Abstract
Sequential polymerization of N-carboxyanhydrides accelerated by nitrogen flow is utilized to generate a novel well-defined diblock copolypeptide (PDI = 1.08), with incorporation of alkyne-functionalized side-chain groups allowing for rapid and efficient thiol-yne click-type modifications, followed by self-assembly into nanopure water to construct a helical polypeptide-based versatile and functional nanoparticle platform.
Polymeric nanoparticles with unique physical and chemical properties at the nanoscale have shown great promise for various biomedical applications by acting as a carrier to package and deliver a wide range of therapeutic, diagnostic and theranostic agents to the desired sites of action.1 In particular, micellar nanoparticles with distinctive core-shell structures have attracted considerable attention due to their tunable sizes and potential modifications in both the core and shell domains by using well-defined and functionalized amphiphilic block copolymers.2 The development of micellar nanoparticles from non-degradable polymers have been well studied and widely used in nanomedicine, while the undesired side effects such as toxicity and immunogenicity have become an inevitable challenge.3 In order to overcome the drawbacks of non-degradable polymers, biodegradable synthetic polymers, such as polyesters,4 polycarbonates,5 polyphosphoesters,6 and polypeptides,7 have been explored as attractive materials to be utilized in biomedical applications due to their intrinsic degradability and low toxicity.8 Compared with other biodegradable synthetic polymers, polypeptides offer additional properties, such as precisely-defined secondary structures derived from chirality and various functionalities introduced by natural and synthetic amino acids that lead to supramolecular hierarchical assemblies, which are often unobtainable from non-polypeptide-based materials.9
Polypeptides consisting of α-amino acids with covalent linkages share significant similarities to natural proteins, and recent synthetic methodology developments have facilitated their extensive utilization in biomedical areas, such as serving as scaffolds for tissue engineering, matrices for drug and gene delivery, and responsive materials for biosensors.10 Amphiphilic block copolypeptides, built from polypeptide blocks with different hydrophilicities, have the capability to self-assemble into well-defined nanostructures in aqueous solutions, including micelles, vesicles and helical cylinders, which can serve as versatile material platforms.11 The most common approach to synthesize block copolypeptides involves sequential ring-opening polymerizations (ROPs) of different amino acid N-carboxyanhydride (NCA) monomers.12 In the last decade, several controlled NCA ROPs have been reported to prepare well-defined block copolypeptides by reducing side reactions that decrease the activity of the terminal amine during polymerizations.13 However, it is still challenging to develop well-defined block copolypeptides with side-chain functionalities via a facile and easily operational strategy.14 Due to the nucleophile- and base-sensitive nature of NCA ROP, the monomers used in the polymerizations are limited to NCAs with inert side-chain groups or NCAs with modified side-chains by suitable protecting groups. The selective protection and deprotection operations in both monomer synthesis and post-polymerization modification processes increase the difficulty to efficiently prepare well-defined functional block copolypeptides.15 Recently, Hammond et al. reported an alkyne-containing NCA monomer, γ-propargyl-l-glutamate (PLG) NCA, by incorporation of an alkynyl functional group onto the side-chain of l-glutamic acid, and performed NCA ROPs to obtain functional homopolypeptides.16 The efficient chemical modifications of alkyne-functionalized polypeptides via either copper-catalyzed azide-alkyne Huisgen 1,3-dipolar cycloaddition or radical-mediated thiol-yne click-type chemistry have advantages including rapid reaction, quantitative conversion and high functional group tolerance with minimal by-products and side reactions.16–17 Until now, well-defined diblock copolypeptides containing a polyPLG segment and other polypeptide blocks have not been reported. It was hypothesized that difficulties with the synthesis and purification of the PLG NCA monomer as well as the long polymerization time during NCA ROP (generally 3 days) may have hampered the efficient preparation of well-defined block copolypeptides, and further prevented the construction of functional polypeptide-based nanomaterials.
