Abstract
The Pharmacogenomics Knowledge Base, PharmGKB, is an interactive tool for researchers investigating how genetic variation affects drug response. The PharmGKB Web site, http://www.pharmgkb.org, displays genotype, molecular, and clinical knowledge integrated into pathway representations and Very Important Pharmacogene (VIP) summaries with links to additional external resources. Users can search and browse the knowledgebase by genes, variants, drugs, diseases, and pathways. Registration is free to the entire research community, but subject to agreement to use for research purposes only and not to redistribute. Registered users can access and download data to aid in the design of future pharmacogenetics and pharmacogenomics studies.
Keywords: PharmGKB, Database, Pharmacogenetics, Pharmacogenomics, Genotype, Phenotype, Pathways, VIP genes, Pharmacogenes
1 Background
In 1999 the National Institutes of Health recognized the need for a freely available collection of high quality genotypic and phenotypic data from pharmacogenetics and pharmacogenomics studies, and announced the funding of the Pharmacogenetics Research Network (PGRN). Its mission: “to enable the formation of a series of multi-disciplinary research groups funded to conduct studies addressing research problems in pharmacogenetics. These groups are united by the purpose of developing and populating a public database, which was envisioned as a tool for all researchers in the field.” [1] This tool is the PharmGKB, the Pharmacogenomics Knowledge Base, with Web site access that provides summaries of pharmacogenomic relationships linked to the data that support them, to be used by the scientific community for pharmacogenetics and pharmacogenomics research (Fig. 1).
Fig. 1.
The PharmGKB homepage, http://www.pharmgkb.org, contains directed search boxes where users can also browse from lists of genes, variants, drugs, or diseases
2 Overview
PharmGKB captures pharmacogenomic relationships in a structured format so that it can be searched, interrelated, and displayed according to the researchers interests, either for manual inspection or to download for further analyses. The knowledge base is valuable both to the researcher who is interested in a specific single nucleotide polymorphism and its influence on a particular drug treatment and to the researcher interested in a disease or drug and looking for candidate genes which may affect disease progression or drug response. At present PharmGKB has over 5,000 variant annotations, with over 900 genes related to drugs and over 600 drugs related to genes [April 2013]. The data contained within the database is curated from a variety of sources to bring together the most relevant features of genes, drugs, and diseases for pharmacogenomics [2]. Some information is imported directly from other trusted standard repositories (such as gene symbols and names from the Human Genome Nomenclature Committee, HGNC [3], drug names and structures from Drugbank [4]); detailed relationship data from the literature is manually curated and described using controlled vocabularies. For genes and drugs where many relationships are known, these are compiled by curators and experts in the field into Very Important Pharmacogene (VIP) summaries and PharmGKB drug pathways and published in an interactive form on the Web site and conventional form in peer reviewed journals [5, 6].
PharmGKB averages around 30,000 visitors per month. Of the more than 5,000 user accounts, approximately 30 % are identified as academic users (.edu), with 30 % from industry (.com) and 8 % from nonprofit or government domains. A user account and agreement to the PharmGKB database license agreement is necessary for downloading data. Data is distributed as zipped up packages of spreadsheets with literature relationships, variant annotations, clinical annotations or pathway relationships. Individualized genotype and phenotype datasets from pharmacogenomics studies of the PGRN can be found under the download tabs of the relevant genes, drugs, and diseases.
PharmGKB exchanges data with Drugbank, dbSNP, the CYP alleles database, and HuGE Navigator. Data is imported from HGNC, Entrez, and UCSC Golden Path. Links are also maintained with a number of other sources as seen under the Downloads/Link Outs tab.
The initial interaction with the Web site is through pages devoted to genes, variants, drugs, diseases, and pathways, with directed searches to make access to these more rapid for focused users (see the hompage, Fig. 1). The data is represented according to a hierarchy and tagged with icons. This enables many facets of the data to be captured and stored in the database but also permits the user to find exactly what they are looking for. The use of standardized vocabulary aids both the sorting and storage of data and supports automated methods of analysis as well as traditional human browsing.
