Summary
The overproduction of nitric oxide (NO) in cells results in nitrosative stress due to the generation of highly reactive species such as peroxynitrite and N2O3. These species disrupt the cellular redox processes through the oxidation, nitration, and nitrosylation of important biomolecules. Microchip electrophoresis (ME) is a fast separation method that can be used to profile cellular nitrosative stress through the separation of NO and nitrite from other redox-active intracellular components such as cellular antioxidants. This paper describes a ME method with electrochemical detection (ME-EC) for the separation of intracellular nitrosative stress markers in macrophage cells. The separation of nitrite, azide (interference), iodide (internal standard), tyrosine, glutathione, and hydrogen peroxide (neutral marker) was achieved in under 40 s using a run buffer consisting of 7.5 to 10 mM NaCl, 10 mM boric acid, and 2 mM TTAC at pH 10.3 to 10.7. Initially, NO production was monitored by the detection of nitrite (NO2−) in cell lysates. There was a 2.5- to 4-fold increase in NO2− production in lipopolysaccharide (LPS)-stimulated cells. The concentration of NO2− inside a single unstimulated macrophage cell was estimatedto be 1.41 mM using the method of standard additions. ME-EC was then used for the direct detection of NO and glutathione in stimulated and native macrophage cell lysates. NO was identified in these studies based on its migration time and rapid degradation kinetics. The intracellular levels of glutathione in native and stimulated macrophages were also compared, and no significant difference was observed between the two conditions.
1. Introduction
NO is involved in several important physiological processes, including neurotransmission, regulation of blood flow, platelet aggregation, and inactivation of pathogens and bacteria.1, 2 NO is produced in cells through the activation of nitric oxide synthase (NOS) and the conversion of L-arginine into L-citrulline.1, 2 There are three isoforms of NOS; namely neuronal, endothelial, and inducible NOS. Activation of inducible NOS (iNOS) is a part of the immune response and leads to the production of large amounts of NO over a long period of time. These elevated NO concentrations can be harmful due to the formation of reactive nitrogen species such as N2O3 and peroxynitrite.1, 2 Both of these species are highly reactive and capable of participating in oxidative stress and nitration/nitrosylation of important biomolecules in vivo.3-5
There are many different types of immune cells in the human body, and macrophages are the primary cell type that is activated as part of an immune response.2, 6 It is also well known that mopnocytes can be differentiated into macrophages, and it has been shown that monocytes in blood can migrate into the intima of a blood vessel and can be differentiated into macrophages during atherosclerosis.7, 8 Macrophages produce NO primarily through the activation of iNOS; however, an uncontrolled or large NO production in these cell types results in cellular nitrosative stress, which has been implicated in many neurodegenerative and cardiovascular diseases.1, 9 The toxic effects of cellular pro-oxidants produced from NO can be mitigated by the presence of antioxidant molecules in the cell.10, 11 Glutathione (GSH) is an antioxidant produced by the cells that can scavenge NO and produce nitrosoglutathione. Also, GSH can react with hydrogen peroxide to form glutathione disulfide. This reaction is catalyzed in the cell by glutathione peroxidase.10, 11 The balance between pro- and antioxidants is important for regulating cellular nitrosative stress.12
There is a wide range of methods reported for the direct and indirect detection of NO. The Griess assay is used extensivelyfor the detection of NO based on a colorimetric reaction with a NO metabolite (NO2-) due to its simplicity. Other analytical methods that have been used for the determination of NO are UV-visible spectroscopy, electron paramagnetic resonance spectroscopy, chemiluminescence, amperometry, and voltammetry.13-17 NO can be directly detected by amperometric detection, and NO biosensors are commonly used in biological applications.16-18 However, despite their low limits of detection (LOD), biosensors can suffer from the presence of interferences and also lack the ability to detect multiple analytes simultaneously.
Another popular approach for NO detection is the use of fluorescent probes such as diaminofluorofluorescein diacetate (DAF-FM DA, specific for NO) and diaminonaphthalene (specific for NO2−).19 However, these probes can cross-react with other species in the sample. Also, microscopic imaging or common spectrometry-based methods cannot distinguish the signal of the desired analyte from those of interferences.19-21 Interferences can be avoided by separating them from the analyte of interest; and therefore, separation-based approaches such as liquid chromatography (LC) and capillary electrophoresis (CE) have become popular for the determination of NO.15, 21
CE has many advantages over liquid chromatographic methods, including very low sample volume requirements, higher separation efficiencies, and faster separations. Therefore, CE has been used for detection of NO from various biological samples.22-26 CE with laser-induced fluorescence (LIF) detection has been extensively used for direct monitoring of NO using selective fluorescent probes.21, 26 Conductivity and UV detection have also been employed with CE for the indirect detection of NO by monitoring its degradation products, nitrite and nitrate.22-25, 27
More recently, microfluidic devices have been employed to detect the production of cellular NO and its metabolites.28-33 These devices have many advantages over classical methods for the study of NO, including the possibility of performing on-chip cell culture, simulating the cellular response in constricted blood vessels, modeling in vivo environments by immobilizing cells on a microchannel, and single cell analysis that can be difficult to achieve using classical methods.28-33 Spectroscopic detection is predominantly used in these devices, and methods for monitoring NO production from erythrocytes,33 endothelial,34 and macrophage cells31 have been reported. Separations with microfluidic devices are most commonly performed using electrophoresis. The use of high field strengths with short channels in the planar format makes it possible to routinely perform subminute separations using this technique. Therefore, this method is especially useful for the detection of chemically labile species since they can be separated and detected before significant degradation occurs.35
Recently, we reported a method for the determination of intracellular NO production in cell lysates using DAF-FM and ME-LIF.36 This method was limited to the determination of NO and could not be used to detect any other RNOS related species. NO can also be detected by amperometric detection, and we also recently reported a ME-EC method for the detection of NO and NO2- produced by NONOate salts.35 Since nitrate (NO3-) is not electroactive, a method using an on-chip Cu2+/Cd reductorcan be used to reduce NO3- to NO2-, which permits the detection of both species by ME-EC.37 Nitrate and nitrite can also be monitored using ME with combined conductivity and amperometric detection.38 In addition to these methods, there are several reports of ME coupled to electrochemical or conductivity detection for determination of nitrite and nitrate.39-41
As described above, the most common method for the quantitation of NO has been capillary or microchip electrophoresis through the detection of its metabolites, nitrite and nitrate, or by reacting NO with a fluorescent probe. In this report, a method that allows the direct detection of NO and its metabolites simultaneously in macrophage cells using ME-EC is described. The electrophoretic method permitssubminute separation of NO, nitrite, and cellular antioxidants as well as potential interferences and other electrochemically active intracellular components (e.g., tyrosine and nitrotyrosine). This approach makes it possible to gather information regarding the overall redox status of the macrophage cells along with NO production. The method was used to investigate NO and intracellular GSH levels in macrophages under native and stimulated conditions. The ME-EC method reported here will be adapted in the future for single cell analysis studies.
