Summary
Yersinia pestis is an arthropod-borne bacterial pathogen that evolved recently from Yersinia pseudotuberculosis, an enteric pathogen transmitted via the fecal-oral route. This radical ecological transition can be attributed to a few discrete genetic changes from a still-extant recent ancestor, thus providing a tractable case study in pathogen evolution and emergence. Here, we determined the precise genetic and mechanistic basis of the evolutionary adaptation of Y. pestis to flea-borne transmission. Remarkably, only four minor changes in the bacterial progenitor, representing one gene gain and three gene losses, enabled transmission by flea vectors. All three loss-of-function mutations enhanced c-di-GMP-mediated bacterial biofilm formation in the flea foregut that greatly increased transmissibility. Our results suggest a step-wise evolutionary model in which Y. pestis emerged as a flea-borne clone, with each genetic change incrementally reinforcing the transmission cycle. The model conforms well to the ecological theory of adaptive radiation.
Introduction
Yersinia pestis is essentially a clonal variant of Yersinia pseudotuberculosis that emerged as the agent of bubonic plague only 1,500 to 6,400 years ago (Achtman et al., 1999; Chain et al., 2004; Cui et al., 2013). Despite their very close phylogenetic relationship, the two species have radically different life histories. Y. pseudotuberculosis is transmitted perorally in contaminated food and water and causes a usually mild, self-limiting enteric disease in a variety of mammals. Y. pestis, uniquely among the enteric group of gram-negative bacteria, relies on a blood-feeding insect, the flea, for transmission and is highly invasive and virulent. Flea-borne transmission usually results in bubonic plague, in which bacteria deposited in the dermis first disseminate through lymphatic vessels to the draining lymph node, where they multiply before spreading systemically to produce an often fatal septicemia. However, fleas sometimes introduce bacteria directly into the vasculature, resulting in primary septicemic plague (Sebbane et al., 2006). The high-density bacteremia that is the end-stage of plague is required to adequately infect new fleas to maintain the transmission cycle (Lorange et al., 2005).
Y. pestis is transmitted by fleas in two different ways. After taking a blood meal from a bacteremic host, fleas have some potential to transmit the very next time they feed. This early-phase transmission phenomenon resembles mechanical transmission but the mechanism is unknown (Eisen et al., 2006; Hinnebusch, 2012). A second, biological transmission mechanism depends on the ability of Y. pestis to multiply in the flea digestive tract and form a biofilm in the proventriculus, the foregut valve that connects the esophagus and midgut. By 1-2 weeks after the infectious blood meal, the biofilm growth can impede and in some cases completely block the inward flow of blood during feeding, resulting in regurgitation of blood contaminated with bacteria into the flea bite site (Bacot, 1915; Bacot and Martin, 1914; Hinnebusch and Erickson, 2008).
Y. pestis acquired two new plasmids during its divergence from Y. pseudotuberculosis, and both contain genes that contributed to evolution of the flea-borne transmission route. The ymt gene on the ∼100 kb pMT1 plasmid encodes a phospholipase D activity that is required for survival of Y. pestis in the flea midgut (Hinnebusch et al., 2002). The pla gene on the ∼10-kb pPCP1 encodes a cell-surface protease/plasminogen activator which, while not required in the flea, is essential for invasiveness from the flea bite site following transmission (Sodeinde et al., 1992).
In addition to gene gain, gene loss has been important to the evolution of Y. pestis. Relative to Y. pseudotuberculosis, the Y. pestis genome contains an abundance of pseudogenes and transposable genetic elements (Chain et al., 2004; Parkhill et al., 2001). These genomic features are typical of bacterial pathogens in the early stages of transition to obligate parasitism (i.e., a recent evolutionary change from a life cycle involving a free-living, environmental stage to one of strict host association) and are thought to be a consequence of the pronounced genetic drift effect in the greatly reduced population that emerges from an ecological transition to host dependence (Moran and Plague, 2004; Parkhill et al., 2003). Thus, although some cases of gene loss likely were adaptive and positively selected during the evolution of flea-borne transmission, it is a challenge to differentiate these few key changes from the many others that are reflective of negative selection or selectively neutral genetic drift. In fact, although Y. pestis is among the most genetically monomorphic of bacterial taxa, pseudogene profiles vary considerably among strains and these differences have been used for phylogenetic typing (Tong et al., 2005).
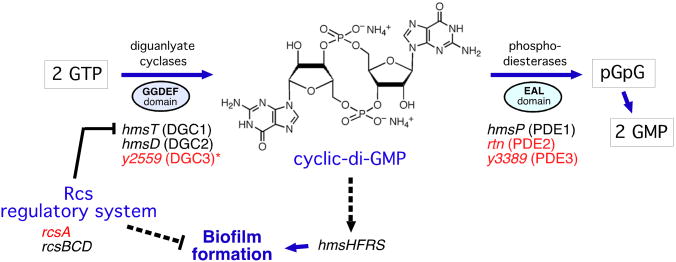
Because the major transmission mechanism of Y. pestis depends on its ability to grow as a biofilm in the proventricular valve in the flea foregut, we concentrated on genes in the two species known to be involved in biofilm development (Figure 1). Y. pseudotuberculosis and Y. pestis both contain a fully-functional hmsHFRS operon that is required to synthesize the extracellular polysaccharide matrix of the biofilm, and Y. pseudotuberculosis is able to form biofilm in some in vitro conditions and on the external cuticle of nematodes (Darby et al., 2002; Erickson et al., 2006; Joshua et al., 2003). Major differences occur, however, in genes that control intracellular levels of cyclic-di-GMP, a bacterial signaling molecule that induces biofilm development (Hennge, 2009). Y. pseudotuberculosis encodes three GGDEF-domain diguanylate cyclases (DGC) that synthesize c-di-GMP and three EAL-domain phosphodiesterases (PDE) that degrade it (Bobrov et al., 2011; Ren et al., 2013; Sun et al., 2011). Notably, two of the three PDE genes (rtn and y3389, hereafter referred to as PDE2 and PDE3) are pseudogenes in Y. pestis; only hmsP (PDE1) is functional (Figure 1). In addition, rcsA, a component of the Rcs gene regulatory system, is a pseudogene in Y. pestis. The Rcs system inhibits biofilm formation, in part by repressing expression of the DGC1 gene hmsT (Sun et al., 2012; Sun et al., 2008).
Figure 1. Comparison of Genes Involved in c-di-GMP Metabolism and Biofilm Formation in Y. pseudotuberculosis and Y. pestis.
Genes indicated in red are pseudogenes in Y. pestis KIM. *The DGC3 gene is functional in most Y. pestis strains. Dashed lines indicate indirect (and undefined) induction or repression mechanisms.