3 Initial Interactions with the PharmGKB Web Site: Gene, Drug, and Disease Pages
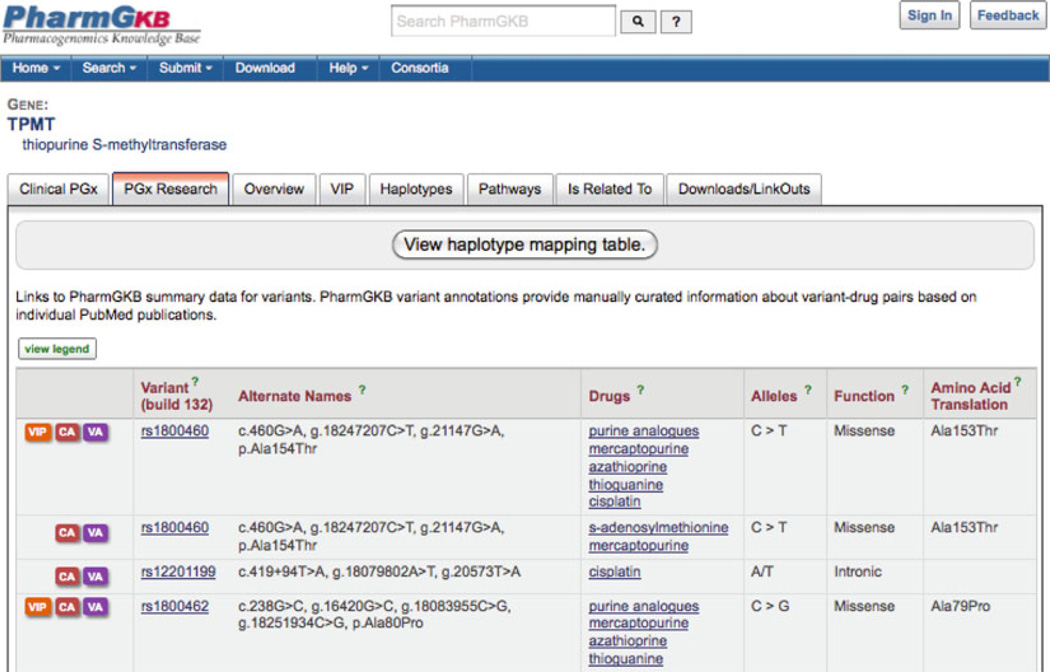
In PharmGKB, genes are catalogued according to the HGNC [3]. In addition alternative names and symbols are also listed and can be submitted by researchers and searched on. The general layout of a gene page is shown in Fig. 2. The data are organized under tabs for clinical pharmacogenomics, pharmacogenomics research, overview, VIP, haplotypes, pathways, related genes, drugs, and diseases, datasets, and downloads or links out. The clinical pharmacogenomics tab displays any dosing guidelines involving the gene published by CPIC (the Clinical Pharmacogenomics Implementation Consortium) [7] and the Royal Dutch Pharmacogenetics Working Group [8]. This tab also has drug labels, high level clinical annotations (described in more detail below) and links to genetic testing sources for the gene. The pharmacogenomics research tab lists genomic variants associated with the gene and the drugs they interact with and links to annotations that describe the relationship between the variants and drugs from individual papers (described in more detail below). The overview page contains the basic data about the gene, standard and alternate names and symbols, and location on the genome. The VIP tab is present for genes where there is considerable knowledge of the pharmacogenomics and a summary has been written (see below for more details and Fig. 2). Pathway tabs link to the curated drug pathways that involve the gene. Related genes, drugs, and diseases are compiled from literature annotations (described below). Download/Link outs provide a mechanism to retrieve primary data files or go to the original source.
Fig. 2.
The TMPT gene page showing genomic variants and related drugs with links to the annotations and haplotypes and tabs for clinical pharmacogenomics, pharmacogenomics research, overview, VIP, haplotypes, pathways, related drugs and diseases, and downloads and links out
Drug and disease pages follow a similar tabbed layout style to the gene pages. Drug information including pharmacological effects, mechanisms of action, and structures was obtained from Drugbank [4] and Pubchem [9]. Additional information and short pharmacogenomics summaries for the top 100 drugs (selected based on a combined list of the most prescribed drugs and the most reported drugs for adverse events) was compiled by PharmGKB curators. Disease information is imported from MeSH [10] and SnoMed [11].