2. Materials and Methods
2.1. Materials and Reagents
The following chemicals and materials were used as received: SU-8 10 photoresist and SU-8 developer (MicroChem Corp., Newton, MA, USA); AZ 1518 photoresist and 300 MIF developer (Mays Chemical Co., Indianapolis, IN, USA); photolithography film mask (50,000 dpi; Infinite Graphics Inc., Minneapolis, MN, USA); N(100) 100 mm (4′) silicon (Si) wafers (Silicon, Inc., Boise, ID, USA); chrome and AZ1518 positive photoresist coated soda lime glass substrate (4″ × 4″ × 0.090″, Nanofilm, Westlake, CA, USA); Pt film-coated glass substrates (2000 Å Pt layer over 200 Å Ti) (The Stanford Nanofabrication Facility, Stanford, CA, USA); Sylgard 184 Silicone Elastomer Kit: Polydimethylsiloxane (Ellsworth Adhesives, Germantown, WI, USA); Titanium (Ti) etchant (TFTN; Transene Co., Danvers, MA, USA); epoxy and 22 gauge Cu wire (Westlake Hardware, Lawrence, KS, USA); silver colloidal paste (Ted Pella, Inc., Redding, CA, USA); acetone, 2-propanol (isopropyl alcohol, IPA), 30% H2O2, H2SO4, HNO3, NaOH, HCl, and Trypan blue (Fisher Scientific, Fair Lawn, NJ, USA); sodium nitrite, boric acid, tetradecyltrimethylammonium bromide (TTAB), tetradecyltrimethylammonium chloride (TTAC), ascorbic acid (AA), tyrosine, reduced glutathione, sodium azide, potassium iodide, NaCl, Lipopolysaccharides from Escherichia coli 0111:B4, and Griess reagent (modified) (Sigma, St. Louis, MO, USA) and buffered oxide etchant (JT Baker, Austin, TX, USA). All water used was ultrapure (18.3 MΩ·cm) (Milli-Q Synthesis A10, Millipore, Burlington, MA, USA).
2.2. PDMS Fabrication
The fabrication of PDMS-based microfluidic devices has been described previously.42 Briefly, SU-8 10 negative photoresist (for electrophoresis channels) was spin-coated on a 4 in diameter Si wafer to a thickness of 15 ± 1 μm using a Cee 100 spincoater (Brewer Science Inc., Rolla, MO, USA). The wafer was then transferred to a programmable hotplate (Thermo Scientific, Asheville, NC, USA) for a soft bake at 65°C for 2 min and then 95°C for 5 min. Microfluidic channel designs were created using AutoCad LT 2004 (Autodesk, Inc., San Rafael, CA, USA) and printed onto a transparency film at a resolution of 50,000 dpi (Infinite Graphics Inc., Minneapolis, MN, USA). The coated wafer was covered with the transparency film mask and exposed (344 mJ/cm2 using an i-line UV flood source (ABM Inc., San Jose, CA, USA)). Following the UV exposure, the wafer was post-baked at 65°C for 2 min and 95°C for 10 min. The wafer was then developed in SU-8 developer, rinsed with IPA, and dried under nitrogen. A final “hard-bake” was performed at 175°C for 2 h. The thickness of the raised photoresist, which corresponds to the depth of the PDMS channels, was measured with a profilometer (Alpha Step-200, Tencor Instruments, Mountain View, CA, USA). PDMS microstructures were made by casting a 10:1 mixture of PDMS elastomer and curing agent, respectively, against the patterned Si master. A simple-T device containing a 5 cm separation channel (from the T intersection to the end of the separation channel) and 0.75 cm side arms was used for these studies. The width and depth of the electrophoresis microchannels were 40 μm and 14 μm, respectively. Holes for the reservoirs were created in the polymer using a 4 mm biopsy punch (Harris Uni-core, Ted Pella Inc., Redding, CA, USA).
2.3. Platinum Electrode Fabrication
All electrochemical measurements were obtained using 15 μm Pt working electrodes. Electrodes were either fabricated using an in-house magnetron sputtering system (AXXIS DC magnetron sputtering system, Kurt J. Lesker Co., Jefferson Hills, PA, USA) or received from the Stanford nanofabrication facility. Details of fabrication of Pt electrodes provided by the Stanford nanofabrication facility were reported previously.43 In the Stanford plates, the Pt electrodes are deposited on top of the glass surface. To obtain better stability, Pt electrodes were fabricated in-house by making a 500–600 nm trench in the glass substrate using a procedure previously reported by our group.44 Briefly, the electrode designs were created using AutoCad LT 2004 (Autodesk, San Rafael, CA, USA) and printed onto a transparency film at a resolution of 50,000 dpi (Infinite Graphics, Minneapolis MN, USA). Then the electrode design was patterned on a chrome and AZ1518 positive photoresist-coated soda lime glass plate. The plate was developed using an AZ®300 MIF (Capitol Scientific, Inc., Austin, TX, USA) solution for 30 s and then baked at 100°C for 10 min on a programmable hotplate (Thermo Scientific, Asheville, NC, USA). Once the photoresist layer was developed, the exposed chrome layer was the shape of the electrode. This chrome layer was then etched using chrome etchant to expose the glass surface underneath. Next, the glass plate was etched for about 5 min using a 10:1 buffered oxide etchant (JT Baker, Austin, TX, USA) to obtain a 500 to 600 nm trench. It has been observed that if the trench is not deep enough (below 400 nm), the Pt-deposited electrodes are not stable under high applied potentials (greater than 1200 mV) and the Pt electrode flakes off the trench during electrophoresis. The plate was washed thoroughly with CaCO3 and water after buffered oxide etching, and the depth of the trench was measured using an Alpha-step 200 profilometer (Tencor Instruments). The plate was dried at 100°C for 10 min and then exposed to an oxygen plasma for 1 min (March Plasmod, Concord, CA, USA). The glass plate was immediately transferred to an AXXIS DC magnetron sputtering system (Kurt J. Lesker Co.). After pumping down the vacuum chamber of the sputtering system to a pressure of 1.0 × 10−6 Torr, a 20-nm Ti layer was deposited (220 V deposition voltage, 40 s deposition time, and 5.0 × 10−3 Torr deposition pressure) and then a Ptlayer was deposited (200 V deposition voltage, 17 to 20 min deposition time, and 5.0 × 10−3 Torr deposition pressure). After metal deposition, the glass plate was washed with acetone to remove the photoresist layer along with all excess Pt. The remaining chrome was then removed from the plate with chrome etchant. The width and height of the resulting Pt electrodes were measured again using an Alpha-step 200 profilometer.
