GSCs and undifferentiated cells maintain high levels of Udd. (A-A’’) Control germarium stained for Udd (green), BrUTP (red) and DNA (blue). GSCs (inset) exhibit high levels of (A’) BrUTP labeling and (A’’) Udd. (B) w1118 germarium stained for Bam (green), BrUTP (red), DNA (blue). (C, C’) Still images from live cell imaging (movies S1 and S2) showing GFP-tagged Udd (green) and mRFP-tagged histone H2Av (red) in a dividing GSC at the times indicated. Arrows point to the dividing GSC and resulting daughters. (D) No heat-shock (no HS) control hs-bam; bam∆86 mutant germarium and (E) heat-shocked (HS) hs-bam; bam∆86 mutant germarium 36 hours after bam induction stained for Udd (green), BrUTP (red) and DNA (blue). (D’, E’) BrUTP labeling alone. Scale bars represent 10 µm.
Udd and rRNA synthesis did not drop in bam∆86 mutant germ cells (Fig. 3D; fig. S10), suggesting that persistently low levels of Pol I transcription during early cyst differentiation correlate with the developmental state of these cells and not with their position relative to the niche. To further test this idea, we overexpressed an inducible bam transgene in a bam∆86 mutant background. Following bam expression, the germ cells differentiated into multicellular cysts and both nucleolar Udd and nascent rRNA production levels decreased (Fig. 3E; fig. S10).
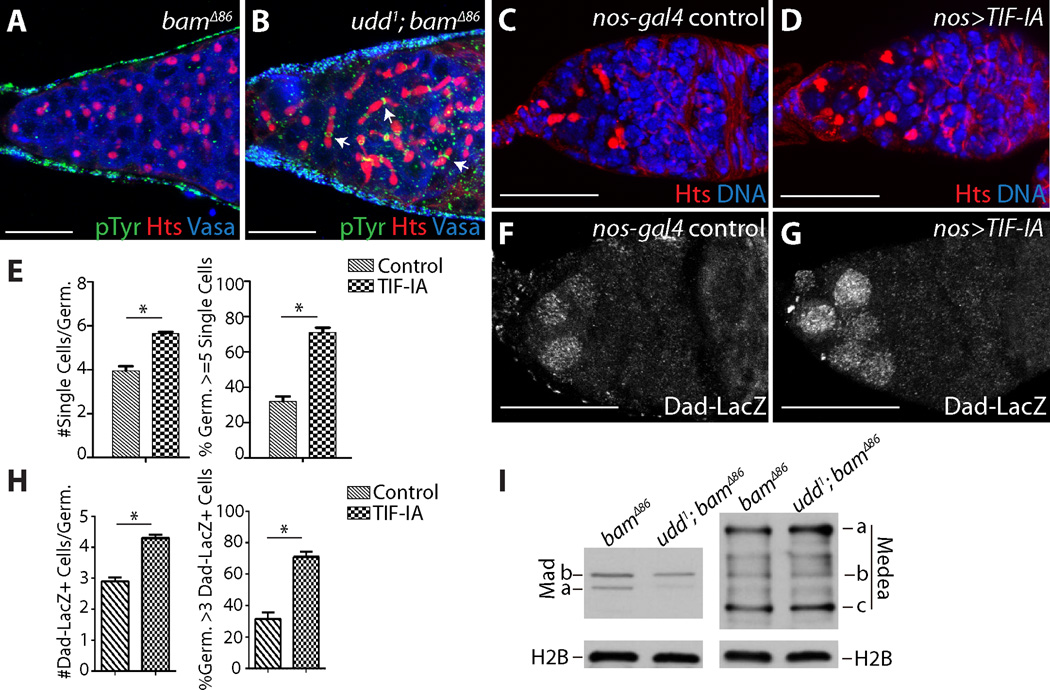
To test the functional significance of reduced rRNA transcription in early differentiating cells, we crossed the udd1 mutation into a bam∆86 mutant background. Although bam∆86 mutant cells remained as single cells with round fusomes (Fig. 4A), udd1 bam∆86 double mutant germaria (94.7%; n=94) contained many four- and eight-cell cysts with branched fusomes and ring canals (Fig. 4B). Mature 16-cell cysts were not observed. RNAi knock-down of TAF1B in a bamRNAi background also resulted in multicellular cyst formation (fig. S13). Consistent with the idea that reduced translation promotes morphological changes that accompany early germline differentiation, knock-down of a rRNA processing factor, ribosomal proteins and a translation initiation factor in a bam loss-of-function background also resulted in multicellular cyst formation (fig. S13). udd1 bam∆86 double mutant germ cells maintain nucleolar Fibrillarin, and fail to down-regulate Sex-lethal and up-regulate A2bp1 despite forming multicellular cysts (figs. S14 and S15).
