Summary
In this issue of Structure, Zehr et al (2014) describe a structure of a three-stranded PhuZ tubulin cytomotive filament determined at 8.6Å resolution. This reveals an assembly mechanism different from that of microtubules, leading to a hypothesis explaining cytomotive-filament dynamics.
The prokaryotic homologs of eukaryotic tubulins can polymerize into thin cytomotive filaments in vivo and in vitro (Lowe and Amos, 2009). The tubulin cytomotive filaments are helical bundles of small numbers of protofilaments (Aylett et al., 2013; Aylett et al., 2010; Kraemer et al., 2012; Pilhofer et al., 2011), in contrast to the microtubule structure in eukaryotic cells containing 13 tubulin protofilaments (tubulin molecule chains). The tubulin cytomotive filaments display nucleotide-dependent behaviors like those of microtubules in eukaryotic cells despite of the difference in high-order architecture. The dynamic properties of cytomotive filaments are thought to be essential to their physiological functions in prokaryotic cells. The mechanism and structural nature underlying prokaryotic tubulin assembly and dynamics were unclear up to now. In this issue, Zehr et al. report the structural map of three-stranded cryomotive filament at 8.6 Å resolution, which clearly reveals secondary structural features (Zehr et al., 2014). The high-quality density map enabled the authors to build a pseudo-atomic model, which has provided good insight into the assembly and dynamics of prokaryotic tubulin filaments.
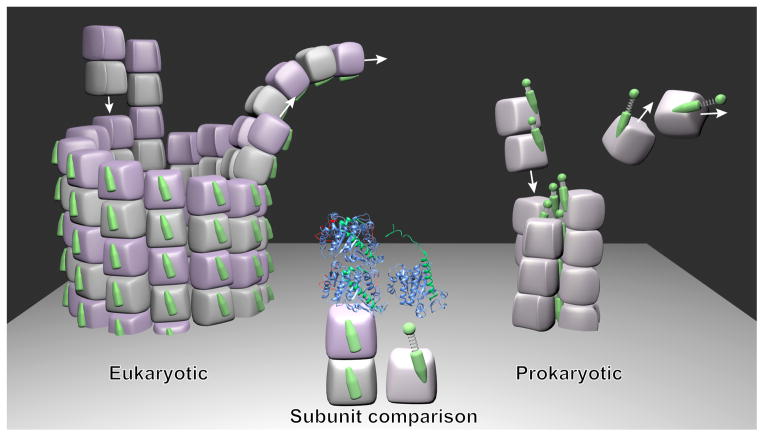
To understand the assembly, let’s first look into the crystal structures of the bacteriophage-encoded PhuZ tubulin-like proteins (Aylett et al., 2013; Kraemer et al., 2012; Oliva et al., 2012). The core structure of the PhuZ tubulins is very similar to that of eukaryotic tubulins. The major difference presents at their C-terminal regions. In PhuZ, the C-terminal region forms an unusually long helix (H11) followed by an extended loop (marked in green on the right side of the Figure 1). In eukaryotic tubulins, however, the H11 is relatively short and the extended loop folds back to the surface of the core structure forming the helices H12′ and H12 (see the left side of the Figure 1 and also see the Figure 3E in (Sui and Downing, 2010)). In the new study Zehr et al. demonstrate the distinct C-terminal regions underlie the architectural and assembly difference between tubulin cytomotive filaments and eukaryotic microtubules.
Figure 1.
Comparison of architecture for the three-stranded PhuZ cytomotive filament and the microtubule. (The figure is not intended to show a typical assembly/disassembly stage but a mixture, in order to display how the building blocks are added to or removed from the ends). The middle of the figure displays the eukaryotic tubulin dimer (on the left, PDB ID: 1JFF) and the PhuZ tubulin monomer (on the right, PDB ID: 3R4V). The C-terminal regions are marked in green. For eukaryotic tubulins polymerized in the microtubule, the C-terminal regions face outwards and are exposed to the environment. For PhuZ subunits polymerized in the three-stranded filaments, the C-terminal regions face inside and are responsible for both the intra-protofilament and inter-protofilament interactions. The tight or loose springs of the PhuZ tubulin subunits represent the compacted or extended forms, respectively.
Zehr et al. first confirmed the intra-protofilament interaction (the head to tail interaction) of the PhuZ tubulin subunits is different from that in eukaryotic microtubules (Aylett et al., 2013; Kraemer et al., 2012; Nogales et al., 1999). Within a protofilament, the helix H11 and the C-terminal loop extend into the adjacent subunit. In addition, Zehr et al. note the elongated C-terminal region also defines the lateral interaction between the protofilaments in the three-stranded PhuZ filament. The elongated C-termini face inwards and hold the protofilaments together as shown in Figure 1 (marked in green with springs on the right). This is completely different to microtubules where the lateral interaction between the protofilaments is defined by the H1′-S2, H2-S3 loops of one tubulin molecule and the M loop of the neighboring tubulin molecule (Sui and Downing, 2010). The C-terminal region of the eukaryotic tubulin forms an additional helix, H12, (marked in green on the left in Figure 1) and faces outwards on the microtubule. This region is responsible for binding with microtubule motors (on β-tubulins) and some microtubule associated proteins (MAPs). The overall architecture of three-stranded PhuZ cytomotive filament resembles an inverted configuration of microtubules with a smaller number of component protofilaments.