2.4. Solution Preparation
All solutions were made using 18.3 MΩ ultrapure water from a Millipore A10 system. Stock solutions of nitrite (NO2−, 10 mM), hydrogen peroxide (H2O2, 10 mM), GSH (10 mM), KI (5 mM), NaN3 (5 mM), and AA (10mM) were all prepared in ultrapure water using appropriate amounts and were stored at 4°C. To dissolve tyrosine (Tyr, 10 mM), the solution was acidified using 1–1.5 M HCl. Subsequent dilutions of each stock solution for use in the microchip system were made in the appropriate run buffer at the time of analysis. For separation and sampling buffer, a boric acid (20 mM) stock solution was prepared and the pH was adjusted to 11 using 10 M or 1 M NaOH solution. The pH-adjusted boric acid buffer was diluted with other buffer constituents in order to obtain a 10 mM boric acid solution. The buffer pH was measured after dilution and before adding surfactant. The buffer pH was 10.3–10.7. TTAC (100 mM) stock solution, NaCl (50 mM) stock solution, and ultrapure water were used for buffer dilution.
2.5. Chip Construction and Electrophoresis Procedure
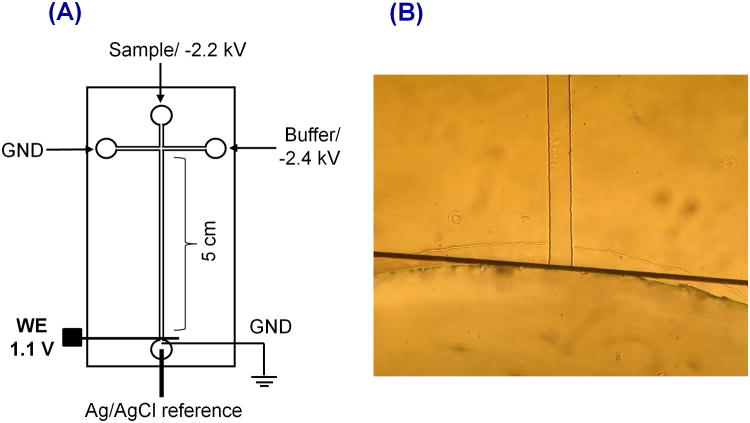
PDMS microchips consisting of a simple-T design with a 5 cm separation channel were used for all studies. Amperometric signals were recorded using a 15 μm Pt working electrode against a Ag/AgCl reference electrode, which was placed in the buffer waste reservoir after the separation ground lead (Figure 1A). The chip containing the separation channel was aligned and reversibly sealed to the glass plate containing the Pt electrode. For in-channel detection, the electrode was placed exactly at the channel end of the separation channel as shown in Figure 1B.
Figure 1.
(A) Schematic of ME-EC setup with in-channel configuration. (B) Electrode alignment
Electrophoretic separations were carried out using reverse polarity with TTAC as the cationic surfactant to modify the channel walls. Two negative high voltage Pt leads (Pt wire) were placed in the sample and buffer reservoirs, while two earth ground Pt leads were placed in the sample waste and buffer waste reservoirs. For sampling, -2200 V was employed, while -2400 V was used for the separation. A gated injection was used to inject the sample, with an injection time between 0.5 and 1 s. Boric acid buffer conditions were evaluated for the separation of nitrite, azide (interference), iodide, tyrosine, GSH, AA, and H2O2. To balance the conductivity difference between the cell lysate and separation buffer, 7.5 to 10 mM NaCl was added to the run buffer. The cells were lysed in buffered solution containing surfactant (10 mM boric acid and 2 mM TTAC) without NaCl.
2.6. Electrochemical Detection
EC detection was accomplished using a modified model of an 8151BP, 8100-K6, or 9051 single- or dual-channel wireless, electrically isolated potentiostat (Pinnacle Technology Inc., Lawrence, KS, USA) operating in a two-electrode format (Pt working; Ag/AgCl reference: Bioanalytical Systems, West Lafayette, IN, USA). The model 8151P, 8100-K6, and 9051 potentiostats have a sampling rate of 5 Hz (Gain = 5,000,000 V/A, Resolution = 30 fA), 10 Hz (Gain = 5,000,000 V/A, Resolution = 27 fA), and 6.5 to 13 Hz (Gain = 5,000,000 V/A, Resolution = 47 fA), respectively. Pinnacle Acquisition Laboratory (PAL or Sirenia) software was used for all data acquisition. The data acquisition is performed via wireless data transmission or Bluetooth from the potentiostat to a computer. A working electrode potential of 1100 mV versus Ag/AgCl reference was used for all experiments.
2.7. Cell Culture and Preparation
RAW 264.7 cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in Dulbecco's Modified Eagle's medium containing 10% (v/v) fetal bovine serum, L-glutamine (2 mM), penicillin (50 IU/mL), and streptomycin (50 μg/mL) (ATCC). The cells were maintained in a humidified environment at 37°C and 5% CO2 and cultured in 25 mL polystyrene culture flasks (Fisher Scientific). Cells were passaged every 2–3 days to avoid overgrowth.
Cell Viability
Cell viability was measured using the Trypan blue (Fisher Scientific) exclusion assay and a hemocytometer cell count (C-Chip disposable hemocytometer, Bulldog Bio, Inc., Portsmouth, NH, USA). The RAW cell suspension was diluted using a 1:1 to 1:3 ratio (based on cell density) with a 0.4% Trypan blue solution. The number of viable cells and the cell density were determined using a 4 mm2 total area hemocytometer. Native RAW cells typically had densities of about 5 million cells in a 25 cm2 flask prior to passaging.
Stimulation Protocol
Stimulation of NO production in cells was accomplished using purified LPS from the Escherichia coli line 0111:B4. A freshly prepared 50 μL aliquot of a 10 μg/mL LPS stock solution was added to healthy RAW 264.7 cells in a 25 cm2 cell culture flask to obtain a 100 ng/mL final LPS concentration and then incubated for 24 h. An unstimulated RAW macrophage cell flask from the same population was incubated under identical conditions and used as a control (native) for each stimulation experiment.