4 Curated Knowledge
Capturing the wealth of pharmacogenomic data already published is a considerable challenge. Most of this information is stored in written natural language text in journal articles or books and not easily retrieved by automated methods. We conduct research into natural language processing (NLP) and ways in which to appropriately aggregate all pharmacogenetics and pharmacogenomics articles in Pubmed [12] but there is still a necessity for human curation to ensure quality data [13].
4.1 Literature Annotations
A basic literature annotation captures the genes, drugs, and diseases involved in a single article from Pubmed and the category (or categories) of evidence that describe the type of relationships measured. Our current process for literature annotation uses NLP to suggest possible genes, drugs, and diseases to the curator [14, 15] but after reading the article the curator decides which are appropriate.
4.2 Genomic Variant Annotations and Very Important Pharmacogenes
In addition to tagging articles for basic relationships curators can also describe in detail the relationships for individual variants and their effects on drug response. The variant is mapped to the dbSNP identifier and controlled vocabularies are used to define the alleles or genotypes observed in the paper and their response to drug, in the particular population studied. Information about the population size, location or race and ethnicity, allele frequencies and statistical measures can be captured and stored in the database. Although time consuming, the benefit of annotating each individual publication in such a detailed manner is that it will allow for all kinds of computational analyses. PharmGKB currently has over 5,000 genomic variant annotations [April 2013].
In addition to the very structured annotations, a more text based, reader-friendly format is provided to summarize the relationships for genes and variants where many there have been many pharmacogenomic studies. These mini-reviews are known as Very Important Pharmacogene summaries or VIPs. PharmGKB currently provides VIPs for 47 genes [April 2013] with a priority list of more to be developed. The list of VIP genes has been used by several groups in a variety of studies to provide a candidate set of genes to work from [16–19]. The NIH Pharmacogenomics Research Network (PGRN) has a longer list of more than 500 genes of relevance to pharmacogenetics which is available at PharmGKB.
4.3 Clinical Annotations
Once there is sufficient evidence available from variant annotations for a given variant and drug combination a clinical annotation is written. This is a summary of the clinical relevance for each of the individual genotypes that may be observed for a given gene variant and drug combination. The PharmGKB’s clinical annotations reflect expert consensus based on clinical evidence and peer-reviewed literature available at the time they are written and are intended only to assist clinicians in decision-making and to identify questions for further research. A strength of evidence score is given for clinical annotations based on the type of study, number of study subjects, and statistical significance reported.
4.4 Pathways
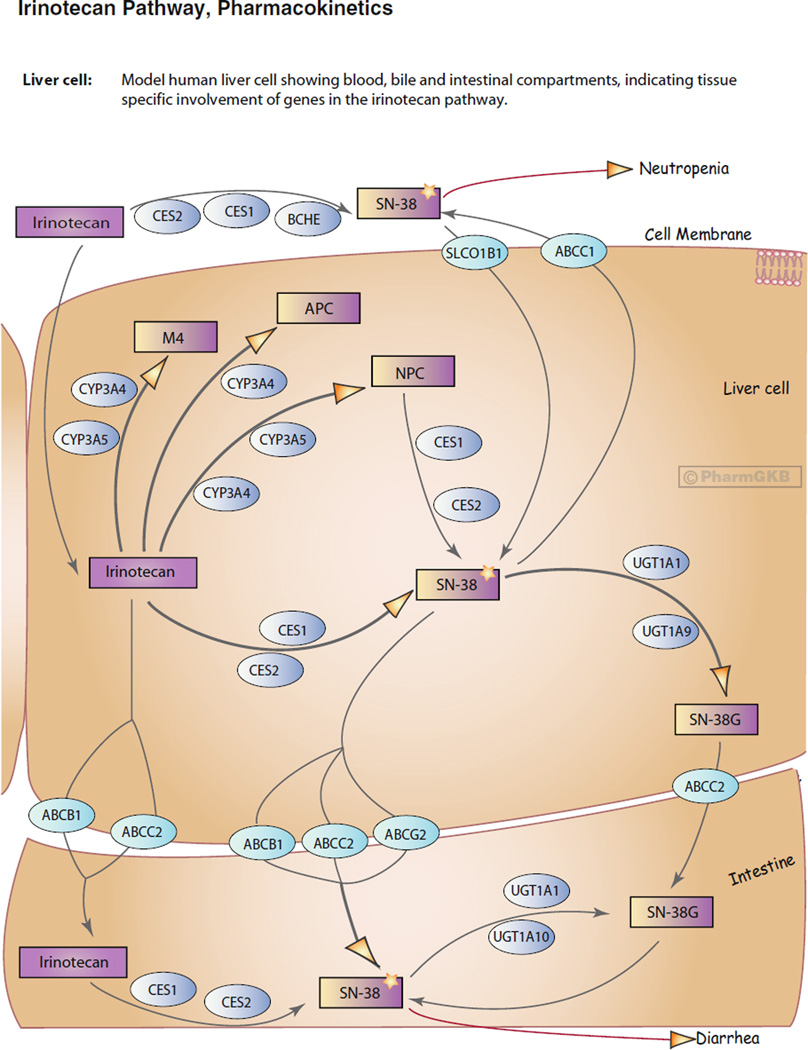