Fig. 4.
Modulating rRNA synthesis influences cyst development and BMP signaling in the germline. (A) bam∆86 and (B) udd1 bam∆86 double mutant germaria stained for Phosphotyrosine (pTyr) (green), Hts (red) and DNA (blue). Arrows point to cysts with branched fusomes. (C) nos-gal4 and (D) nos>Tif-IA stained for Hts (red) and DNA (blue). (E) Quantification of single cells upon Tif-IA over-expression. (F) nos-gal4 and (G) nos>TIF-IA stained for Dad-LacZ. (H) Quantification of Dad-LacZ expression upon Tif-IA over-expression. (I) Western blots of bam∆86 and udd1 bam∆86 ovarian extracts probed for Mad, Medea and histone H2B proteins. (C,D and F,G) Two copies of nos-gal4 were present. Scale bars represent 20 µm. (*) Denotes significance of p<0.0001.
The udd bam double mutant phenotype suggested that attenuation of Pol I activity promotes some of the early steps of germ cell differentiation. We speculated that increasing rRNA transcription in stem cell daughters exiting the niche might delay their ability to initiate cyst formation. Over-expression of TIF-IA, a conserved factor that bridges divergent Pol I regulatory factors with the Pol I transcriptional complex, results in greater rRNA transcription (19). Although we were unable to drive robust TIF-IA expression (fig. S16), low levels of TIF-IA over-expression resulted in a modest but significant increase in the number of single germ cells with round fusomes within germaria and the percentage of germaria containing over five single undifferentiated cells (compare Fig. 4C to 4D; fig. S16). These cells continued to express Dad-LacZ, a hallmark of BMP signal transduction and GSC identity (20) (Fig. 4E and 4F; fig. S16). We compared the levels of two downstream components of the BMP pathway, Mad and Medea (21), in bam∆86 and udd1 bam∆86 double mutants. Disruption of udd resulted in reduced levels of Mad but not Medea or histone H2B, indicating that modulation of rRNA transcription affects the expression of specific proteins that regulate cell-fate decisions within the GSC lineage (Fig. 4G; fig. S16E). Down-regulation of Mad in response to reduced rRNA transcription likely acts in concert with other mechanisms that extinguish BMP signaling in GSC daughters displaced away from the stem cell niche (22, 23).
Besides TIF-IA and dMyc (19, 24), few regulators of Drosophila Pol I have been characterized. The identification of a Drosophila SL1-like complex provides insights into the mechanisms that regulate rRNA transcription in a developmental context (fig. S16E). Seminal work has shown that specific cellular structures asymmetrically segregate during stem cell divisions in Drosophila and mice (18, 25–27). Results presented here indicate rRNA transcriptional machinery also partitions unevenly during certain cell divisions. These data reveal distinct levels of ribosome biogenesis, once considered a generally constitutive process, modulate the expression of specific proteins that direct cell fate decisions, growth and proliferation within an in vivo stem cell lineage more rapidly or to a greater extent than others. Of note, the direction of asymmetric enrichment of ribosome biogenesis factors may be reversed in other lineages, especially in those stem cells destined to enter a quiescent state. These findings may have important implications for human ribosome related diseases (28, 29).
Supplementary Material
Acknowledgments
The authors thank L. Raftery, Z. Chen, A. Spradling, J. Pradel, N. Perrimon, the Bloomington Stock Center and the Iowa Developmental Studies Hybridoma Bank for providing reagents. J. Huynh and P. R. Hiesinger provided imaging advice. N. Conrad, J. Jiang and P. R. Hiesinger provided comments. Supported by NIH (R01GM086647 and R01GM045820) and E.E. and Greer Garson Fogelson Endowment (UTSW Medical Center).
Footnotes
Udd (Flybase ID CG18316; accession number NM_136509), TAF1B (Flybase ID CG6241; accession number NM_141725), and Taf1C-like (Flybase ID CG10496; accession number NM_137741) sequences have been deposited into GenBank previously.