Sample Preparation
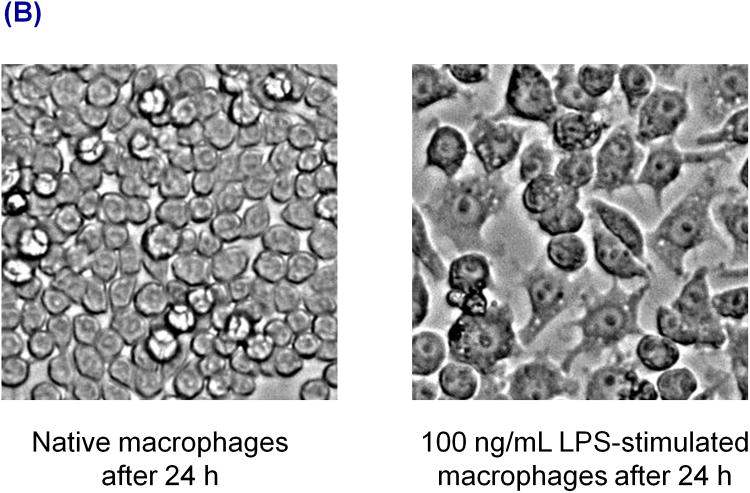
The protocol used for cell analysis is shown in the Figure 2A. Cells were grown in 25 cm2 polystyrene flasks until they reached approximately 80% confluence. At 80% confluence level, there are around 5 million RAW cells in the flask. These cells were stimulated using LPS and, after the stimulation period (24 h with a 100 ng/mL final LPS concentration, Figure 2B), cells were harvested using a scraper and centrifuged at 3500 rpm for 2.5 min to make a live cell pellet. Before centrifugation, 250 μL of the cell solution was taken out for cell counting. The supernatant medium was then removed, leaving only the cell pellet. Then the cell pellet was washed with 10 mM phosphate buffered saline at pH 7.4. Next the cell pellet was lysed using a lysis buffer containing 10 mM boric acid and 2 mM TTAC at pH 10.3 to 10.7. Both the high pH and surfactant assisted with the immediate lysis of cells. Higher molecular weight compounds such as proteins and cell membranes were removed by centrifugation of the lysate for 2–7 min using a 3 kDa molecular weight cut-off filter (VWR International, West Chester, PA, USA). The filtered lysate was then loaded into the sample reservoir of the microchip.
Figure 2.
(A) Diagram of the stimulation and sample preparation protocol for RAW 264.7 macrophage cells prior to ME-EC and Griess assay analyses. (B) Images of RAW 264.7 macrophage cells after 24 h without stimulation (left) and with LPS stimulation (right).
For the standard addition studies, four 25 cm2 cell flasks with the same passage number were harvested and lysed using 1 mL of 10 mM boric acid with 2 mM TTAC at pH 10.3 (for 1 cell flask, 250 μl of buffer was used). The lysate was divided into five portions, and the internal standard was appropriately added to ensure a final concentration 10 μM. Standard addition concentrations of 15, 30, 60, and 120 μM nitrite were chosen and the required nitrite volume from a 1 mM nitrite standard was added to the cell lysates. Before the addition of iodide and nitrite, an equal volume of solution was removed from the cell lysate.
Griess Assay Protocol
The Griess assay was performed using 96-well plates and a plate reader (Molecular Devices, Spectra Max M5, Sunnyvale, CA, USA). To perform the assay, 100 μL of the filtered cell lysate was added into 100 μL of Griess reagent, left to react for 15 min, after which the absorbance at 540 nm was recorded using the plate reader. A buffer background was always employed for these measurements. For nitrite quantitation, a calibration curve was prepared using nitrite standards from 1 to 50 μM. Cell counts were taken before lysing the cells, and the final nitrite concentration was calculated, taking into account the cell counts.
3. Results and Discussion
3.1. Microchip Electrophoresis with Electrochemical Detection
There are two primary electrode configurations that are used for ME under reverse polarity conditions. The electrode can be placed either slightly inside the channel (in-channel) or outside the channel (end-channel). The advantage of the in-channel configuration is it allows higher resolution between closely migrating species, which cannot be separated by end-channel configuration due to band broadening.43 Therefore, faster separations and shorter analysis times can be obtained using the in-channel configuration. Also, we have observed an increase in peak height, better sensitivity, and a higher number of theoretical plates with the in-channel configuration compared to the end-channel configuration.43 However, an important consideration with in-channel detection is that one must take into account the working electrode potential shift that occurs due to the separation voltage when an electrode is placed inside the channel. To minimize this effect in these experiments, the working electrode was placed exactly at the channel end, which still preserves the higher resolution and separation efficiencies characteristic of in-channel detection that are necessary for these studies, but minimizes the potential shift at the working electrode (Figure 1B).43
3.2. Separation Buffer Optimization
The analytes of interest in our studies of nitrosative stress included NO, nitrite (a metabolite of NO), GSH (cellular antioxidant), AA (cellular antioxidant), and tyrosine (amino acid, which is nitrated in the presence of ONOO−). We have previously reported the separation and detection of several of these analytes (nitrite, ascorbic acid, tyrosine, glutathione, and H2O2) by ME-EC as compounds that could potentially interfere with the quantitation of NO and nitrite in macrophage cell lysates.43 For the macrophage cell lysate studies described here, the same separation conditions (10 mM boric acid with 2 mM TTAB) with slight modifications were utilized.
Internal Standard, Surfactant, and Interferences
To quantitate the compounds in the cell lysates and increase the precision of the analytical method, iodide was incorporated as an internal standard and, therefore, had to be taken into consideration during the separation optimization procedures. In our previous studies, TTAB was used to reverse the EOF. In these studies, TTAB was replaced with TTAC, where the counter ion is Cl− instead of Br−. It was found that bromide can be oxidized to Br2 at around 1200 mV versus Ag/AgCl, leading to an increase in background current at the EC detector. Bromide, chloride, and nitrite have similar electrophoretic mobilities and, hence, migrate closely. We observed a vacancy peak close to the nitrite peak during initial cell studies due to high Cl− content. Another species that needed to be separated from the cell lysate components was azide. The molecular weight cut-off filters used for cell lysate filtration were found to contain a small amount of this compound, which is used as an anti-microbial agent. Under these separation conditions, azide migrated between nitrite and iodide but did not interfere with either measurement.
Conductivity Issues
During the initial analysis of the cell lysates, it was observed that the sampling current was always higher than the separation current and the high conductivity samples suppressed the nitrite peak due to destacking.45 A similar suppression in the nitrite signal has been reported in CE when a high conductivity sample was analyzed.22 To reduce the amount of salt and matrix components present in biological samples prior to CE analysis, solid-phase microextraction,24 acetonitrile addition (acetonitrile lowers the sample conductivity),22 dialysis,46 and pre-electrophoresis separation47 have been widely employed.