Historically many pharmacogenetic studies have focused on single genes involved in drug side affects, there is now a growing interest in how pathways of interacting genes can affect both drug metabolism and drug response. PharmGKB pathways are drug-centered, depicting candidate genes for pharmacogenetics and pharmacogenomics studies, they provide the means to connect separate data sets to represent the current knowledge as a cohesive snapshot. The diagrams have information content in the shape and color of the icons that represent whether the component is a gene, a drug, a metabolic intermediate, and so on. This information is captured in the database in a Biopax [20] compatible format that can be downloaded and used in pathway analysis packages. The Web-displayed pathways are interactive and clicking on a gene icon opens a window with the gene page, clicking on a drug opens a window of a drug page, etc. The Irinotecan Pathway is shown in Fig. 3 as an example. We currently have 99 curated pathways [April 2013], many of which have been published in peer reviewed journals [21–35].
Fig. 3.
The Irinotecan Pathway, view of a model human liver cell showing blood, bile, and intestinal compartments, indicating tissue-specific involvement of genes in the irinotecan pathway. Drugs are depicted by purple boxes, transporter genes by turquoise ovals, genes coding for metabolic enzymes by blue ovals. http://www.pharmgkb.org/do/serve?objId=PA2001&objCls=Pathway
A summary is provided to describe in words the content of the graphic, its particular view and limitations, and additional, perhaps ill-defined or controversial, data that was not included in this representation. The pathways are generated by collaboration of investigators to link data, either novel or in the public domain, centered on a particular drug. The representation is a consensus of the opinions of the authors. Currently these pathways are constructed by hand as graphic images. They are then converted by a curator into gpml, GenMapp pathway markup language, a BioPax compatible format, and stored in the knowledgebase.
5 Future Directions
Since the year 2000, the PharmGKB has become the “go to” site for pharmacogenetics and pharmacogenomics knowledge [36, 37]. In response to assessment of the field and feedback from users, the priorities for the next 5 years include:
Supporting data-sharing consortia in which multiple investigators pool their data in collaboration with PharmGKB to answer specific questions that require large datasets, not typically available to single research groups.
Developing algorithms for text mining in order to identify appropriate pharmacogenomics literature, and begin the process of extracting the key genes, variations, drugs, and phenotypes that form the basis for our curator annotations.
Creating algorithms for the analysis of rare variations that emerge from whole exome and whole genome sequencing efforts. Most of the efforts to date in pharmacogenomics have focused on the analysis of common variants, but the era of genome sequencing has made it clear that a primary challenge will be interpreting rare or novel variations found in individual genomes.
Helping lead the clinical implementation and impact of pharmacogenomics knowledge in clinical settings. The contents of PharmGKB can provide a base of peer-reviewed information from which clinical guidelines can be constructed.
Studying the molecular and cellular mechanisms of drug response in order to provide the knowledgebase required to understand the systemic effects of drugs, their side effects, and their unexpected interactions.