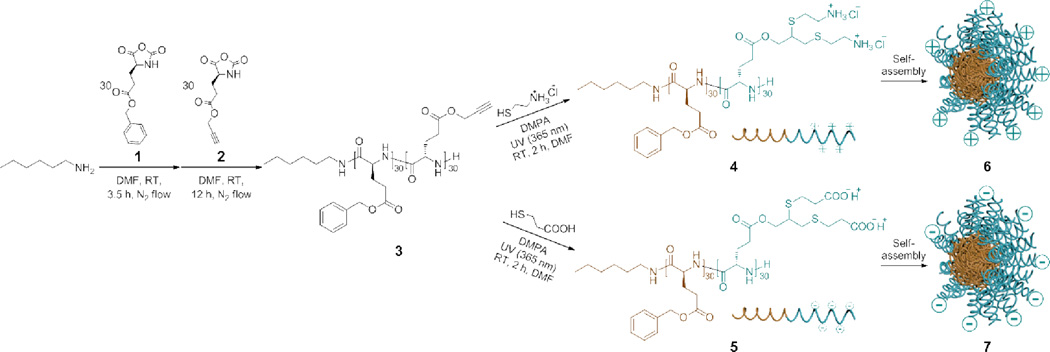
We had anticipated that our facile and easily operational synthetic strategy to construct well-defined polypeptides by conducting NCA ROP under normal Schlenk techniques, with the rate of polymerization being controlled by a straightforward nitrogen flow method,18 could be utilized to prepare functional multi-block polypeptides. Diblock copolymers with incorporation of a copolypeptide segment as one block and the other block being poly(ethylene glycol) (PEG) by statistical ring-opening copolymerization of different NCA monomers from a PEG macroinitiator had been prepared via this strategy,19 and recently, the approach was shown to allow for the preparation of a series of di- and triblock polymers comprised of PEG and peptide segments, designed as multi-anchor systems for the functionalization of gold with PEG.20 Herein, a functional and helical diblock copolypeptide, poly(γ-benzyl-l-glutamate)-block-poly(γ-propargyl-l-glutamate) (PBLG-b-PPLG 3), was prepared by this nitrogen flow method within 16 h by one-pot sequential ROPs of two NCA monomers, BLG 1 and PLG 2 NCAs. Subsequent modification of the diblock copolypeptide was conducted by a thiol-yne click-type reaction to attach charged and functional side-chain moieties onto the PPLG segment. The obtained functionalized diblock copolypeptides were observed to assemble into micellar nanoparticles in aqueous solution with corresponding surface charges and expected helical conformations derived from the secondary sub-structure of the polypeptides.
The diblock copolypeptide, PBLG-b-PPLG 3, was prepared by sequentially adding 1 and 2 into a solution of n-hexylamine in anhydrous N,N-dimethylformamide (DMF) (Scheme 1), which provided a facile and straightforward approach to construct well-defined diblock copolypeptides. This strategy extends from our previous study, in which the propagating chain-ends of the polypeptides in the nitrogen flow method had the capability to undergo chain extension by sequentially introducing additional aliquots of NCA monomers into the reaction mixture.18 The required amount of 1 in anhydrous DMF was added into n-hexylamine, the initiator, with the monomer : initiator = 30 : 1 and allowed to proceed at room temperature, using standard Schlenk techniques. An aliquot of 2 in anhydrous DMF with the monomer : initiator = 30 : 1 was introduced into the reaction mixture via syringe after 3.5 h, when the monomer conversion of 1 had reached 99%, as determined by attenuated total reflectance-Fourier transform infrared spectroscopy (ATR-FTIR) using the intensity of NCA anhydride absorption at 1788 cm−1. The conversion of 2 was greater than 99% after another 12 h, and the reaction mixture was then precipitated into diethyl ether to isolate PBLG30-b-PPLG30 3 as a white solid product. Due to the accelerated polymerization rate of the nitrogen flow method, 3 was prepared in a rapid (16 h) and atom-efficient manner (each monomer conversion > 99%, yield > 85%) with a well-defined structure, as confirmed by the observed GPC analyses of a peak shift from 24.9 to 22.8 min elution time from the first to second block growth profiles and a narrow molecular weight distribution (PDI = 1.08) for 3 (Fig. S1 in ESI†). It is worth mentioning that the polymerizations under continuous nitrogen flow did not require solvent purification and the PLG monomer could be purified simply by repeated precipitations, which reduced the time and effort required for scale-up to gram-scale polymerizations.