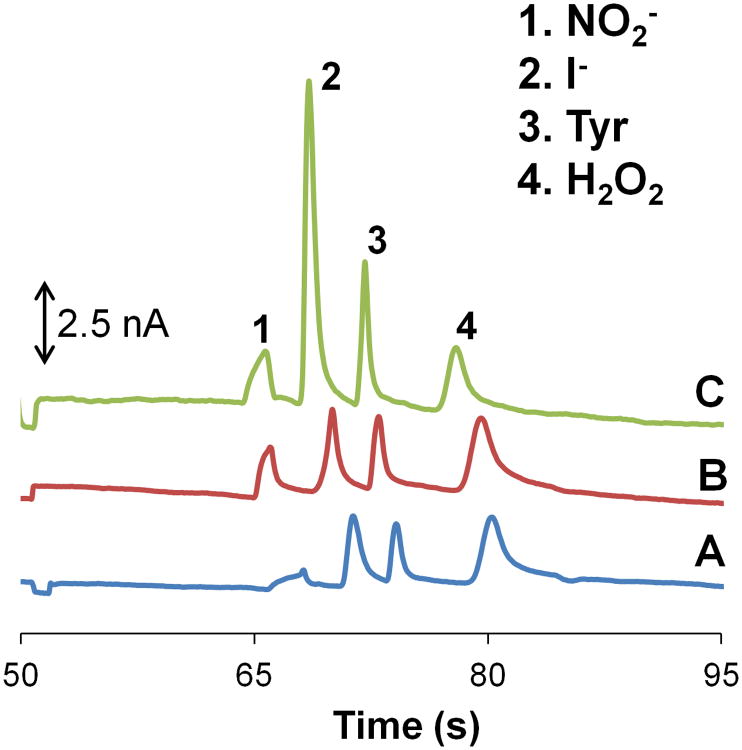
An alternative approach to avoid nitrite destacking is to increase the conductivity of the separation buffer by using sodium chloride. Figure 3A shows the nitrite peak suppression that occurs when standards are prepared in a high conductivity buffer (10 mM boric acid with 2 mM TTAC and 10 mM NaCl at pH 10.3) and the separation buffer consists of a low conducting buffer (10 mM boric acid with 2 mM TTAC at pH 10.3). In contrast, Figure 3B illustrates that the addition of 7.5 mM NaCl to the separation buffer causes an approximately 3-fold increase in the nitrite signal. This can then be compared to a case where both the sample buffer and separation buffer are low conductivity buffers (10 mM boric acid with 2 mM TTAC at pH 10.3) (Figure 3C). In this last case, the nitrite signal is similar to that seen in Figure 3B. These experiments confirmed the destacking of nitrite in high conductivity samples. All three electropherograms used for the comparison studies were recorded with the same microchip, working electrode, and working electrode potential.
Figure 3.
Electropherograms of a standard containing 100 μM nitrite, 10 μM iodide (internal standard), 50 μM tyrosine, and 200 μM hydrogen peroxide (neutral marker) using a 10 mM boric acid and 2 mM TTAC buffer at pH 10.3 while varying the sample and run buffer conductivities. (A) High conductivity sample buffer (10 mM NaCl) and normal separation buffer. (B) High conductivity sample buffer (10 mM NaCl) and high conductivity separation buffer (7.5 mM NaCl). (C) No change to the conductivity of the sample and separation buffer.
3.3. Detection of Nitrite from Macrophage Cell Lysates
RAW 264.7 macrophage cells are known to produce large amounts of NO through the activation of iNOS. LPS, an endotoxin in negative gram bacteria and an external stimulant, can be used to activate iNOS.48, 49 It has been reported that RAW 264.7 macrophage cells produce significantly higher amounts of NO in the presence of LPS.48, 49 In these studies, a LPS concentration of 100 ng/mL over 24 h was used for cell stimulation (Figure 2A). A substantial difference in physical appearance between native and LPS-stimulated cells was observed, as can be seen in Figure 2B.
To compare intracellular nitrite produced in stimulated and native macrophage cells, bulk cell lysates were prepared as shown in Figure 2A, and analyzed by ME-EC. The Griess assay was also performed to compare with the results obtained with ME-EC. To confirm that NO production was due solely to an increase in iNOS activity, a separate set of cells was exposed to L-NAME, which is a known inhibitor of iNOS, before LPS stimulation and analyzed via Griess assay. These results were compared to those from native and LPS-stimulated cell lysate samples with the same passage number. Each flask contained around 5 million cells, which were lysed in 250 μL of borate buffer (10 mM boric acid with 2 mM TTAC at pH 10.3 to 10.7) in order to minimize the sample conductivity (Figure 3A).
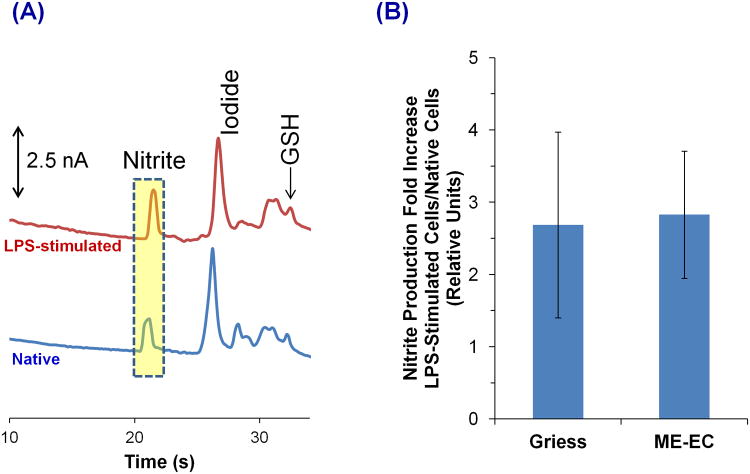
Figure 4A shows the electropherograms obtained for native and LPS-stimulated cell lysates using our ME-EC device. The migration times for the first two peaks in the native cell electropherogram were similar to those for nitrite and iodide standards, and the peak identities were confirmed by spiking with standards. Azide was also spiked to further ensure that the nitrite peak does not comigrate with azide during cell studies.
Figure 4.
(A) Comparison of LPS-stimulated (top) and native (bottom) RAW 264.7 macrophage cell lysates using ME-EC. (B) Comparison of the ME-EC method and the Griess assay for determining the increase in nitrite concentration resulting from a 24 h LPS stimulation relative to the nitrite concentration produced from native cells. The sample was prepared in 10 mM boric acid and 2 mM TTAC buffer at pH 10.3 and the separation was achieved with a 10 mM boric acid, 7.5 mM NaCl and 2 mM TTAC buffer at pH 10.3.
3.4. Comparison of Nitrite Production in Macrophage Cell Lysates using ME-EC and Griess Assay
Three different pairs of native and LPS-stimulated cell lysates were analyzed by ME-EC and the Griess assay, respectively, for the comparison of nitrite concentrations. Both methods were used to determine nitrite production increase in LPS-stimulated cells versus native cells (Figure 4B). A t-test was performed to compare the two sets of data (Greiss versus ME-EC), and it was found that these two series exhibited no statistical difference at a 90% confidence level. This shows that the nitrite level detected with ME-EC is similar to that seen in the results of the Griess assay.