Supplementary Materials
Materials and Methods
Figs. S1-S16
Movie Legends
References (30–43)
Movies S1–S8
References
- 1.Spradling A, Drummond-Barbosa D, Kai T. Nature. 2001;414:98. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]
- 2.Morrison SJ, Spradling AC. Cell. 2008;132:598. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Cuevas M, Matunis EL. Development. 2011;138:2861. doi: 10.1242/dev.056242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirilly D, Xie T. Cell Res. 2007;17:15. doi: 10.1038/sj.cr.7310123. [DOI] [PubMed] [Google Scholar]
- 5.Lin H, Yue L, Spradling AC. Development. 1994;120:947. doi: 10.1242/dev.120.4.947. [DOI] [PubMed] [Google Scholar]
- 6.de Cuevas M, Spradling AC. Development. 1998;125:2781. doi: 10.1242/dev.125.15.2781. [DOI] [PubMed] [Google Scholar]
- 7.Perrin L, et al. J Cell Sci. 1998;111 ( Pt 18):2753. doi: 10.1242/jcs.111.18.2753. [DOI] [PubMed] [Google Scholar]
- 8.Knutson BA, Hahn S. Science (New York, N.Y. 2011;333:1637. doi: 10.1126/science.1207699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naidu S, Friedrich JK, Russell J, Zomerdijk JC. Science (New York, N.Y. 2011;333:1640. doi: 10.1126/science.1207656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zomerdijk JC, Beckmann H, Comai L, Tjian R. Science (New York, N.Y. 1994;266:2015. doi: 10.1126/science.7801130. [DOI] [PubMed] [Google Scholar]
- 11.Comai L, et al. Science (New York, N.Y. 1994;266:1966. doi: 10.1126/science.7801123. [DOI] [PubMed] [Google Scholar]
- 12.Comai L, Tanese N, Tjian R. Cell. 1992;68:965. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- 13.Seifarth W, et al. Mol Gen Genet. 1991;228:424. doi: 10.1007/BF00260636. [DOI] [PubMed] [Google Scholar]
- 14.Giordano E, Peluso I, Senger S, Furia M. J Cell Biol. 1999;144:1123. doi: 10.1083/jcb.144.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohlstein B, McKearin D. Development. 1997;124:3651. doi: 10.1242/dev.124.18.3651. [DOI] [PubMed] [Google Scholar]
- 16.McKearin DM, Spradling AC. Genes Dev. 1990;4:2242. doi: 10.1101/gad.4.12b.2242. [DOI] [PubMed] [Google Scholar]
- 17.McKearin D, Ohlstein B. Development. 1995;121:2937. doi: 10.1242/dev.121.9.2937. [DOI] [PubMed] [Google Scholar]
- 18.Fichelson P, et al. Nature cell biology. 2009;11:685. doi: 10.1038/ncb1874. [DOI] [PubMed] [Google Scholar]
- 19.Grewal SS, Evans JR, Edgar BA. J Cell Biol. 2007;179:1105. doi: 10.1083/jcb.200709044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song X, et al. Development. 2004;131:1353. doi: 10.1242/dev.01026. [DOI] [PubMed] [Google Scholar]
- 21.Sutherland DJ, Li M, Liu XQ, Stefancsik R, Raftery LA. Development. 2003;130:5705. doi: 10.1242/dev.00801. [DOI] [PubMed] [Google Scholar]
- 22.Harris RE, Pargett M, Sutcliffe C, Umulis D, Ashe HL. Dev Cell. 2011;20:72. doi: 10.1016/j.devcel.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia L, et al. Cell. 2010;143:978. doi: 10.1016/j.cell.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 24.Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA. Nature cell biology. 2005;7:295. doi: 10.1038/ncb1223. [DOI] [PubMed] [Google Scholar]
- 25.Cheng J, et al. Nature. 2008;456:599. doi: 10.1038/nature07386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Science (New York, N.Y. 2007;315:518. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, et al. Nature. 2009;461:947. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hannan KM, Sanij E, Rothblum LI, Hannan RD, Pearson RB. Biochim Biophys Acta. 2013;1829:342. doi: 10.1016/j.bbagrm.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu JM, Ellis SR. Blood. 2006;107:4583. doi: 10.1182/blood-2005-12-4831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.