Scheme 1.
Synthetic route of diblock copolypeptide PBLG30-b-PPLG30 3 via one-pot sequential ROPs of BLG 1 and PLG 2 NCA monomers, followed by post-polymerization modifications to prepare positively-charged diblock copolypeptide 4 or negatively-charged diblock copolypeptide 5 via radical-mediated thiol-yne click-type chemistry with corresponding 2-aminoethanethiol hydrochloride or 3-mercaptopropionic acid, and schematic representation of the further self-assembly of 4 and 5 into micellar cationic 6 and anionic 7 nanoparticles, respectively.
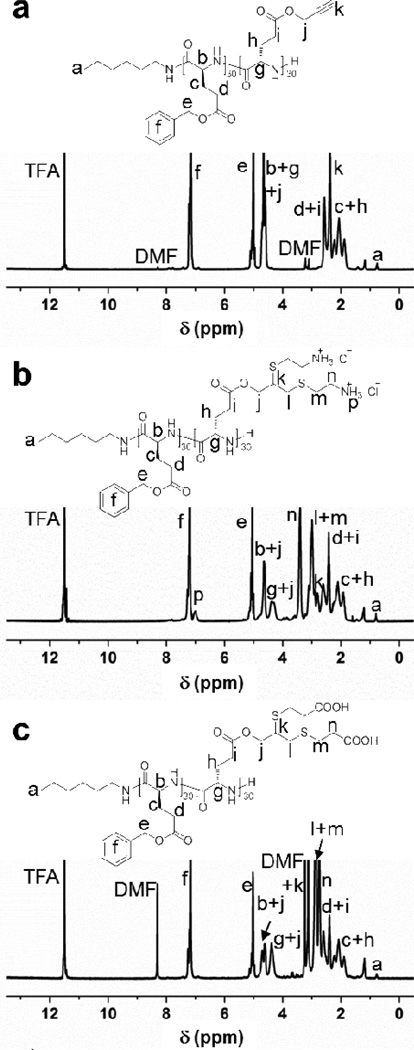
The radical-mediated thiol-yne click-type reaction is a robust and versatile approach that tolerates a variety of functional groups, such as carboxylic acid or amino groups, to rapidly and efficiently functionalize the alkynyl groups without the use of a metal catalyst.21 In order to graft charged side-chain moieties onto the backbones of copolypeptides in the construction of amphiphilic block copolypeptides, the thiol-yne click chemistry was applied by coupling two thiol reagents with one alkynyl group of each repeat unit within the PPLG block to achieve a double addition product with 1,2-regioselectivity. With the aim of avoiding chain-chain coupling or crosslinking, as well as ensuring high efficiency, a total amount of 20 equivalents of thiol reagents relative to the alkynyl groups were used in the thiol-yne click reaction under UV irradiation with 2,2-dimethoxy-2-phenylacetophenone (DMPA) as the photoinitiator. The commercially-available 2-aminoethanethiol hydrochloride and 3-mercaptopropionic acid were utilized to synthesize the corresponding positively-charged 4 and negatively-charged 5 diblock copolypeptides, followed by their self-assembly into cationic 6 and anionic 7 micellar nanoparticles, respectively (Scheme 1). In addition, the linkers between the side-chain charged groups along the polypeptide backbones (10 and 11 σ-bonds) were expected to minimize the effect of side-chain repulsion and maintain the helical conformation of 4 and 5.22 The obtained 1,2-dithioester products were characterized by 1H NMR spectroscopy by using deuterated trifluoroacetic acid (TFA-D) as the solvent, which was capable of breaking the strong hydrogen-bonding between the polypeptide segments and maintaining the diblock copolypeptides in the solution state (Fig. 1). The diastereotopic splitting of the methylene protons (j in both Fig. 1b and 1c), corresponding to the 1,2-regioselectivity from the thiol-yne reaction, together with the observation of other functional groups demonstrated the successful installation of the charged and functionalized thiol reagents onto the backbone of 3.