Zehr et al. propose a C-terminal “spring model” to explain the dynamic assembly and disassembly of PhuZ cytomotive filaments (Zehr et al., 2014). In this hypothesis, the elongated C-terminal regions are in a compacted form and tightly hold the adjacent PhuZ tubulin molecules longitudinally in the GTP state (See the Figure 7 in (Zehr et al., 2014)). The compacted PhuZ subunits are assembled into the twisted protofilament in the cytomotive filament. After hydrolysis, the compacted subunits cannot revert to an extended form within the helical filament lattice. Therefore, energy as strain is stored resulting in highly dynamic metastable filaments. This intriguing hypothesis can explain the dynamic behavior for cytomotive filaments with the same concept to the “GTP cap model” for the dynamic microtubules.
There is other important information in the paper: The nucleus for the PhuZ tubulin polymerization is proposed to contain six monomers (organized into a trimer of dimers) based on the growth kinetics measured by right-angle light scattering. From the structural model, the authors identified conserved salt-bridge forming residues. Polymerization characterization after a series of point mutations confirmed their importance in PhuZ cytomotive filament assembly. This in turn demonstrates the credibility of the proposed structural model determined with a density map revealing secondary structural features.
The new study of three-stranded PhuZ filament provides structural insight into the assembly of prokaryotic tubulin filaments. Can we derive from it a universal understanding that the C-terminal region defines the lateral interaction in all types of prokaryotic tubulin filaments? While this paper was in press, the same research group published a separate paper showing that the two-stranded filament of the bacterial tubulin TubZ-Bt can transit into a four-stranded filament upon GTP hydrolysis (Montabana and Agard, 2014). In that paper, the C-terminal regions of TubZ-Bt face inwards in a two-stranded filament, which is consistent with the architecture of the three-stranded PhuZ filament structure. However, in the reported stable four-stranded TubZ-Bt filament, the protofilaments are rotated outwards exposing the C-terminal regions on the outside, an orientation similar to that in the microtubule. Based on the two papers, the lateral interactions in various cryomotive filaments of prokaryotic tubulins can be complex. Nevertheless, the changeable lateral interaction indicates that intra-protofilament interaction should be more stable than the inter-protofilament interaction in cytomotive filaments of prokaryotic tubulins.
Because both the protofilament number and the lateral interaction changed with nucleotide states for the TubZ-Bt filaments, the “spring model” from the PhuZ tubulin family may not represent a comprehensive view for all the prokaryotic tubulin filaments. With excellent electron microscopes and a state-of-the-art direct electron detector, better structural details of these cytomotive filaments are anticipated, that may help to develop a detailed and integrated understanding about prokaryotic tubulin assembly.
Purified prokaryotic tubulins can polymerize into a variety of filamentous structures under different conditions. Another remaining question therefore becomes which of the reported in vitro structures naturally present in the bacteria. Resolving this issue will help to clarify if both forms of the reported lateral interactions play physiological functional roles in bacteria.
Acknowledgments
The work was supported by the NIH grants GM101026 and GM097010.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aylett CHS, Izoré T, Amos LA, Löwe J. Journal of Molecular Biology. 2013;425:2164–2173. doi: 10.1016/j.jmb.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylett CHS, Wang Q, Michie KA, Amos LA, Löwe J. Proc Natl Acad Sci U S A. 2010;107:19766–19771. doi: 10.1073/pnas.1010176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer James A, Erb Marcella L, Waddling Christopher A, Montabana Elizabeth A, Zehr Elena A, Wang H, Nguyen K, Pham Duy Stephen L, Agard David A, Pogliano J. Cell. 2012;149:1488–1499. doi: 10.1016/j.cell.2012.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Amos LA. Int J Biochem Cell Biol. 2009;41:323–329. doi: 10.1016/j.biocel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- Montabana EA, Agard DA. Proc Natl Acad Sci U S A. 2014;111:3407–3412. doi: 10.1073/pnas.1318339111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- Oliva MA, Martin-Galiano AJ, Sakaguchi Y, Andreu JM. Proc Natl Acad Sci U S A. 2012;109:7711–7716. doi: 10.1073/pnas.1121546109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilhofer M, Ladinsky MS, McDowall AW, Petroni G, Jensen GJ. PLoS biology. 2011;9:e1001213. doi: 10.1371/journal.pbio.1001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui H, Downing KH. Structure. 2010;18:1022–1031. doi: 10.1016/j.str.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr EA, Kraemer JA, Erb ML, Coker JKC, Montabana EA, Pogliano J, Agard DA. Structure. 2014;22(4):539–548. doi: 10.1016/j.str.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]