Replacing the Y. pestis rcsA pseudogene with its fully functional Y. pseudotuberculosis homolog results in greatly reduced ability to block fleas, indicating that this gene loss enhanced transmissibility (Sun et al., 2008). However, the converse exchange was not sufficient to enable Y. pseudotuberculosis to block fleas. Our goal in this study was to identify the specific genetic changes required for the transition to the arthropod-borne life style, characterize their phenotypic effects on flea infection and transmissibility, and experimentally recapitulate the evolutionary pathway leading to flea-borne transmission.
Results
The Evolutionary Starting Point
Y. pseudotuberculosis, the progenitor of Y. pestis, causes enteric infection in rodents that usually progresses no farther than the mesenteric lymph nodes. However, bacteremia can develop from high infectious oral doses, or in immunocompromised animals or those with hemochromatosis (serum iron overload) (Carniel et al., 2006). Therefore, because many rodents harbor a permanent ectoparasitic flea fauna that feed on them daily, Y. pseudotuberculosis occasionally would be ingested by fleas feeding on a bacteremic host. To examine this evolutionary starting point, we fed Xenopsylla cheopis rat fleas on blood containing wild-type Y. pseudotuberculosis. The bacteria produce an initial toxic response in fleas immediately after the infectious blood meal (Erickson et al., 2007); but this quickly subsides, and wild-type Y. pseudotuberculosis was able to establish a chronic infection of the flea hindgut (Figure 2B, 3B, C), persisting in modest numbers at that location for at least five weeks after the single infectious blood meal without causing detectible morbidity to the fleas and continuously being shed in flea feces. Thus, Y. pseudotuberculosis causes a benign lower digestive tract infection in fleas (Figure 2B) as well as mammals.
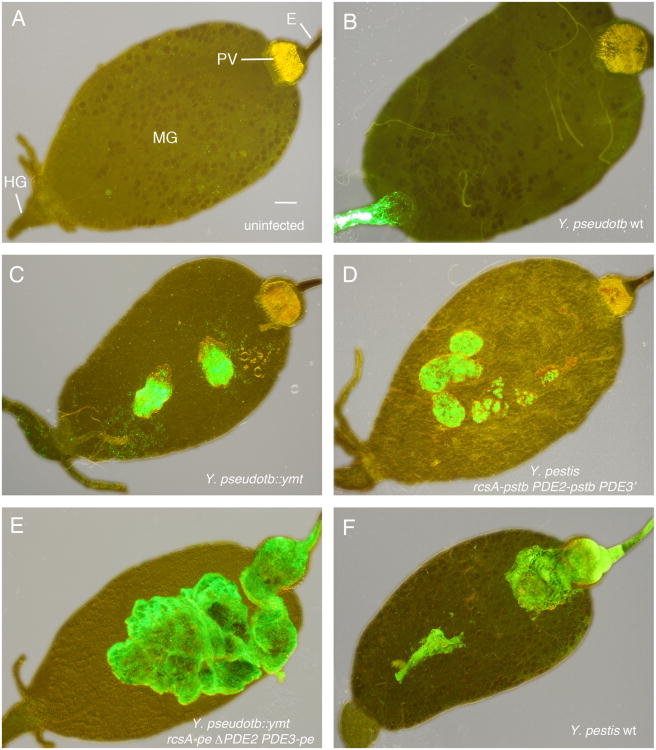
Figure 2. Effect of Four Genetic Changes on the Flea Infection Phenotype of Y. pestis and Y. pseudotuberculosis.
Digestive tracts dissected from an uninfected flea (A) and from fleas 1-3 weeks after infection with the Y. pseudotuberculosis or Y. pestis strains indicated (B-F). All bacteria strains expressed green fluorescent protein and images were taken using combined phase contrast and fluorescent microscopy. Strain descriptions are in Table S1; -pstb indicates the Y. pseudotbuerculosis functional allele and -pe the Y. pestis pseudogene allele. E, esophagus; PV, proventriculus; MG, midgut; HG, hindgut. Scale bar = 0.1 mm.
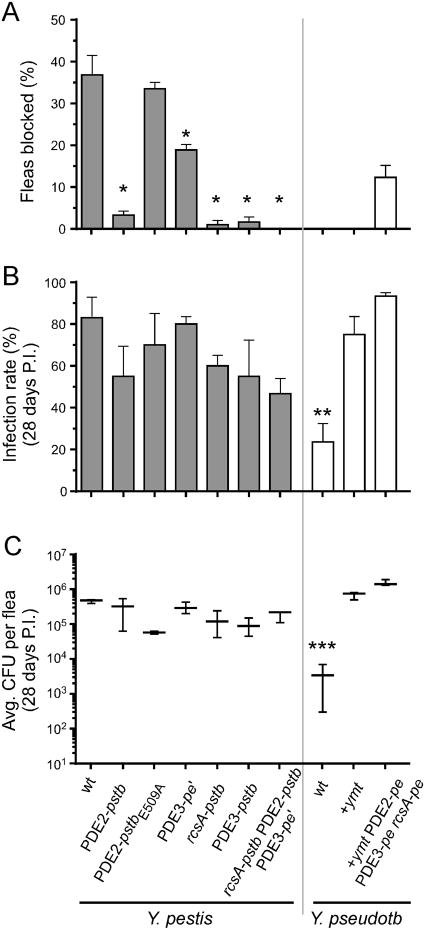
Figure 3. Restoration of Three Y. pestis Pseudogenes Eliminates the Ability to Produce Proventricular Blockage in Fleas.
(A) Percentages of fleas that developed proventricular blockage during the 4-wk period after feeding on blood containing the Y. pestis or Y. pseudotuberculosis strain indicated. (B, C) Percentages of fleas still infected (B) and average bacterial load per infected flea (C) 4 wks after the infectious blood meal. The PDE2-pstbE509A allele has a single nucleotide site-specific mutation in the EAL catalytic domain that eliminates enzyme activity, other allele notations are as in Figure 1 and Table S1. The mean and SEM (A, B) or range (C) of three independent experiments are shown; data for the Y. pestis rcsA-pstb strain are from (Sun et al., 2008). *, P < 0.0001 by Fisher's exact test; **, P = 0.007 and ***, P = 0.003 by Kruskal-Wallis test with Dunn's multiple comparison post-test.