Fig. 1.
1H NMR spectra of diblock copolypeptide 3 (a), charged diblock copolypeptides 4 (b) and 5 (c) after thiol-yne modifications.
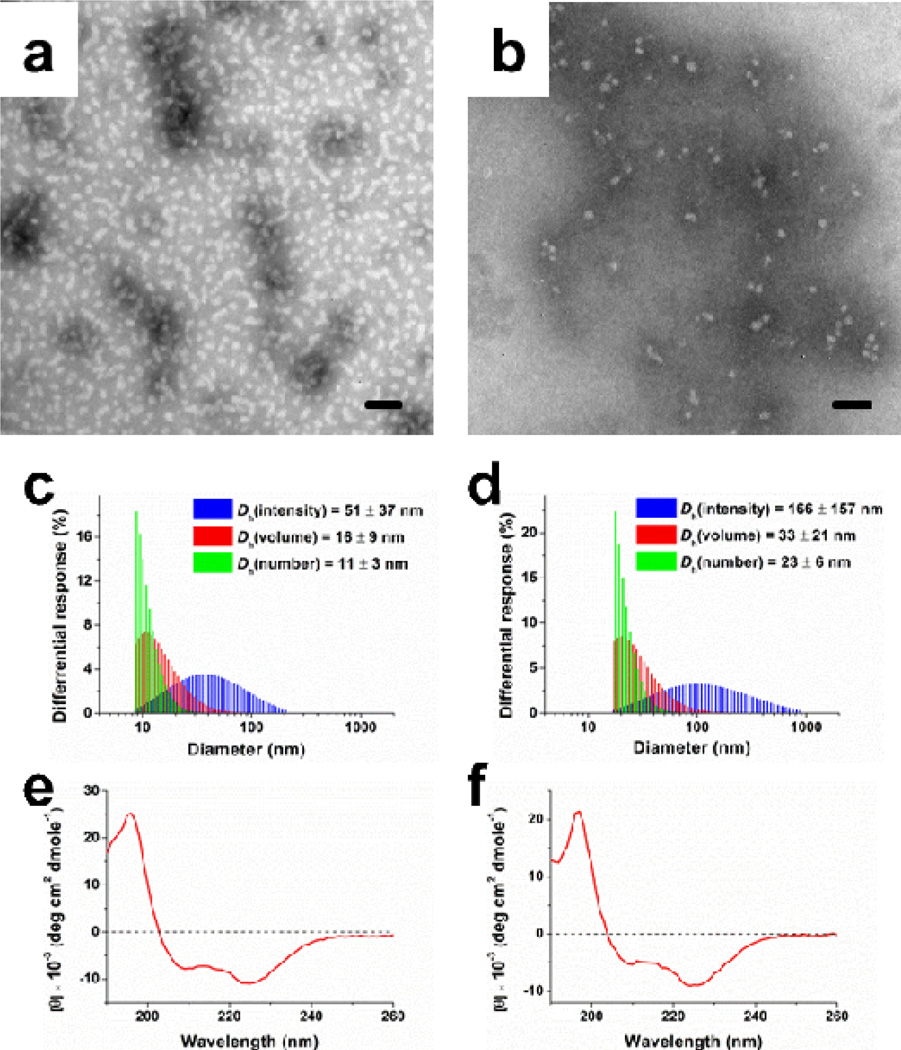
The self-assembly behaviors of the two amphiphilic diblock copolypeptides were evaluated by either direct re-suspension of lyophilized polymer 4 into nanopure water or a nanoprecipitation method by adding the organic solution of polymer 5 into nanopure water. The positively-charged diblock copolypeptide 4 was directly suspended into nanopure water at a concentration of 1 mg/mL, followed by sonication for 10 min at room temperature, resulting in spontaneous formation of cationic nanoparticles 6. However, the negatively-charged diblock copolypeptide 5 was unable to be directly suspended into water, rather, the corresponding anionic nanoparticles 7 were prepared by adding a DMF solution of the polymer into nanopure water, followed by further dialysis against nanopure water. The morphologies of the resulting nanoparticles were characterized by transmission electron microscopy (TEM) and dynamic light scattering (DLS). DLS analyses revealed that 4 underwent assembly with high uniformity of the resulting nanoparticles, giving a number-averaged hydrodynamic diameter of 11 ± 3 nm for 6 (Fig. 2c), whereas the assemblies from 5 were larger and of broader size distribution, 23 ± 6 nm for 7 (Fig. 2d). The monomodal distributions in DLS for both 6 and 7 suggested that the nanoparticle assemblies were well-defined, which were also observed in TEM images (Fig. 2a and 2b, respectively). However, TEM imaging reveals that the morphologies are not spherical, but more likely of a globular nature. Moreover, upon storage, elongation of the nanoassembly morphologies occurred to produce rod-like or cylindrically-shaped nanostructures. These unique long-term assembly processes are under further evaluation. The surface charge densities were characterized via zeta potential analyses in nanopure water at pH 5.6. Zeta potential values of 28 ± 2 mV (6) and −62 ± 1 mV (7) indicated the cationic and anionic surface characteristics of the self-assembled nanoparticles (Fig. S2 in ESI†).
Fig. 2.
TEM images for cationic 6 (a) and anionic 7 (b) nanoparticles. The scale bars in both TEM images are 50 nm. (c) DLS results of cationic nanoparticles 6: Dh (intensity) = 51 ± 37 nm, Dh (volume) = 16 ± 9 nm and Dh (number) = 11 ± 3 nm. (d) DLS results of anionic nanoparticles 7: Dh (intensity) = 166 ± 157 nm, Dh (volume) = 33 ± 21 nm and Dh (number) = 23 ± 6 nm. CD spectra of 4 (e) and 5 (f) in nanopure water at a polymer concentration of 0.1 mg/mL.
