Abstract
We describe an ultra-high-throughput screening platform enabling discovery and/or engineering of natural product antibiotics. The methodology involves creation of hydrogel-in-oil emulsions in which recombinant microorganisms are co-emulsified with bacterial pathogens; antibiotic activity is assayed by use of a fluorescent viability dye. We have successfully utilized both bulk emulsification and microfluidic technology for the generation of hydrogel microdroplets that are size-compatible with conventional flow cytometry. Hydrogel droplets are ~25 pL in volume, and can be synthesized and sorted at rates exceeding 3,000 drops/s. Using this technique, we have achieved screening throughputs exceeding 5 million clones/day. Proof-of-concept experiments demonstrate efficient selection of antibiotic-secreting yeast from a vast excess of negative controls. In addition, we have successfully used this technique to screen a metagenomic library for secreted antibiotics that kill the human pathogen Staphylococcus aureus. Our results establish the practical utility of the screening platform, and we anticipate that the accessible nature of our methods will enable others seeking to identify and engineer the next generation of antibacterial biomolecules.
Keywords: antibiotics, drug discovery, in vitro compart-mentalization, high-throughput screening, metagenomics, microfluidics
Introduction
The increasing frequency of multidrug resistant bacterial infections in hospital and community acquired settings is a widely recognized and growing threat to public health. Current estimates of U.S. healthcare costs associated with hospital-acquired infections are in the range of $16 billion/year (Hassan et al., 2010). Staphylococcus aureus is a particularly pervasive bacterial pathogen that underlies the majority of healthcare associated blood stream infections and infective endocarditis (Garau et al., 2009) and is a major causative agent of ventilator associated and other nosocomial pneumonias (Athanassa et al., 2008; Bamberger and Boyd, 2005). S. aureus infections are marked by a high incidence of antibiotic resistance: a full two-thirds of hospital associated infections, and ~50% of community acquired S. aureus infections are now methicillin-resistant (MRSA) (Taubes, 2008). The emergence of multidrug-resistance in S. aureus and other infectious bacteria underscores an increasingly desperate need for next generation antibiotics capable of combating these dangerous pathogens.
Despite the urgent need for new antibacterials, the question of where best to search for these compounds remains open. Historically, antimicrobial agents have been discovered as natural products (β-lactams, aminoglycosides, polyketides, cyclic liproproteins, etc.) secreted by ecological competitors of bacterial pathogens. After an initial boon from such natural product drug discovery programs, all but a few pharmaceutical companies have now reduced or eliminated their antimicrobial pipelines due in part to diminishing returns from screening culturable microbes (Taubes, 2008). Importantly, the majority of microbial species cannot be cultured using standard laboratory techniques, and these organisms likely produce a vast reservoir of bioactive compounds that have yet to be efficiently mined (Curtis et al., 2002; Daniel, 2004; Zotchev et al., 2012).
Recent advances in culture-independent DNA capture, amplification, sequencing, cloning, and expression have renewed interest in natural product drug discovery. Specifically, biotechnologists can now access genetic determinants of natural product biosynthesis without the need to culture fastidious bacterial species. The application of this technology, termed metagenomics (Handelsman et al., 1998), follows one of two general formulas. In the bioinformatics approach, sequenced environmental DNA (eDNA) is mined for new natural product genes or biosynthetic gene clusters based on homology searches using known genes or operons. Alternatively, expression libraries from eDNA may be functionally screened to identify entirely novel gene products and pathways (Iqbal et al., 2012). Panning metagenomic DNA libraries presents the opportunity to interrogate hundreds of microbial genomes simultaneously, but accessing this enormous diversity in an efficient manner has required commensurate advances in data analysis and high-throughput techniques.
Sequence-based techniques such as in silico cloning and next generation sequencing have enabled efficient bioinfor-matic screening of metagenomic libraries. For example, metagenomic sequence mining has yielded novel enzymes able to produce analogs of antibiotics and chemotherapeutics (Banik and Brady, 2008; Ziemert et al., 2010). Such sequence-based approaches are limited, however, by the fact that they require known genes or biosynthetic pathways as a basis for comparison; truly novel molecular scaffolds cannot be identified using sequence-based metagenomic screening. In contrast, functional screening of metagenomic expression libraries can enable isolation of novel bioactive molecules having no known homologous structures or sequences, although this approach has its own limitations. In particular, the dearth of sufficiently high-throughput screening methods restricts the molecular diversity that can be sampled and thus constrains new discoveries to those genes that happen to be among the small fraction of clones interrogated. Nonetheless, successful function-based screening of metagenomic libraries has been demonstrated (Brady et al., 2004; Gillespie et al., 2002). A representative work flow involves plating Escherichia coli expression hosts on top agar overlays followed by 5–7 days of growth and visual inspection to identify colonies yielding zones of inhibition (Brady, 2007). In the absence of robotics and advanced image processing, the practical throughput for such a scheme is ~250,000–500,000 clones per week. In light of the limited throughput and intense manual labor, the successful identification of novel bioactive molecules from these efforts is all the more impressive. One is left to contemplate, however, the breakthrough compounds yet be discovered for lack of a more powerful screening platform.