The First Adaptive Step: Gene Gain
Y. pseudotuberculosis infection of the flea digestive tract was confined to the hindgut; the midgut and proventriculus were never colonized. In contrast, Y. pestis colonizes the flea midgut and proventriculus, but never the hindgut (Figure 2F). Survival of Y. pestis in the flea midgut requires a phospholipase D activity encoded by ymt, a gene on a plasmid acquired by Y. pestis after it diverged from Y. pseudotuberculosis (Hinnebusch et al., 2002). As predicted, therefore, transformation of Y. pseudotuberculosis with the ymt gene was sufficient to extend infectivity from the flea hindgut to the midgut (Figure 2C). The ability of Ymt+ Y. pseudotuberculosis to exploit the midgut niche resulted in an increase in the infection rate and bacterial load per flea to levels equal to those achieved by Y. pestis (Figure 3B, C), consistent with previous results (Hinnebusch et al., 2002). This strain remained unable to block fleas (Figure 3A), and in most cases the infection was confined to the midgut and hindgut. In some fleas, however, the proventriculus was also colonized to some extent.
Effect of Three Gene Losses on Flea Infection and Blockage Phenotypes
Because of the known importance of proventricular biofilm formation in the flea to transmissibility, we examined the phenotypic consequences of differences between the two species in genes predicted to be involved in biofilm development (Figure 1). In Y. pestis and other bacteria, biofilm formation is directly correlated with intracellular c-di-GMP concentration. We have shown previously that the Rcs regulatory system negatively regulates biofilm formation in Y. pseudotuberculosis because it represses hmsT, one of the c-di-GMP synthesizing DGC genes (Sun et al., 2012; Sun et al., 2008). The Y. pestis rcsA gene is nonfunctional, and replacing it with the functional Y. pseudotuberculosis homolog results in greatly reduced biofilm formation both in vitro and in the flea (Sun et al., 2012; Sun et al., 2008). In addition, bioinformatics and biochemical analyses have confirmed that two of the three PDE genes that function to degrade c-di-GMP in Y. pseudotuberculosis are pseudogenes in Y. pestis (Bobrov et al., 2011; Sun et al., 2011) (Figure 1, Table 1).
Table 1. Loss of Function Mutations in Y. pestis that Enabled Flea-borne Transmission.
| Gene | Gene IDa | Mutation in Y. pestis | Effect | Occurrenceb |
|---|---|---|---|---|
| PDE2 (rtn) | YPTB1308 y2909 | frameshift (6A→7A) in ORF | N-terminal truncation eliminates signal peptide sequence and phosphodiesterase activity; increased c-di-GMP levels | all Y. pestis |
| PDE3c | YPTB3308 y3389 | a) point mutation (C→T) in promoter region (PDE3-pe' allele) | decreased transcription (cf. Fig. S3B) | some Y. pseudotuberculosis (e.g. YPIII) and Y. pestis Branch 0 strains Pestoides A (0.PE4b) and Microtus 91001 (0.PE4c) |
| b) PDE3-pe' mutation plus point mutation (nonsense) in ORF (PDE3-pe allele) | N-terminal truncation eliminates transmembrane domain and phosphodiesterase activity; increased c-di-GMP levels | all Y. pestis except Branch 0 strains Pestoides A (0.PE4b) and Microtus 91001 (0.PE4c) | ||
| c) deletion | no PDE3 phosphodiesterase activity | Y. pestis Branch 0 strains Pestoides F (0.PE2a) and Angola (0.PE3a) | ||
| rcsA | YPTB2486 y1741 | 30-bp internal duplication in ORF* | 10 amino acid tandem repeat insertion, eliminates RcsAB repression of hmsT diguanylate cyclase gene | all Y. pestis except Branch 0 strain Pestoides A (0.PE4b) |
Gene annotation numbers for Y. pseudotuberculosis IP32953 and Y. pestis KIM homologs
Cf. Figure 5;
See Figure S3A for PDE3 allele details
Two Branch 1 Y. pestis strains, 1.ANTa (Antiqua) and 1.ANTb (UG05), have a transposon (Tn) insertion in rcsA instead of an internal duplication.
Derepression of a synthetic DGC gene due to the rcsA mutation and loss of function of two degradative PDE genes would be predicted to increase and maintain c-di-GMP levels and result in enhanced biofilm development. Thus, we investigated whether these genetic changes were important for the evolution of flea-borne transmission. X. cheopis fleas were infected with Y. pestis allelic exchange strains in which the rcsA, PDE2, and PDE3 pseudogenes had been replaced with their functional Y. pseudotuberculosis equivalents, and conversely, with Y. pseudotuberculosis strains in which the three native genes had been replaced with the non-functional Y. pestis versions (Table S1). Y. pestis blocks the proventriculus of 25-40% of X. cheopis fleas during the four-week period after an infectious blood meal, whereas Y. pseudotuberculosis is completely unable to block fleas, even when transformed with ymt (Figure 3A). As reported previously for rcsA (Sun et al., 2008), individual replacement of the Y. pestis PDE2 and PDE3 pseudogenes with the functional Y. pseudotuberculosis versions resulted in a large decrease in blockage rate, an effect that depended on the PDE catalytic domain EAL (Figure 3A). Y. pestis carrying the intermediate PDE3-pe' allele present in two Pestoides isolates from central Asia (Table 1; Figure S3) also had a significantly reduced blockage rate. Replacement of all three pseudogenes with their ancestral homologs completely eliminated the ability of Y. pestis to produce proventricular-blocking biofilm in the fleas (Figure 2D, 3A). Conversely, replacing these three functional genes in Y. pseudotuberculosis with their Y. pestis pseudogene counterparts and addition of ymt rendered Y. pseudotuberculosis able to colonize and block the proventriculus (Figure 2E, 3A).
The ability to form proventricular-blocking biofilm in the flea strongly correlates with the in vitro pigmentation (Pgm) phenotype, the formation of red-pigmented colonies on Congo red agar, because both depend upon the extracellular polysaccharide matrix synthesized by the hmsHFRS gene products (Jarrett et al., 2004; Lillard et al., 1997). As predicted, transformation of Y. pestis with the functional PDE2 or PDE3 homologs from Y. pseudotuberculosis resulted in decreased intracellular levels of c-di-GMP and greatly attenuated the Pgm phenotype and ability to form biofilm in vitro (Figure S1, S2). These in vitro results are consistent with the negative effect the same gene replacements had on proventricular blockage in the flea (Figure 2, 3), and were also dependent on the phosphodiesterase catalytic domain.