Due to the distances between the charged side-chain groups and the polypeptide backbones, helical conformations were expected for the charged block polypeptides 4 and 5. FTIR and circular dichroism (CD) studies were employed to confirm the α-helical conformations of the polypeptide backbones. For both charged block copolypeptides, the absorbances in the FTIR spectra (Fig. S3 in ESI†) at ~1650 cm−1 (amide I region) and 1547 cm−1 (amide II region) were attributed to α-helical conformations.23 In the CD measurement, the polymer concentration of the nanoparticle solution in nanopure water was diluted to 0.1 mg/mL. The one positive band at 196 nm as well as two negative bands at 208 and 222 nm in the CD spectra indicated α-helical structures for both 4 and 5 (Fig. 2e and 2f, respectively). The critical micelle concentration (CMC) of 4 and 5 were measured to be 0.19 mg/mL and 0.10 mg/mL, which means that the CD measurements conducted at 0.1 mg/mL observed α-helical conformations primarily for free polymer chains. However, the negative bands were maintained (while the intensity of the positive band was off-scale) when the solution concentrations were increased to above the CMC values, suggesting that the α-helical conformations were largely maintained for the polypeptide backbones in the self-assembled cationic 6 and anionic 7 micellar nanoparticles.24 Due to the significant role of α-helical conformation in the regulation of protein folding and biological activity, its presence in these polypeptide nanoassemblies is an attractive property to impart biological activity, such as cell penetration efficiency, for the polypeptide-based nanoparticles for biomedical applications.22
In summary, a polypeptide-based versatile and functional polymer and nanoparticle platform with reactive, positively- or negatively-charged functionalities was developed from a sequential ROP and chemical transformation strategy. Well-defined diblock copolypeptide, PBLG-b-PPLG, with the incorporation of the alkynyl functional groups was prepared by performing facile and straightforward one-pot sequential ROPs of BLG and PLG NCA monomers with accelerated polymerization rates by nitrogen flow. The clickable alkynyl groups were then modified to install different charges and side-chain functionalities onto the polypeptide backbones with diverse thiol reagents by photoinitiated, radical-medicated thiol-yne click-type chemistry. The resulting amphiphilic block copolypeptides were self-assembled by either direct re-suspension or a nanoprecipitation method into nanopure water to form globular nanoparticles with distinct surface charges, as shown from DLS, TEM and zeta potential analyses. The distances between charged side-chain groups and polypeptide backbones rendered the charged block copolypeptides to maintain the helical conformations in aqueous solution, which was demonstrated by a combination of FTIR and CD characterizations. Currently, this polypeptide-based nanoparticle platform is being applied in various nanomedical applications, such as drug and gene delivery systems.