The nitrite concentration varied from one sample to another due to the samples having different cell counts. Therefore, the cell counts were taken into account in both the Griess assay and ME-EC studies when calculating the final nitrite concentrations. The nitrite production in a single cell was estimated by assuming that the volume of a macrophage is approximately 0.5 pL. The Griess assay results show that the average intracellular concentrations of nitrite in single unstimulated and LPS-stimulated macrophage cells are 0.63 ± 0.16 mM (0.31 ± 0.08 fmol/cell) and 1.69 ± 1.06 mM (0.84 ± 0.53 fmol/cell), respectively.
In the case of ME-EC analysis, an external calibration curve could not be used for the quantitation of nitrite due to the nitrite peak suppression. Therefore, the method of standard additions was used, employing iodide as an internal standard. Two different ME-EC setups were used for the analysis of these samples, and two standard addition calibration curves of the nitrite/iodide response vs. standard addition concentration were plotted. These plotsyielded R2 values of 0.987 and 0.973 resulting in values for intracellular nitrite of 0.58 and 0.83 fmol/cell, respectively. This resulted in an average estimated intracellular nitrite concentration for a single native macrophage cell of 1.41 mM.The average nitrite level in single LPS-stimulated cells was then estimated using the nitrite production increase in LPS-stimulated cells relative to that in native cells, which is a 2.83-fold increase (Figure 4B). Consequently, LPS-stimulated cells have a nitrite concentration of approximately 4.00 mM (1.99 fmol/cell). Goto et al. reported similar levels for extracellular nitrite production (1 fmol/cell) in single LPS-stimulated macrophage cells using the Griess reagent and a microfluidic device.31
3.5. Direct Detection of NO and Other Electroactive Species in Macrophage Cells
NO Detection
The reason for employing ME-EC in these studies is the ability to directly detect NO, its metabolite NO2−, and other cellular electroactive species (e.g., cellular antioxidents) simultaneously. The overall goal is to implement this in a single cell analysis system in the future. Detection of all these species cannot be achieved with the Griess assay or LIF detection alone.
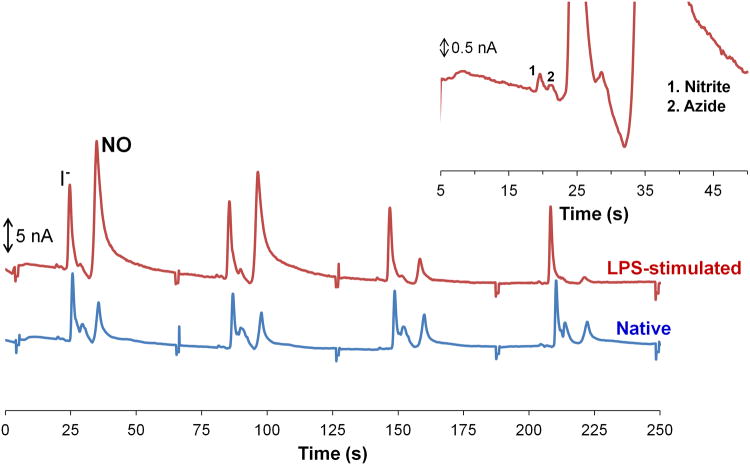
When detecting NO in cell lysates, sample preparation steps were shortened to minimize NO degradation and evaporation. Cells were quickly lysed (10–20 s), and the lysate was centrifuged for only 2 min. Figure 5 shows electropherograms obtained for native and LPS-stimulated cell lysates following this procedure. It can be seen that the height of the peak that migrates at approximately 30 s decreases over time compared to the internal standard peak. The migration time of the decreasing peak is close to the neutral marker (32.3 ± 2.1 s), and the quick disappearance of this peak over several injections suggests that the compound is unstable. Since cells produce NO following LPS stimulation due to the induction of iNOS, this peak is most likely NO. The disappearance of this peak is probably due to loss of the gas through the open reservoirs on the microchip or permeation through the PDMS. Nitrite was also detected during these studies, but the nitrite peak is very small compared to the NO peak (Figure 5 inset), which confirms that NO disappears from the wells quickly before degradation occurs. When the sample preparation time was lengthened, this peak disappeared.
Figure 5.
Detection of NO in cell lysate. LPS-stimulated cell lysate (top) and native cell lysate (bottom). Inset is a magnified portion of the LPS-stimulated cell lysate. The sample was prepared in 10 mM boric acid and 2 mM TTAC buffer at pH 10.3 and the separation was achieved with a 10 mM boric acid, 7.5 mM NaCl and 2 mM TTAC buffer at pH 10.3.
We previously reported a ME method for the detection of NO generated using diethylammine NONOate (DEA/NO) and proline NONOate (PROLI/NO) salts.35 The migration time of NO in those studies is comparable to the migration time of the decaying peak in the cell lysates considering the slight variation in chip-to-chip migration times that is expected in PDMS-based systems.35 It can be seen in the native cell lysate that the last peak does not decay at the same rate as the unstable NO peak seen in LPS-stimulated cell lysate. This indicates that the peak observed in the native cell sample is contaminated with a more stable electroactive species. This species was found to be an interfering filter component that migrates close to the neutral marker (Figure 5). Therefore, the NO peak observed in these studies is contaminated.
Currently, the NO peak cannot be used for a quantitative comparison of native and stimulated cells due to the necessity for further peak identification, experimental variability, the presence of an interference due to the filters, and, most importantly, the fact that the peak decreases quickly over time due to evaporation and degradation. However, detection of NO will be better accomplished using a single cell analysis microfluidic device where cells are lysed inside the device and the content is immediately analyzed. Since the cell lysis procedure is automated, a single cell cytometric device would provide better precision. Furthermore, a single cell cytometric device eliminates the cell lysate filtering step.