Four Gene Changes Enable Flea-borne Transmission
The ultimate test was to directly determine the effect of these four genetic differences on flea-borne transmission dynamics. After feeding on blood containing isogenic Y. pestis or Y. pseudotuberculosis strains, infected fleas were maintained and fed twice weekly for four weeks. One of these weekly feedings was on sterile blood in an artificial feeding device. After a one-hour feeding period, blood and washings recovered from the feeding device were plated to determine the number of CFUs transmitted. Low numbers of bacteria were transmitted by fleas three days after being infected with Y. pestis, or with the Y. pseudotuberculosis strains containing just ymt or all four specific Y. pestis-related genetic changes (Figure 4). This represents early-phase transmission, which does not require biofilm formation (Vetter et al., 2010). Later, transmission efficiency by the proventricular biofilm-related mechanism was orders of magnitude greater. The Y. pseudotuberculosis with all four changes was transmitted as efficiently as Y. pestis, correlating with the ability of this strain to block fleas (Figure 3A, 4). Fleas never transmitted wild-type Y. pseudotuberculosis, even by the early-phase mechanism, whereas Y. pseudotuberculosis with just the addition of ymt was transmitted both early (day 3) and to a limited extent later as well, up to the final time point 31 days after infection, even though this strain did not block fleas. Presumably this is because the significant proventricular colonization in some fleas (Figure 4) impeded normal blood feeding enough for some regurgitative transmission to occur. Indeed, complete blockage is not essential for transmission; incompletely blocked fleas are also competent vectors (Bacot, 1915). Fleas infected with Y. pestis rcsA-pstb PDE2-pstb PDE3-pe' did not transmit beyond the early phase, in contrast to those infected with Y. pseudotuberculosis::ymt.
Figure 4. Four Genetic Changes Enable Transmissibility by Fleabite.
The number of CFU transmitted by fleas infected with the bacteria indicated was quantified at weekly intervals after a single infectious blood meal. Day 3 represents biofilm-independent early-phase transmission (EPT); later timepoints represent proventricular biofilm-dependent transmission. Numbers in parentheses associated with data points are the number of fleas that fed, and of those, the number that were completely or partially blocked. The maximum extent of proventricular infection achieved by the strains is shown at right.
Genetic Changes that Enabled Flea-borne Transmission Occurred Early During the Emergence of Y. pestis
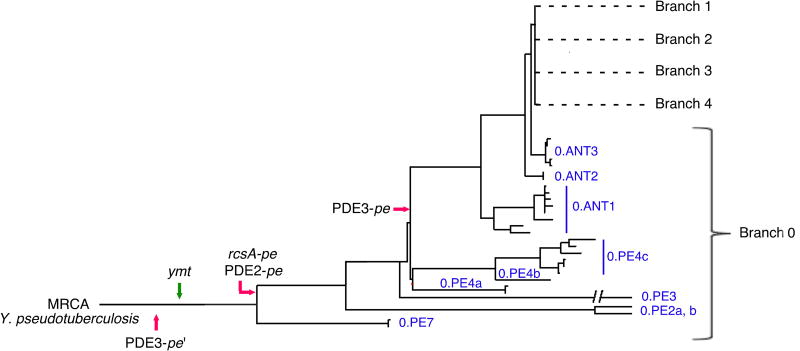
Y. pestis has a clonal population structure with limited genetic diversity. Phylogenetic analyses of 133 strains collected from around the world indicate that Y. pestis emerged in central China < 6,400 years ago (Cui et al., 2013; Harbeck et al., 2013). The genealogy is rooted by the Y. pseudotuberculosis progenitor at the base of Branch 0, which includes the ancestral Y. pestis Pestoides group (0.PE) from central Asia (Cui et al., 2013; Morelli et al., 2010) (Figure 5). Branches 1-4 represent more recent lineages that spread from the Asian homeland to cause historically documented plague pandemics beginning about 1,500 years ago (Cui et al., 2013; Harbeck et al., 2013). The four genetic changes that enabled transmission by fleas preceded or occurred very early in the emergence of Y. pestis (present in the root Branch 0) and have been stably maintained since then (Figure 5). This genealogy indicates that flea-borne transmission has been a component of Y. pestis ecology since it first emerged.
Figure 5. Early Occurrence and Fixation of Genetic Changes that Enabled Flea-borne Transmission During the Emergence of Y. pestis.
The earliest documented appearance in available genome sequences of gain of function (green arrow) and loss-of-function (red arrows) gene changes in the genealogy of Y. pestis is indicated. All changes are present in the Pestoides (0.PE) group in the root Branch 0 that are most closely related to the most recent common ancestor (MRCA), Y. pseudotuberculosis. The four changes in Y. pestis KIM are found in 0.ANT as well as all of the evolutionarily more recent Branch 1 and 2 strains. Genome sequences of the following were available for this analysis: Branch 0 strains 0.PE2a (Pestoides F), 0.PE3a (Angola), 0.PE4b (Pestoides A), 0.PE4c (91001), 0.ANT2a (B42003004) [The 0.PE7 strain contains ymt but the rcsA, PDE2, and PDE3 sequences were not accessible]; Branch 1 strains 1.ANTa (Antiqua), 1.ANT1b (UG05), 1.ANT1b (CP000308), 1.IN3g (E1979001), 1.ORI1 (A1122), 1.ORI3c (IP275), 1.ORI1d (CA88), 1.ORI1e (CO92), 1.ORI1g (FV-1), 1.ORI2u (F1991016), 1.ORI3b (MG05); Branch 2 strains 2.ANT1c (Nepal), 2.MED1a (KIM), 2.MED2c (K1973002), and 9 other unclassified biovar Orientalis strains. The figure and nomenclature are adapted from (Cui et al., 2013).
Discussion
We propose a sequential step-wise model for the evolution of flea-borne transmission in the genus Yersinia (Figure 6) that conforms to the ecological theory of adaptive radiation, the leading expository model for rapid evolutionary divergence and speciation (Schluter, 2000; Simpson, 1953). The model also incorporates epidemiologic theory on the evolutionary emergence of novel pathogens (André and Day, 2005; Antia et al., 2003; Arinaminpathy and McLean, 2009; Sokurenko et al., 2006; Woolhouse et al., 2005). The adaptive radiation theory stresses ecological opportunity, in which a “key innovation” in a progenitor allows its lineage to exploit a previously unoccupied environmental niche. Further adaptation is characterized by fitness tradeoffs, as genetic changes that increase fitness or are neutral in the new environment may be deleterious in the original source environment. Adaptive changes eventually lead to niche specialization, diversifying selection, and the emergence of new species. What is usually lacking in commonly accepted examples of adaptive divergent evolution, however, is knowledge of the specific key genetic changes involved and the mechanisms by which they increased fitness (Elena and Lenski, 2003; Kassen, 2009). In the present case, the availability of the closely related common ancestor, the limited sequence diversity in the recently emerged Y. pestis clade, and the importance of c-di-GMP in regulating biofilm formation, a phenotype critical to the Y. pestis transmission mechanism, enabled us to identify the genetic basis of adaptation to the flea-borne transmission route. We were able to directly demonstrate the fitness consequences in the flea environment of candidate adaptive genetic changes by using isogenic strains with defined mutations, ultimately showing that Y. pestis became transmissible by flea vectors after only four minor genetic changes from its Y. pseudotuberculosis progenitor.