Finally, we will evaluate how these and other activities impact the requirements for the PharmGKB Web site, and consider its evolution from a purely research repository of knowledge to a more integrated research and clinical resource for personalized medicine.
Acknowledgments
The authors would like to acknowledge Dorit Berlin, Michelle Whirl Carrillo, John Conroy, Adrien Coulet, Sean David, Katrina Easton, Ray Fergerson, Yael Garten, Li Gong, Mei Gong, Winston Gor, Joan Hebert, Tina Hernandez-Boussard, Micheal Hewett, Amy Hodge, Laura Hodges, Daniel Holbert, Tiffany Jung, Mark Kiuchi, Steve Lin, Feng Liu, Xing Jian Lou, Charity Lu, Andrew MacBride, Ellen McDonagh, Diane Oliver, Connie Oshiro, Ryan Owen, Daniel Rubin, Katrin Sangkuhl, Farhad Shafa, Ravi Shankar, Rebecca Tang, TC Truong, Ryan Whaley, Mark Woon, and Tina Zhou for their contributions to building the PharmGKB.
The PharmGKB is financially supported by NIH/NIGMS (R24GM61374).
References
- 1.NIH. Goals for the PGRN. http://www.nigms.nih.gov/Research/FeaturedPrograms/PGRN/
- 2.Altman RB, Klein TE. Challenges for biomedical informatics and pharmacogenomics. Annu Rev Pharmacol Toxicol. 2002;42:113–133. doi: 10.1146/annurev.pharmtox.42.082401.140850. [DOI] [PubMed] [Google Scholar]
- 3.Povey S, et al. The HUGO Gene Nomenclature Committee (HGNC) Hum Genet. 2001;109:678–680. doi: 10.1007/s00439-001-0615-0. [DOI] [PubMed] [Google Scholar]
- 4.Wishart DS, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eichelbaum M, et al. New feature: pathways and important genes from PharmGKB. Pharmacogenet Genomics. 2009;19:403. doi: 10.1097/FPC.0b013e32832b16ba. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sangkuhl K, et al. PharmGKB: understanding the effects of individual genetic variants. Drug Metab Rev. 2008;40:539–551. doi: 10.1080/03602530802413338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Relling MV, Klein TE. CPIC: clinical pharmacogenetics implementation consortium of the pharmacogenomics research network. Clin Pharmacol Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swen JJ, et al. Pharmacogenetics: from bench to byte-an update of guidelines. Clin Pharmacol Ther. 2011;89:662–673. doi: 10.1038/clpt.2011.34. [DOI] [PubMed] [Google Scholar]
- 9.Bolton E, Wang Y, Thiessen PA, Bryant SH. Annual Reports in Computational Chemistry. Washington, DC: American Chemical Society; 2008. PubChem: integrated platform of small molecules and biological activities. [Google Scholar]
- 10.(US), N. L. o. M. MeSH Browser. http://www.nlm.nih.gov/mesh/MBrowser.html.