Comparison of Glutathione Levels in Native and Stimulated Cells
Other electroactive species such as tyrosine and GSH were also detected in macrophage cell lysates. However, electropherograms of native and LPS-stimulated cell lysates showed a very small peak or no peak for AA, which agreed with previous ME-LIF studies.36 Macrophages do not naturally produce AA and an AA free media was used for cell culture. Previous studies reported undetectable levels of AA in RAW macrophage cells.50
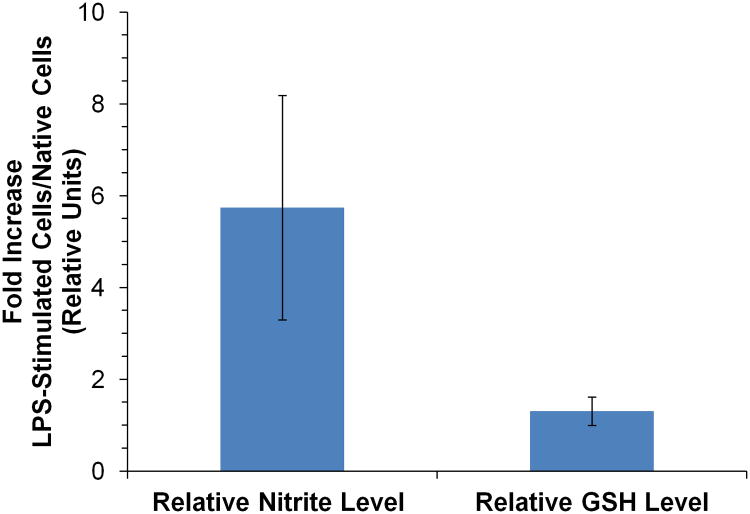
The relative GSH and nitrite levels for three separate LPS-stimulated cell lysates were compared to that of a native cell lysate with the same passage number using the same ME-EC conditions used for nitrite detection. As before, it was found that the nitrite level in LPS-stimulated cells was increased 5.74 ± 2.44 times relative to the native cell lysates. However, the GSH levels showed no significant change (1.30 ± 0.31) when the cells were stimulated with LPS (Figure 6). Hothersall et al. also observed that GSH levels were not changed when macrophage cells were stimulated with LPS alone. However, they have shown that the GSH level changed when the cells were stimulated with LPS and interferon gamma.51
Figure 6.
Comparison of the nitrite and glutathione (GSH) levels as a result of LPS stimulation relative to that of the native cell lysate. The sample was prepared in 10 mM boric acid and 2 mM TTAC buffer at pH 10.3 and the separation was achieved with a 10 mM boric acid, 10 mM NaCl and 2 mM TTAC buffer at pH 10.7.
Conclusion
In this paper, a ME-EC method was optimized for the detection of nitrite, NO, and other electroactive species within macrophage cell lysates. ME-EC makes it possible to obtain more information regarding the overall cellular redox state of the cell. It also provides a separation of interfering species from the analytes of interest that cannot be achieved using classical methods such as the Griess assay and fluorescence imaging. Initially, NO production was detected through the detection of nitrite using a ME-EC device. The results obtained for nitrite production between LPS-stimulated and native cell lysates using ME-EC were compared to those from the Griess assay. Then this method was used for the direct detection of NO and other electroactive species in the cell lysate. An unstable species, which had many of the chemical and physical properties of NO, was detected during these studies. However, the NO peak cannot currently be used for a quantitative comparison of native and stimulated cells. The detection of NO will be better accomplished using a single cell analysis microfluidic device where cells are lysed inside the device and the content immediately analyzed. We have already reported a single cell chemical cytommetric device for NO detection from Jurkat cells using a NO-selective fluorophore.29 The ultimate goal is to use ME-EC to measure multiple redox-active species in a single cell as an indication of nitrosative stress.
Acknowledgments
This research was funded by the following NIH grants: R01NS042929 and R21 NS061202 and COBRE grant P20GM103638. The authors thank Pinnacle Technologies, Inc., Lawrence, KS, USA for the development and loan of the isolated potentiostats. M.K.H. was a recipient of an American Heart Association postdoctoral fellowship. G.C. was supported by the International Internship Program, University of Catania, Italy. J.M.S. has received support from the Madison and Lila Self Graduate Fellowship, University of Kansas, Lawrence, KS, USA. The authors would also like to thank Prof. José Alberto Fracassi da Silva, Pann Pichetsurnthorn, Keelan Trull, Diogenes Meneses do Santos, and Emilie R. Mainz for assistance with some of the experiments; Prof. Christian Schöneich for helpful discussions; Ryan Grigsby for help with microchip fabrication; and Nancy Harmony for editorial support.
References
- 1.Pacher P, Beckman JS, Liaudet L. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacMicking J, Xie Q, Nathan C. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 3.Calabrese V, Cornelius C, Rizzarelli E, Owen JB, Dinkova-Kostova AT, Butterfield DA. Antioxid Redox Signal. 2009;11:2717–2739. doi: 10.1089/ars.2009.2721. [DOI] [PubMed] [Google Scholar]
- 4.Kröncke KD, Fehsel K, Kolb-Bachofen V. Nitric Oxide. 1997;1:107–120. doi: 10.1006/niox.1997.0118. [DOI] [PubMed] [Google Scholar]
- 5.Moller MN, Li Q, Lancaster JR, Jr, Denicola A. IUBMB Life. 2007;59:243–248. doi: 10.1080/15216540701311147. [DOI] [PubMed] [Google Scholar]
- 6.Martinez FO, Helming L, Gordon S. Annu Rev Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 7.Gordon S, Taylor PR. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 8.Lilly LS. Pathophysiology of Heart Disease : A Collaborative Project of Medical Students and Faculty. 5th. Lippincott Williams & Wilkins; Baltimore, MD: 2011. [Google Scholar]
- 9.Minghetti L, Levi G. Prog Neurobiol. 1998;54:99–125. doi: 10.1016/s0301-0082(97)00052-x. [DOI] [PubMed] [Google Scholar]
- 10.Valko M, Leibfritz D, Moncol J, Cronin MTD, Mazur M, Telser J. Int J Biochem Cell Biol. 2006;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Zhang H, Forman HJ. Semin Cell Dev Biol. 2012;23:722–728. doi: 10.1016/j.semcdb.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulz JB, Lindenau J, Seyfried J, Dichgans J. Eur J Biochem. 2000;267:4904–4911. doi: 10.1046/j.1432-1327.2000.01595.x. [DOI] [PubMed] [Google Scholar]
- 13.Amatore C, Arbault S, Koh ACW. Anal Chem. 2010;82:1411–1419. doi: 10.1021/ac902486x. [DOI] [PubMed] [Google Scholar]