Figure 6. Model of the Evolutionary Route to Flea-borne Transmission of Y. pestis.
A sequence of genetic changes and their incremental effect on transmissibility is diagramed (see text). Black arrows indicate transmission of bacteria between rodent, flea, and the external environment and their weight indicates ecological importance; dashed arrows indicate occasional stochastic transmission. Green arrows, gene gains; red arrows, gene losses. PDE3′ refers to the gene with the promoter region point mutation; PDE3″ to the gene with both the point and stop codon mutations (cf. Table 1 and Figure S3). pMT1 and pPCP1 refer to the two Y. pestis-specific plasmids that encode ymt and pla. The informal Y. pre-pestis nomenclature is adopted from (Carniel, 2003).
To begin, occasional uptake by fleas was not a dead-end sink for Y. pseudotuberculosis because the bacteria colonize the hindgut and are continuously excreted. Thus, fleas may be an occasional reservoir host for Y. pseudotuberculosis, but not a vector (except in the sense that grooming of fur soiled with contaminated flea feces may have provided another peroral infection route). Acquisition of ymt was likely the precipitating key innovation, enabling the Y. pseudotuberculosis recipient to stably colonize a new niche, exploiting the abundant resources and scant competition in the flea midgut to reproduce to large numbers (Figure 2C, 3C). Although primarily shed in feces throughout the flea's lifetime, some vector potential by flea bite existed even at this stage, via early-phase transmission and limited proventricular biofilm mechanisms (Figure 4). Thus, rare stochastic transmission events from flea to rodent would be predicted. Since the LD50 of Y. pseudotuberculosis to mice is <10 bacteria when injected intravenously (Carniel et al., 2006), bacteria transmitted directly into the vasculature by fleas could cause a disease analogous to primary septicemic plague, with the potential for subsequent cycling of bacteria back into naive fleas. This sporadic incidence and long, stuttering transmission chains, while probably insufficient to sustain an enzootic cycle, nevertheless increase the probability of further pathogen evolution and disease emergence (Antia et al., 2003; Arinaminpathy and McLean, 2009).
The next innovations greatly enhanced this rudimentary transmissibility. Sequential loss of rcsA, PDE2 and PDE3 functions would have additively improved transmission efficiency by increasing proventricular biofilm and blockage (Figure 3). Thus, if the R0 (here defined as the average number of secondary cases vectored by fleas from a septicemic rodent) of Y. pseudotuberculosis after the single addition of ymt (R0(1)) was nonzero but less than one, the increased transmissibility after each of the three sequential gene losses would incrementally increase the R0(1) value [R0(1) < R0(2) < R0(3) < R0(4)]. After just these four changes, flea-borne transmissibility was nearly equivalent to that of modern Y. pestis (Figure 3, 4), although disease incidence would be limited by the requirement for intravascular transmission.
Other dominant themes in adaptive radiation theory are the emergence of a specialist descendant from a generalist ancestor and rapid speciation. These are well exemplified by the recent divergence of Y. pestis, now restricted primarily to a rodent-flea parasitic life cycle. Ecological specialization often incurs fitness tradeoffs— genetic changes that permit adaptation to a new environment can be accompanied by loss of fitness in the original environment. Y. pestis Ymt belongs to a widespread phospholipase D superfamily but is most similar to the homolog of Photorhabdus luminescens, an environmental gram-negative bacterium that parasitizes insects and other invertebrates (Rudolph et al., 1999). Gain of ymt probably did not adversely affect the fitness of Y. pseudotuberculosis in its normal environments and may have been advantageous in encounters with soil invertebrates. Loss of the three genes that regulate c-di-GMP probably did have fitness costs, assuming that Y. pseudotuberculosis biofilm-regulatory pathways are adapted to its environmental life stage, and were likely to be selectively maintained only in the nascent Y. pestis lineage as it became less dependent on the original source habitat (the external environment).
In addition to rcsA, PDE2, and PDE3, one of the three DGC genes of Y. pseudotuberculosis (DGC3) is a pseudogene in ∼25% of Y. pestis strains, including the KIM strain used in this study (Figure 1). However, restoration of this gene in either KIM6+ wild-type or PDE3-pe' strains did not affect their flea infection and blockage phenotype or their biofilm-forming ability in vitro (data not shown). Furthermore, strains with and without a functional DGC3 are found in all Y. pestis lineages (Branches 0-4) and pseudogenization resulted from a variety of different mutational events in the strains in which it occurred. These observations suggest that loss of DGC3 was selectively neutral in the flea, and that this gene is more important for the environmental life stage of Y. pseudotuberculosis.
Loss or alteration of rcsA, PDE2, and PDE3 gene function resulted from simple frameshift, insertion, or point mutations that preceded or occurred early in the emergence of Y. pestis (Table 1, Figure 5). Loss of function of the Y. pestis PDE3 gene appears to have occurred in two stages. A transition mutation (C to T) in the promoter region 171 bp upstream of the gene is present in all Y. pestis and some Y. pseudotuberculosis strains; in addition, with the exception of two Branch 0 Pestoides strains, the Y. pestis PDE3 is truncated by a stop codon (Table 1). In the Y. pestis background, the C to T change alone resulted in significantly decreased expression of the gene in vitro (Figure S3). The Y. pestis rcsA, PDE2, and PDE3 mutations would be expected to have a high reversion rate. However, because they increased transmissibility, a potent selective force, the rcsA and PDE mutations were likely subject to strong Darwinian (positive) selection during the initial adaptation to the new route of transmission. As each transmission event represents a bottleneck with small numbers of bacteria regurgitated from the proventriculus, these changes were fixed early in the emergence of Y. pestis and have been selectively maintained ever since (Figure 5). With time, these and other Y. pestis pseudogenes would be expected to undergo further degradation and eventual complete loss (Moran and Plague, 2004). Evidence for this can be seen for the PDE3 gene, in which mutational loss occurred in two stages and which subsequently was deleted entirely in the 0.PE2a and 0.PE3 strains (Table 1, Figure 5). In addition, two Branch 1 Y. pestis strains have a transposon insertion in rcsA instead of the 30-bp internal repeat (Table 1). The Tn disruption likely occurred in the already mutated rcsA, and since tandem duplications have a high reversion rate unless under selective pressure, the original 30-bp repeat mutation was presumably deleted. A functional rcsA allele (rcsA-pstb) is present in the 0.PE4b strain. Since the 0.PE2a and 0.PE3 strains have the rcsA pseudogene allele (rcsA-pe), the 30-bp repeat may also have been subsequently deleted in 0.PE4b.