Supplementary Material
Acknowledgments
This work was supported in part from the National Heart Lung and Blood Institute of the National Institutes of Health as a Program of Excellence in Nanotechnology (HHSN268201000046C), the National Science Foundation (DMR-1105304), and the Welch Foundation through the W. T. Doherty-Welch Chair in Chemistry (A-0001). The microscopy & imaging center (MIC) at Texas A&M University is also gratefully acknowledged. The authors thank Professor Tadhg P. Begley for access to the CD spectrometer.
Footnotes
Electronic Supplementary Information (ESI) available: Experimental section and Fig. S1–S3. See DOI: 10.1039/c000000x/
Notes and references
- 1.(a) Yan Y, Such GK, Johnston APR, Best JP, Caruso F. ACS Nano. 2012;6:3663. doi: 10.1021/nn3016162. [DOI] [PubMed] [Google Scholar]; (b) Mout R, Moyano DF, Rana S, Rotello VM. Chem. Soc. Rev. 2012;41:2539. doi: 10.1039/c2cs15294k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Zhang F, Elsabahy M, Zhang S, Lin LY, Zou J, Wooley KL. Nanoscale. 2013;5:3220. doi: 10.1039/c3nr34320k. [DOI] [PubMed] [Google Scholar]
- 2.Elsabahy M, Wooley KL. Chem. Soc. Rev. 2012;41:2545. doi: 10.1039/c2cs15327k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohane DS, Langer R. Chem. Sci. 2010;1:441. [Google Scholar]
- 4.Robin MP, Mabire AB, Damborsky JC, Thom ES, Winzer-Serhan UH, Raymond JE, O'Reilly RK. J. Am. Chem. Soc. 2013;135:9518. doi: 10.1021/ja403587c. [DOI] [PubMed] [Google Scholar]
- 5.Tempelaar S, Mespouille L, Coulembier O, Dubois P, Dove AP. Chem. Soc. Rev. 2013;42:1312. doi: 10.1039/c2cs35268k. [DOI] [PubMed] [Google Scholar]
- 6.Shen Y, Zhang S, Zhang F, Loftis A, Pavía-Sanders A, Zou J, Fan J, Taylor J-SA, Wooley KL. Adv. Mater. 2013;25:5609. doi: 10.1002/adma.201302842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naik SS, Ray JG, Savin DA. Langmuir. 2011;27:7231. doi: 10.1021/la200882f. [DOI] [PubMed] [Google Scholar]
- 8.Zou J, Zhang F, Zhang S, Pollack SF, Elsabahy M, Fan J, Wooley KL. Adv. Healthcare Mater. 2014;3:441. doi: 10.1002/adhm.201300235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong H, Shu JY, Dube N, Ma YF, Tirrell MV, Downing KH, Xu T. J. Am. Chem. Soc. 2012;134:11807. doi: 10.1021/ja3048128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Deming TJ. Nat. Mater. 2010;9:535. doi: 10.1038/nmat2789. [DOI] [PubMed] [Google Scholar]; (b) Huang J, Heise A. Chem. Soc. Rev. 2013;42:7373. doi: 10.1039/c3cs60063g. [DOI] [PubMed] [Google Scholar]; (c) Tang HY, Li YC, Lahasky SH, Sheiko SS, Zhang DH. Macromolecules. 2011;44:1491. [Google Scholar]