- 14.Tsikas D. J Chromatogr B. 2007;851:51–70. doi: 10.1016/j.jchromb.2006.07.054. [DOI] [PubMed] [Google Scholar]
- 15.Tsikas D. Anal Biochem. 2008;379:139–163. doi: 10.1016/j.ab.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 16.Bedioui F, Griveau S. Electroanal. 2013;25:587–600. [Google Scholar]
- 17.Hetrick EM, Schoenfisch MH. Annu Rev Anal Chem. 2009;2:409–433. doi: 10.1146/annurev-anchem-060908-155146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trouillon R. Biol Chem. 2013;394:17–33. doi: 10.1515/hsz-2012-0245. [DOI] [PubMed] [Google Scholar]
- 19.Gomes A, Fernandes E, Lima JLFC. J Fluoresc. 2006;16:119–139. doi: 10.1007/s10895-005-0030-3. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Kim WS, Hatcher N, Potgieter K, Moroz LL, Gillette R, Sweedler JV. J Biol Chem. 2002;277:48472–48478. doi: 10.1074/jbc.M209130200. [DOI] [PubMed] [Google Scholar]
- 21.Ye X, Rubakhin SS, Sweedler JV. Analyst. 2008;133:423–433. doi: 10.1039/b716174c. [DOI] [PubMed] [Google Scholar]
- 22.Friedberg MA, Hinsdale ME, Shihabi ZK. J Chromatogr A. 1997;781:491–496. doi: 10.1016/s0021-9673(97)00583-9. [DOI] [PubMed] [Google Scholar]
- 23.Morcos E, Wiklund NP. Methods Mol Biol. 2004;279:21–34. doi: 10.1385/1-59259-807-2:021. [DOI] [PubMed] [Google Scholar]
- 24.Boudko DY. Methods Mol Biol. 2004;279:9–19. doi: 10.1385/1-59259-807-2:009. [DOI] [PubMed] [Google Scholar]
- 25.Boudko DY, Cooper BY, Harvey WR, Moroz LL. J Chromatogr B. 2002;774:97–104. doi: 10.1016/s1570-0232(02)00219-2. [DOI] [PubMed] [Google Scholar]
- 26.Kim WS, Ye X, Rubakhin SS, Sweedler JV. Anal Chem. 2006;78:1859–1865. doi: 10.1021/ac051877p. [DOI] [PubMed] [Google Scholar]
- 27.Govindaraju K, Toporsian M, Ward ME, Lloyd DK, Cowley EA, Eidelman DH. J Chromatogr B. 2001;762:147–154. doi: 10.1016/s0378-4347(01)00358-9. [DOI] [PubMed] [Google Scholar]
- 28.Hunter RA, Privett BJ, Henley WH, Breed ER, Liang Z, Mittal R, Yoseph BP, McDunn JE, Burd EM, Coopersmith CM, Ramsey JM, Schoenfisch MH. Anal Chem. 2013;85:6066–6072. doi: 10.1021/ac400932s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metto EC, Evans K, Barney P, Culbertson AH, Gunasekara DB, Caruso G, Hulvey MK, Fracassi da Silva JA, Lunte SM, Culbertson CT. Anal Chem. 2013;85:10188–10195. doi: 10.1021/ac401665u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogel PA, Halpin ST, Martin RS, Spence DM. Anal Chem. 2011;83:4296–4301. doi: 10.1021/ac2004746. [DOI] [PubMed] [Google Scholar]
- 31.Goto M, Sato K, Murakami A, Tokeshi M, Kitamori T. Anal Chem. 2005;77:2125–2131. doi: 10.1021/ac040165g. [DOI] [PubMed] [Google Scholar]
- 32.Hulvey MK, Martin RS. Anal Bioanal Chem. 2009;393:599–605. doi: 10.1007/s00216-008-2468-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halpin ST, Spence DM. Anal Chem. 2010;82:7492–7497. doi: 10.1021/ac101130s. [DOI] [PubMed] [Google Scholar]
- 34.Letourneau S, Hernandez L, Faris AN, Spence DM. Anal Bioanal Chem. 2010;397:3369–3375. doi: 10.1007/s00216-010-3687-7. [DOI] [PubMed] [Google Scholar]
- 35.Gunasekara DB, Hulvey MK, Lunte SM, Fracassi da Silva JA. Anal Bioanal Chem. 2012;403:2377–2384. doi: 10.1007/s00216-012-5810-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mainz ER, Gunasekara DB, Caruso G, Jensen DT, Hulvey MK, Fracassi da Silva JA, Metto EC, Culbertson AH, Culbertson CT, Lunte SM. Anal Methods. 2012;4:414–420. [Google Scholar]
- 37.Kikura-Hanajiri R, Martin RS, Lunte SM. Anal Chem. 2002;74:6370–6377. doi: 10.1021/ac0204000. [DOI] [PubMed] [Google Scholar]
- 38.Vázquez M, Frankenfeld C, Coltro WKT, Carrilho E, Diamond D, Lunte SM. Analyst. 2010;135:96–103. doi: 10.1039/b908985c. [DOI] [PubMed] [Google Scholar]
- 39.Shiddiky MJA, Lee KS, Son J, Park DS, Shim YB. J Agric Food Chem. 2009;57:4051–4057. doi: 10.1021/jf900230x. [DOI] [PubMed] [Google Scholar]
- 40.Troška P, Chudoba R, Danč L, Bodor R, Horčičiak M, Tesařová E, Masár M. J Chromatogr B. 2013;930:41–47. doi: 10.1016/j.jchromb.2013.04.042. [DOI] [PubMed] [Google Scholar]
- 41.Noblitt SD, Schwandner FM, Hering SV, Collett JL, Jr, Henry CS. J Chromatogr A. 2009;1216:1503–1510. doi: 10.1016/j.chroma.2008.12.084. [DOI] [PubMed] [Google Scholar]
- 42.Hulvey MK, Frankenfeld CN, Lunte SM. Anal Chem. 2010;82:1608–1611. doi: 10.1021/ac902821v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gunasekara DB, Hulvey MK, Lunte SM. Electrophoresis. 2011;32:832–837. doi: 10.1002/elps.201000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott DE, Grigsby RJ, Lunte SM. ChemPhysChem. 2013;14:2288–2294. doi: 10.1002/cphc.201300449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boden J, Bächmann K. J Chromatogr A. 1996;734:319–330. [Google Scholar]
- 46.Haddad PR, Doble P, Macka M. J Chromatogr A. 1999;856:145–177. doi: 10.1016/s0021-9673(99)00431-8. [DOI] [PubMed] [Google Scholar]
- 47.Timerbaev AR, Fukushi K, Miyado T, Ishio N, Saito K, Motomizu S. J Chromatogr A. 2000;888:309–319. doi: 10.1016/s0021-9673(00)00539-2. [DOI] [PubMed] [Google Scholar]
- 48.Lorsbach RB, Murphy WJ, Lowenstein CJ, Snyder SH, Russell SW. J Biol Chem. 1993;268:1908–1913. [PubMed] [Google Scholar]
- 49.Held TK, Weihua X, Yuan L, Kalvakolanu DV, Cross AS. Infect Immun. 1999;67:206–212. doi: 10.1128/iai.67.1.206-212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Badrakhan CD, Petrat F, Holzhauser M, Fuchs A, Lomonosova EE, de GH, Kirsch M. J Biochem Biophys Methods. 2004;58:207–218. doi: 10.1016/j.jbbm.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 51.Hothersall JS, Cunha FQ, Neild GH, Norohna-Dutra AA. Biochem J. 1997;322:477–481. doi: 10.1042/bj3220477. [DOI] [PMC free article] [PubMed] [Google Scholar]