In this study, we have concentrated on interactions with the flea and the mechanisms of vector-borne transmissibility. All Y. pestis strains, even those closest to the Y. pseudotuberculosis progenitor, contain at minimum ymt, the PDE2 pseudogene, and the PDE3-pe' mutation (Figure 5). Thus, the genealogy suggests that Y. pestis emerged as a flea-borne clone. Some Branch 0 Pestoides strains are associated with latent enzootic disease and limited host range compared to Branch 1-4 strains responsible for plague epidemics (Bearden et al., 2009). Dependence on the flea for transmission would have imposed selective pressure for more invasive clones better able to disseminate from the flea bite site and produce the high threshhold bacteremia level required to infect fleas (Lorange et al., 2005). These further evolutionary changes that enabled bubonic plague progression after fleabite transmission into the dermis would have greatly increased disease incidence and R0 enough to permit enzootic and epizootic plague (Figure 6). Some of these changes that modified pathogenesis in the mammal have been identified, and they also include both gene gain (the cell-surface protease Pla) and gene loss (Bearden et al., 2009; Cui et al., 2013; Derbise et al., 2007; Sodeinde et al., 1992). The Y. pseudotuberculosis strain with the four changes was transmitted by fleas just as well as Y. pestis (Figure 4) but produced complete blockage in fleas at only about half the rate as Y. pestis (Figure 3A), so other minor differences that affect the phenotype in the flea also remain to be identified. For example, Y. pestis is no longer able to colonize the flea hindgut (which would be a dead-end sink for Y. pestis now), presumably due to loss of an adhesin still present in Y. pseudotuberculosis.
The adaptive evolutionary pathway that led to flea-borne transmission was based on only a few minor genetic changes that progressively expanded colonization of the flea digestive tract from the hindgut to the midgut and then to the foregut in the form of proventricular biofilm, setting up direct transmission without the necessity of an environmental life stage. Three of these changes affected c-di-GMP signaling pathways, extending the pre-existing biofilm-forming capability of Y. pseudotuberculosis to the flea environment. Such regulatory mutations that alter the expression of existing biosynthetic pathways are a common feature of adaptive radiation and ecological specialization in bacteria because they can have profound phenotypic effects (MacLean, 2005). In one of the first laboratory experiments of bacterial evolution, Pseudomonas fluorescens radiated rapidly into different morphotypes able to colonize new niches in a spatially heterogeneous environment (Rainey and Travisano, 1998). One pellicle-forming niche specialist resulted from mutations in c-di-GMP metabolic pathways that enhanced biofilm formation (McDonald et al., 2009), exactly analogous to Y. pestis evolution that enabled biofilm-dependent colonization of the flea foregut. In another related example, the ability of Vibrio fischeri to colonize its invertebrate host depended on gene gain that upregulated biofilm development (Mandel et al., 2009). The evolutionary history of Y. pestis provides an example of how just a few genetic changes can alter infection phenotypes, in this case leading to arthropod-borne transmission and the emergence of plague; and how gene loss can be as critical to the evolutionary process as gene acquisition (Bliven and Maurelli, 2012).
Experimental Procedures
Bacteria and Mutagenesis
Bacterial strains and their genotypes are listed in Table S1. The Y. pestis KIM6+ parent strain is referred to as wild-type (relative to the isogenic mutant strains) but lacks the 70-kb pCD1 virulence plasmid, which does not affect the flea infection phenotype (Hinnebusch et al., 1996). All gene replacements and chromosomal insertion of ymt were performed by two-step allelic exchange (Donnenberg and Kaper, 1991; Sun et al., 2008; Sun et al., 2011). Deletion of the Y. pseudotuberculosis PDE2 (rtn) gene and insertion of a chloramphenicol resistance gene in its place was performed using lambda Red recombinase mutagenesis (Datsenko and Wanner, 2000; Sun et al., 2008). Strains overexpressing PDE alleles were made by first cloning a wild-type copy of each gene and its native promoter, PCR-amplified from either Y. pestis or Y. pseudotuberculosis using specific primers listed in Table S2, into the plasmid vectors pUC18 or pLG339. Recombinant plasmids were then transformed into Y. pestis by electroporation. Mutated versions of the PDE genes in which the EAL- or GGDEF-encoding domains were replaced by noncatalytic AAL- or GGAAF-encoding domains were made by site-specific mutagenesis after cloning (Sun et al., 2011). To construct the PDE3-peR allele, the promoter region and N-terminus of PDE3-pstb was ligated to a C-terminal fragment of PDE3-pe at a conserved HindIII site just downstream of the PDE3-pe stop codon. The PDE3-peR allele was then introduced into Y. pestis by allelic exchange. Genotypes of all strains were verified by nucleotide sequencing.
Flea Infections
Xenopsylla cheopis rat fleas were infected by allowing them to feed on blood containing ∼5 × 108 bacteria/ml in an artificial feeding device and examined for four weeks as described (Derbise et al., 2010; Hinnebusch et al., 1996; Rebeil et al., 2013; Sun et al., 2011). Fleas (∼50 males and 50 females) that took an infectious blood meal were maintained by allowing them to feed on uninfected mice on days 2, 6, 9, 13, 16, 20, 23, and 27. Immediately after each of these feedings, all fleas were individually examined under a dissecting microscope to determine how many had taken a normal blood meal (indicated by the presence of fresh red blood filling the midgut) and how many were blocked (indicated by the presence of fresh red blood only in the esophagus, anterior to the proventriculus). The infection rate and average bacterial load per flea at 1 hour and 28 days after infection were determined by CFU counts from additional samples of 20 fleas that were individually triturated and plated on BHI agar (Derbise et al., 2010; Hinnebusch et al., 1996; Rebeil et al., 2013; Sun et al., 2011). Three independent flea infection experiments were performed for each strain. Flea blockage data were analyzed by Fisher's exact test; flea infection rate and bacterial load data by Kruskal-Wallis test with Dunn's multiple comparison post-test. All experiments involving animals were approved by the Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, National Institutes of Health Animal Care and Use Committee and were conducted in accordance with all National Institutes of Health guidelines.