- 11.Organisation, I. H. T. S. D. SNOMED CT. http://www.ihtsdo.org/snomed-ct/
- 12.Rubin DL, et al. A statistical approach to scanning the biomedical literature for pharmacogenetics knowledge. J Am Med Inform Assoc. 2005;12:121–129. doi: 10.1197/jamia.M1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altman RB, et al. Indexing pharmacogenetic knowledge on the World Wide Web. Pharmacogenetics. 2003;13:3–5. doi: 10.1097/00008571-200301000-00002. [DOI] [PubMed] [Google Scholar]
- 14.Garten Y, Altman RB. Pharmspresso: a text mining tool for extraction of pharmacogenomic concepts and relationships from full text. BMC Bioinformatics. 2009;10(Suppl 2):S6. doi: 10.1186/1471-2105-10-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coulet A, et al. Using text to build semantic networks for pharmacogenomics. J Biomed Inform. 2010;43:1009–1019. doi: 10.1016/j.jbi.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, et al. Interethnic comparisons of important pharmacology genes using SNP databases: potential application to drug regulatory assessments. Pharmacogenomics. 2010;11:1077–1094. doi: 10.2217/pgs.10.79. [DOI] [PubMed] [Google Scholar]
- 17.Sissung TM, et al. Clinical pharmacology and pharmacogenetics in a genomics era: the DMET platform. Pharmacogenomics. 2010;11:89–103. doi: 10.2217/pgs.09.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamazon ER, et al. A pharmacogene database enhanced by the 1000 Genomes Project. Pharmacogenet Genomics. 2009;19:829–832. doi: 10.1097/FPC.0b013e3283317bac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng J, et al. Compilation of a comprehensive gene panel for systematic assessment of genes that govern an individual’s drug responses. Pharmacogenomics. 2010;11:1403–1425. doi: 10.2217/pgs.10.99. [DOI] [PubMed] [Google Scholar]
- 20.Demir E, et al. The BioPAX community standard for pathway data sharing. Nat Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desta Z, et al. Antiestrogen pathway (aromatase inhibitor) Pharmacogenet Genomics. 2009;19:554–555. doi: 10.1097/FPC.0b013e32832e0ec1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorn CF, Klein TE, Altman RB. Codeine and morphine pathway. Pharmacogenet Genomics. 2009;19:556–558. doi: 10.1097/FPC.0b013e32832e0eac. [DOI] [PubMed] [Google Scholar]
- 23.Yang J, et al. Etoposide pathway. Pharmacogenet Genomics. 2009;19:552–553. doi: 10.1097/FPC.0b013e32832e0e7f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsh S, et al. Platinum pathway. Pharmacogenet Genomics. 2009;19:563–564. doi: 10.1097/FPC.0b013e32832e0ed7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sangkuhl K, Klein TE, Altman RB. Selective serotonin reuptake inhibitors pathway. Pharmacogenet Genomics. 2009;19:907–909. doi: 10.1097/FPC.0b013e32833132cb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaza G, et al. Thiopurine pathway. Pharmacogenet Genomics. 2010;20:573–574. doi: 10.1097/FPC.0b013e328334338f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong L, Altman RB, Klein TE. Bisphosphonates pathway. Pharmacogenet Genomics. 2011;21:50–53. doi: 10.1097/FPC.0b013e328335729c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maitland ML, et al. Vascular endothelial growth factor pathway. Pharmacogenet Genomics. 2010;20:346–349. doi: 10.1097/FPC.0b013e3283364ed7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sangkuhl K, Klein TE, Altman RB. Clopidogrel pathway. Pharmacogenet Genomics. 2010;20:463–465. doi: 10.1097/FPC.0b013e3283385420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sangkuhl K, et al. Platelet aggregation pathway. Pharmacogenet Genomics. 2011;21(8):516–521. doi: 10.1097/FPC.0b013e3283406323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oshiro C, et al. Taxane Pathway. Pharmacogenet Genomics. 2009;19:979–983. doi: 10.1097/FPC.0b013e3283335277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikkelsen TS, et al. PharmGKB summary: methotrexate pathway. Pharmacogenet Genomics. 2011;21(10):679–686. doi: 10.1097/FPC.0b013e328343dd93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sangkuhl K, Klein TE, Altman RB. PharmGKB summary: citalopram pharmacokinetics pathway. Pharmacogenet Genomics. 2011;21(11):769–772. doi: 10.1097/FPC.0b013e328346063f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorn CF, et al. Doxorubicin pathways: pharmacodynamics and adverse effects. Pharmacogenet Genomics. 2011;21(7):440–446. doi: 10.1097/FPC.0b013e32833ffb56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thorn CF, et al. PharmGKB summary: fluoropyrimidine pathways. Pharmacogenet Genomics. 2011;21:237–242. doi: 10.1097/FPC.0b013e32833c6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sim SC, Altman RB, Ingelman-Sundberg M. Databases in the area of pharmacogenetics. Hum Mutat. 2011;32:526–531. doi: 10.1002/humu.21454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorn CF, Klein TE, Altman RB. Pharmacogenomics and bioinformatics: PharmGKB. Pharmacogenomics. 2010;11:501–505. doi: 10.2217/pgs.10.15. [DOI] [PMC free article] [PubMed] [Google Scholar]