Fundamentally, the success of any biological high-throughput screen relies on detection of a desired property or phenotype, in this case bactericidal activity, and linkage of that phenotype to the causative genotype. Toward this end, inverted emulsions have been used extensively to partition library populations in picoliter-compartmentalized bioreactors (Aharoni et al., 2005; Griffiths and Tawfik, 2003; Levy et al., 2005). More recently, microfluidic devices have proven increasingly useful for in vitro compartmentalization (IVC). So-called drop-based microfluidics is a powerful methodology for co-encapsulating a genetic determinant or recombinant microbe with a reporter molecule, usually a fluorogenic probe for the desired activity (Agresti et al., 2010; Fallah-Araghi et al., 2012; Paegel and Joyce, 2010). The surfactant-stabilized reaction compartments, isolated from each other in an immiscible oil phase, can be incubated in bulk and then interrogated independently, thereby maintaining the genotype-phenotype linkage. While this form of chemical compartmentalization is undeniably powerful, it is most compatible with on-chip sorting (Mazutis et al., 2013), and requires customized, elaborate set-ups that are not widely available to most academic researchers.
Here, we employ an alternative based on hydrogel microdroplets (GMD) within which we immobilize both a recombinant host and a screening target. By virtue of immobilization within a solid hydrogel matrix, our GMD maintain the genotype-phenotype link whether in emulsion or in a continuous aqueous phase. Importantly, we fabricate the GMD at a size compatible with conventional fluorescence-activated cell sorting (FACS). Agarose hydrogel droplets have been used previously for combinatorial co-culture of mammalian cells (Tumarkin et al., 2011), single-cell RT-PCR for transcription profiling (Zhang et al., 2012), and even flow cytometric determination of antibiotic minimum inhibitory concentration (Eun et al., 2010). We describe a novel GMD application in which a recombinant antibiotic-producing microbe, here Saccharomyces cerevisiae or E. coli, is co-encapsulated with a target pathogen, here S. aureus. Following growth of the host and induction of the recombinant expression cassette, GMD in which the target pathogen has been killed are stained with a fluorogenic viability probe. The fluorescent GMD, encoding bactericidal agents, are efficiently isolated by high speed FACS. We validate our methods using active and inactive bacteriolytic enzymes, and we further demonstrate the concept by screening a small metagenomic library. In both cases, we isolate clones producing a lytic hydrolase specific for S. aureus. We anticipate that our GMD-FACS screen will allow more efficient metagenomic library screening for antibiotics, and will likewise enable more ambitious biomolecular engineering projects seeking to combat drug-resistant bacterial pathogens.
Materials and Methods
Strains and Plasmids
S. cerevisiae expression strain BJ5464 [MATalpha ura3–52 trp1 leu2-delta1 his3-delta200 pep4::HIS3 prb1-delta1.6R can1 GAL] (Jones, 1991) was obtained from ATCC (208288). Plasmids were maintained in E. coli strain DH5a [F— Ф80lacZΔM15 Δ (lacZYA-argF) U169 recA1 endA1 hsdR17 (rK−, mK+) phoA supE44 λ– thi-1 gyrA96 relA1]. Screening of native staphylococcal plasmid libraries was performed in E. coli strain BL21(DE3) [F– ompT hsdSB(rB–, mB–) gal dcm (DE3)] using the pET26b(+) expression vector (Novagen, Darmstadt, Germany). S. aureus USA400 was a generous gift from Ambrose Cheung (Dartmouth College). To make an S. cerevisiae strain that constitutively expresses yEGFP (yeastenhanced green fluorescent protein), the yEGFP-Kan construct was PCR amplified from plasmid pKT0127 (Sheff and Thorn, 2004) (Addgene, Cambridge, MA) with the following oligonucleotides 5’-cactctagagccaccatgtctaaaggtgaagaattattc-3’ (XbaI), and 5’-gcagggtacctcgatgaattcgagctcg-3’ (KpnI), enabling the amplicon to be subcloned into p416-GPD (Mumberg et al., 1995) using standard restriction enzyme cloning to create vector pYIc-yEGFP-Kan . By amplifying this plasmid with oligonucleotides containing >40bp of homology to the S. cerevisiae genome, a constitutively expressed yEGFP construct with all regulatory elements for selection and expression can be targeted to any location in the genome. We chose to integrate into the ura3 locus, which is already mutated in the BJ5464 strain. The following oligonucleotides were used in a PCR reaction to amplify yEGFP-Kan from pYIc-yEGFP-Kan : 5’-ctacatataaggaacgtgctgctactcatcctagtcctgtgagctcagtttatcattatc-3’; and 5’-tccaatgaagcacacaagtttgtttgcttttcgtgcatgatcgatgaattcgagctcg-3’. The amplicon was purified after agarose gel electrophoresis and transformed into BJ5464 by electroporation. Yeast were recovered for 3 h in YPD, and then plated on YPD agar + 300 mg/mL G418. GFP fluorescence of isolated clones was confirmed by fluorescence microscopy. A clonal population was isolated and archived as BJ5464::yEGFP. Growth rate of BJ5464::yEGFP was equal to the BJ5464 strain in all growth media tested. Unless stated otherwise, DNA modifying enzymes were from New England Biolabs.