Flea-borne Transmission
For transmission experiments, fleas were infected as described above. At different times after infection, ∼200 fleas were allowed to feed on sterile blood for 90 min. Blood was removed, the feeder was washed with PBS, and the external surface of the feeding membrane was disinfected with ethanol. Blood, pooled washes, and the triturated membrane were spread onto blood agar plates to determine the colony-forming units (CFU) transmitted by the fleas. Fleas were examined microscopically as described above to determine how many fed, and of those, how many were blocked or partially blocked. The infection rate and average bacterial load was determined from a sample of 20 fleas that fed. Day 3 was the first feeding opportunity after the infectious blood meal and represents early-phase transmission. Fleas were allowed to feed twice-weekly thereafter— once on an uninfected mouse and once for the weekly transmission test three days later— for one month.
Supplementary Material
Highlights.
Y. pseudotuberculosis causes only a benign lower digestive tract infection in fleas
Gain of ymt encoding a phospholipase D activity allows flea mid gut infection
Loss of three functional genes enhances biofilm formation in the flea to facilitate transmissibility
Four gene changes occurring early in Y. pestis evolution enabled flea-borne transmission
Acknowledgments
This work was supported by the Division of Intramural Research, NIAID, NIH and grant IRT13007 from the Program for Changjiang Scholars and Innovative Research Team in University to Y-C. S. We thank J. Callison for molecular biology assistance; K. Lawrence for help with HPLC, A. Athman for help with preparing figures; and T. Schwan, M. Gottesman, J. Shannon, J. Spinner and I. Chouikha for review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- André JB, Day T. The effect of disease life history on the evolutionary emergence of novel pathogens. Proc Royal Soc B. 2005;272:1949–1956. doi: 10.1098/rspb.2005.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antia R, Regoes RR, Koella JC, Bergstrom CT. The role of evolution in the emergence of infectious diseases. Nature. 2003;426:658–661. doi: 10.1038/nature02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arinaminpathy N, McLean AR. Evolution and emergence of novel human infections. Proc Royal Soc B. 2009;276:3937–3943. doi: 10.1098/rspb.2009.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacot AW. Further notes on the mechanism of the transmission of plague by fleas. J Hyg. 1915;14:774–776. [PMC free article] [PubMed] [Google Scholar]
- Bacot AW, Martin CJ. Observations on the mechanism of the transmission of plague by fleas. J Hyg. 1914;13:423–439. [PMC free article] [PubMed] [Google Scholar]
- Bearden SW, Sexton C, Pare J, Fowler JM, Arvidson CG, Yerman L, Viola RE, Brubaker RR. Attenuated enzootic (pestoides) isolates of Yersinia pestis express active aspartase. Microbiology. 2009;155:198–209. doi: 10.1099/mic.0.021170-0. [DOI] [PubMed] [Google Scholar]
- Bliven KA, Maurelli AT. Antivirulence genes: insights into pathogen evolution through gene loss. Infect Immun. 2012;80:4061–4070. doi: 10.1128/IAI.00740-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrov AG, Kirillina O, Ryjenkov DA, Waters CM, Price PA, Fetherston JD, Mack D, Goldman WE, Gomelsky M, Perry RD. Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol Microbiol. 2011;79:533–551. doi: 10.1111/j.1365-2958.2010.07470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carniel E. Evolution of pathogenic Yersinia, some lights in the dark. Adv Exp Med Biol. 2003;529:3–12. doi: 10.1007/0-306-48416-1_1. [DOI] [PubMed] [Google Scholar]
- Carniel E, Autenrieth I, Cornelis G, Fukushima H, Guinet F, Isberg R, Pham J, Prentice M, Simonet M, Skurnik M, et al. Yersinia enterocolitica and Yersinia pseudotuberculosis. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes. New York: Springer; 2006. pp. 270–398. [Google Scholar]
- Chain PSG, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, Georgescu AM, Vergez LM, Land ML, Motin VL, et al. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 2004;101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Yu C, Yan Y, Li D, Li Y, Jombart T, Weinert LA, Wang Z, Guo Z, Xu L, et al. Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc Natl Acad Sci USA. 2013;110:577–582. doi: 10.1073/pnas.1205750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby C, Hsu JW, Ghori N, Falkow S. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature. 2002;417:243–244. doi: 10.1038/417243a. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbise A, Chenal-Francisque V, Huon C, Fayolle C, Demeure CE, Chane-Woon-Ming B, Medigue C, Hinnebusch BJ, Carniel E. Delineation and analysis of chromosomal regions specifying Yersinia pestis. Infect Immun. 2010;78:3930–3941. doi: 10.1128/IAI.00281-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbise A, Chenal-Francisque V, Pouillot F, Fayolle C, Prevost MC, Medigue C, Hinnebusch BJ, Carniel E. A horizontally acquired filamentous phage contributes to the pathogenicity of the plague bacillus. Mol Microbiol. 2007;63:1145–1157. doi: 10.1111/j.1365-2958.2006.05570.x. [DOI] [PubMed] [Google Scholar]
- Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen RJ, Bearden SW, Wilder AP, Montenieri JA, Antolin MF, Gage KL. Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc Natl Acad Sci USA. 2006;103:15380–15385. doi: 10.1073/pnas.0606831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena SF, Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nature Rev Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- Erickson DL, Jarrett CO, Wren BW, Hinnebusch BJ. Serotype differences and lack of biofilm formation characterize Yersinia pseudotuberculosis infection of the Xenopsylla cheopis flea vector of Yersinia pestis. J Bacteriol. 2006;188:1113–1119. doi: 10.1128/JB.188.3.1113-1119.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson DL, Waterfield NR, Vadyvaloo V, Long D, Fischer ER, ffrench-Constant RH, Hinnebusch BJ. Acute oral toxicity of Yersinia pseudotuberculosis to fleas: implications for the evolution of vector-borne transmission of plague. Cell Microbiol. 2007;9:2658–2666. doi: 10.1111/j.1462-5822.2007.00986.x. [DOI] [PubMed] [Google Scholar]
- Harbeck M, Seifert L, Hansch S, Wagner DM, Birdsell D, Parise KL, Wiechmann I, Grupe G, Thomas A, Keim P, et al. Yersinia pestis DNA from skeletal remains from the 6(th) century AD reveals insights into Justinianic Plague. Plos Pathog. 2013;9:e1003349. doi: 10.1371/journal.ppat.1003349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennge R. Principles of c-di-GMP signalling in bacteria. Nature Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- Hinnebusch BJ. Biofilm-dependent and biofilm-independent mechanisms of transmission of Yersinia pestis by fleas. Adv Exp Med Biol. 2012;954:237–243. doi: 10.1007/978-1-4614-3561-7_30. [DOI] [PubMed] [Google Scholar]