- 11.Lalatsa A, Schatzlein AG, Mazza M, Thi BHL, Uchegbu IF. J. Control. Release. 2012;161:523. doi: 10.1016/j.jconrel.2012.04.046. [DOI] [PubMed] [Google Scholar]
- 12.(a) Kricheldorf HR. Angew. Chem. Int. Ed. 2006;45:5752. doi: 10.1002/anie.200600693. [DOI] [PubMed] [Google Scholar]; (b) Hadjichristidis N, Iatrou H, Pitsikalis M, Sakellariou G. Chem. Rev. 2009;109:5528. doi: 10.1021/cr900049t. [DOI] [PubMed] [Google Scholar]; (c) Hehir S, Cameron NR. Polym. Int. 2014 [Google Scholar]; (d) Zou J, Zhang F, Chen Y, Raymond JE, Zhang S, Fan J, Zhu J, Li A, Seetho K, He X, Pochan DJ, Wooley KL. Soft Matter. 2013;9:5951. doi: 10.1039/C3SM50582K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Deming TJ. Nature. 1997;390:386. doi: 10.1038/37084. [DOI] [PubMed] [Google Scholar]; (b) Aliferis T, Iatrou H, Hadjichristidis N. Biomacromolecules. 2004;5:1653. doi: 10.1021/bm0497217. [DOI] [PubMed] [Google Scholar]; (c) Lu H, Cheng JJ. J. Am. Chem. Soc. 2007;129:14114. doi: 10.1021/ja074961q. [DOI] [PubMed] [Google Scholar]
- 14.Habraken GJM, Heise A, Thornton PD. Macromol. Rapid. Commun. 2012;33:272. doi: 10.1002/marc.201100730. [DOI] [PubMed] [Google Scholar]
- 15.Quadir MA, Martin M, Hammond PT. Chem. Mater. 2013;26:461. [Google Scholar]
- 16.Engler AC, Lee HI, Hammond PT. Angew. Chem. Int. Ed. 2009;48:9334. doi: 10.1002/anie.200904070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang YG, Zeng YH, Yang JW, Zeng ZH, Zhu FM, Chen XD. Chem. Commun. 2011;47:7509. doi: 10.1039/c1cc12177d. [DOI] [PubMed] [Google Scholar]
- 18.Zou J, Fan J, He X, Zhang S, Wang H, Wooley KL. Macromolecules. 2013;46:4223. doi: 10.1021/ma4007939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan J, Zou J, He X, Zhang F, Zhang S, Raymond JE, Wooley KL. Chem. Sci. 2014;5:141. doi: 10.1039/C3SC52504J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obeid R, Armstrong T, Peng X, Busse K, Kressler J, Scholz C. J. Polym. Sci., Part A: Polym. Chem. 2014;52:248. [Google Scholar]
- 21.Xi W, Scott TF, Kloxin CJ, Bowman CN. Adv. Funct. Mater. 2014 [Google Scholar]
- 22.Gabrielson NP, Lu H, Yin LC, Li D, Wang F, Cheng JJ. Angew. Chem. Int. Ed. 2012;51:1143. doi: 10.1002/anie.201104262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo SW, Lee HF, Huang WJ, Jeong KU, Chang FC. Macromolecules. 2009;42:1619. [Google Scholar]
- 24.Greenfield NJ, Fasman GD. Biochemistry. 1969;8:4108. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.