- Hinnebusch BJ, Erickson DL. Yersinia pestis biofilm in the flea vector and its role in the transmission of plague. Curr Top Microbiol Immunol. 2008;322:229–248. doi: 10.1007/978-3-540-75418-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch BJ, Perry RD, Schwan TG. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science. 1996;273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- Hinnebusch BJ, Rudolph AE, Cherepanov P, Dixon JE, Schwan TG, Forsberg Å. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science. 2002;296:733–735. doi: 10.1126/science.1069972. [DOI] [PubMed] [Google Scholar]
- Jarrett CO, Deak E, Isherwood KE, Oyston PC, Fischer ER, Whitney AR, Kobayashi SD, DeLeo FR, Hinnebusch BJ. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J Inf Dis. 2004;190:783–792. doi: 10.1086/422695. [DOI] [PubMed] [Google Scholar]
- Joshua GWP, Karlyshev AV, Smith MP, Isherwood KE, Titball RW, Wren BW. A Caenorhabditis elegans model of Yersinia infection: biofilm formation on a biotic surface. Microbiology. 2003;149:3221–3229. doi: 10.1099/mic.0.26475-0. [DOI] [PubMed] [Google Scholar]
- Kassen R. Toward a general theory of adaptive radiation. Insights from microbial experimental evolution. Ann NY Acad Sci. 2009;1168:3–22. doi: 10.1111/j.1749-6632.2009.04574.x. [DOI] [PubMed] [Google Scholar]
- Lillard JW, Fetherston JD, Pedersen L, Pendrak ML, Perry RD. Sequence and genetic analysis of the hemin storage (hms) system of Yersinia pestis. Gene. 1997;193:13–21. doi: 10.1016/s0378-1119(97)00071-1. [DOI] [PubMed] [Google Scholar]
- Lorange EA, Race BL, Sebbane F, Hinnebusch BJ. Poor vector competence of fleas and the evolution of hypervirulence in Yersinia pestis. J Inf Dis. 2005;191:1907–1912. doi: 10.1086/429931. [DOI] [PubMed] [Google Scholar]
- MacLean RC. Adaptive radiation in microbial microcosms. J Evol Biol. 2005;18:1376–1386. doi: 10.1111/j.1420-9101.2005.00931.x. [DOI] [PubMed] [Google Scholar]
- Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. A single regulatory gene is sufficient to alter bacterial host range. Nature. 2009;458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MJ, Gehrig SM, Meintjes PL, Zhang XX, Rainey PB. Adaptive divergence in experimental populations of Pseudomonas fluorescens. IV. Genetic constraints guide evolutionary trajectories in a parallel adaptive radiation. Genetics. 2009;183:1041–1053. doi: 10.1534/genetics.109.107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Plague GR. Genomic changes following host restriction in bacteria. Curr Opin Gen Dev. 2004;14:627–633. doi: 10.1016/j.gde.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, Wagner DM, Feldkamp M, Kusecek B, Vogler AJ, Li Y, et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nature Gen. 2010;42:1140–1143. doi: 10.1038/ng.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Holden MT, Churcher CM, Bentley SD, Mungall KL, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nature Gen. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MTG, Prentice MB, Sebhaihia M, James KD, Churcher C, Mungall KL, et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- Rebeil R, Jarrett CO, Driver JD, Ernst RK, Oyston PC, Hinnebusch BJ. Induction of the Yersinia pestis PhoP-PhoQ regulatory system in the flea and its role in producing a transmissible infection. J Bacteriol. 2013;195:1920–1930. doi: 10.1128/JB.02000-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren GX, Yan HQ, Zhu H, Guo XP, Sun YC. HmsC, a periplasmic protein, controls biofilm formation via repression of HmsD, a diguanylate cyclase in Yersinia pestis. Environ Microbiol. 2013 Nov 5; doi: 10.1111/1462-2920.12323. [DOI] [PubMed] [Google Scholar]
- Rudolph AE, Stuckey JA, Zhao Y, Matthews HR, Patton WA, Moss J, Dixon JE. Expression, characterization, and mutagenesis of the Yersinia pestis murine toxin, a phospholipase D superfamily member. J Biol Chem. 1999;274:11824–11831. doi: 10.1074/jbc.274.17.11824. [DOI] [PubMed] [Google Scholar]
- Schluter D. The Ecology of Adaptive Radiation. Oxford: Oxford University Press; 2000. [Google Scholar]
- Sebbane F, Jarrett CO, Gardner D, Long D, Hinnebusch BJ. Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc Natl Acad Sci USA. 2006;103:5526–5530. doi: 10.1073/pnas.0509544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG. The Major Features of Evolution. New York: Columbia University Press; 1953. [Google Scholar]
- Sodeinde OA, Subrahmanyam YV, Stark K, Quan T, Bao Y, Goguen JD. A surface protease and the invasive character of plague. Science. 1992;258:1004–1007. doi: 10.1126/science.1439793. [DOI] [PubMed] [Google Scholar]
- Sokurenko EV, Gomulkiewicz R, Dykhuizen DE. Source-sink dynamics of virulence evolution. Nature Rev Microbiol. 2006;4:548–555. doi: 10.1038/nrmicro1446. [DOI] [PubMed] [Google Scholar]
- Sun YC, Guo XP, Hinnebusch BJ, Darby C. The Yersinia pestis Rcs phosphorelay inhibits biofilm formation by repressing transcription of the diguanylate cyclase gene hmsT. J Bacteriol. 2012;194:2020–2026. doi: 10.1128/JB.06243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YC, Hinnebusch BJ, Darby C. Experimental evidence for negative selection in the evolution of a Yersinia pestis pseudogene. Proc Natl Acad Sci USA. 2008;105:8097–8101. doi: 10.1073/pnas.0803525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YC, Koumoutsi A, Jarrett C, Lawrence K, Gherardini FC, Darby C, Hinnebusch BJ. Differential control of Yersinia pestis biofilm formation in vitro and in the flea vector by two c-di-GMP diguanylate cyclases. PLoS One. 2011;6:e19267. doi: 10.1371/journal.pone.0019267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z, Zhou D, Song Y, Zhang L, Pei D, Han Y, Pang X, Li M, Cui B, Wang J, et al. Pseudogene accumulation might promote the adaptive microevolution of Yersinia pestis. J Med Microbiol. 2005;54:259–268. doi: 10.1099/jmm.0.45752-0. [DOI] [PubMed] [Google Scholar]
- Vetter SM, Eisen RJ, Schotthoefer AM, Montenieri JA, Holmes JL, Bobrov AG, Bearden SW, Perry RD, Gage KL. Biofilm formation is not required for early-phase transmission of Yersinia pestis. Microbiology. 2010;156:2216–2225. doi: 10.1099/mic.0.037952-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse MEJ, Haydon DT, Antia R. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol Evol. 2005;20:238–244. doi: 10.1016/j.tree.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.