Construction of Lysozyme and Lysostaphin Expression Constructs
Recombinant S. cerevisiae expressing human lysozyme (LYZ) in frame with the a-mating factor was cloned into the S. cerevisiae centromeric expression vector p416-GAL1 (Mumberg et al., 1994), as described in Scanlon et al. (2009), and transformed into BJ5464. The coding sequence for the mature peptide of lysostaphin (amino acids 1–246) was amplified by PCR from pRG5 (ATCC: 67076), and cloned into the XhoI/BamHI sites of the p416-GAL1-aMF vector, using standard restriction enzyme cloning, and then transformed into BJ5464::yEGFP.
Preparation of Bulk Emulsions
Overnight cultures of S. aureus and yeast were grown in baffled shake flasks at 30° C in dextrose growth media + casamino acids (DGMC-URA): 0.67% yeast nitrogen base (Difco), 0.77 g/L CSM-URA (MP Biomedicals, Solon, OH), 2% dextrose, 1% casamino acids, 100 mM potassium phosphate, pH 7.1. On the day of the experiment, 40 OD600nm units of yeast were centrifuged, resuspended in fresh galactose induction media (GIM-URA): 0.67% yeast nitrogen base, 0.77 g/L CSM-URA, 3% galactose, 100 mM potassium phosphate, pH 7.1, and incubated for 3 h at 30°C, with shaking at 250 rpm. Yeast and S. aureus were washed twice in sterile PBS (2.7 mM KCl, 1.5 mM KH2PO4, 136.9 mM NaCl, 8.9 mM Na2HPO4, pH 7.4) and resuspended at a concentration of ^100 OD600nm. Low melt agarose (SeaPlaque Agarose, Lonza, Allendale, NJ) was used for creation of hydrogel-in-oil emulsions. Agarose was dissolved in GIM-URA by heating 0.3 g in 20 mL GIM-URA at 70°C for 5 min, and then cooled to 50°C. Exactly 5mL of liquid agarose was filtered with a 0.22 mm syringe-filter, and maintained at 42°C. S. cerevisiae and S. aureus were added to the agarose at a final OD600nm of 2.0, and 1.0, respectively. Meanwhile, 23.64 mL of light mineral oil (Fisher: O121-4) was mixed with 0.36 mL of Span80, heated to 42°C in a 50 mL beaker, and vigorously agitated with a magnetic stirbar. Three mL of the agarose-microbe mixture was added drop-wise to the stirring liquid, and emulsified for exactly 5 min. At this point, the entire beaker was transferred to a cold water bath, held at 10°C for 4 min with gentle stirring. Under these conditions, the pore size of the gelled agarose is estimated to be on the order of 200–300 nm (Narayanan et al., 2006). Emulsions were broken by washing three times with surfactant-free mineral oil: the emulsion was centrifuged at 900g for 5 min, the supernatant (containing very small agarose droplets) was pipetted off, and the pellet was resuspended with 25 mL of mineral oil. After three washes, the agarose microdrops were pelleted, the mineral oil was removed, and the pellet was resuspended in PBS. Hydrogel droplets were washed two more times with PBS to remove traces of mineral oil, and then filtered using nylon mesh. First, droplets were poured over a sterilized 30mm nylon mesh (Millipore: NY3004700). Droplets that were retained on the top of the filter were recovered in PBS and poured through a 40 mm nylon mesh filter (Fisher: 08–771-1), and resuspended in 50 mL GIM-URA media. The hydrogel microdroplets are then incubated for 15–17 h at 30°C, and prepared for FACS analysis.
Construction of Metagenomic Library
Three strains of Staphylococcus were obtained from the US Department of Agriculture ARS Culture Collection (NRRL): S. simulans (B-2628), S. arlettae (14764), and S. equorum (14765). Starter cultures were grown in 3 mL of TSB, diluted 1:1,000 into fresh TSB, and grown overnight at 37°C, 250 rpm in baffled shake flasks. Native plasmids from these species were purified using FosmidMAX DNA Purification Kit (Epicentre, Madison, WI) by using a modified lysis procedure; after centrifuging the overnight cultures, the pellets were resuspended in 3 mL of Fosmid Max Buffer #1 and treated with 120 mg lysostaphin and 1.2 mg of lysozyme for 45 min at 37°C. After lysis, plasmid purification was performed according to manufacturer’s instructions. The purified native staphylococcal plasmid DNA was treated with DNaseI in order to make random double-stranded breaks. In 100 µL of DNaseI reaction buffer at 15°C, 5 mg of native plasmid DNA was mixed with 0.002 units of DNaseI (New England Biolabs, Ipswich, MA), and reacted for 2.5–20 min. At various time points, the DNaseI reactions were stopped by adding 200 µL of DNA binding buffer; the resulting DNA fragments were isolated by spin column purification (Zymo Research, Irvine, CA, DNA Clean and Concentrator kit). Reactions were run on 1% agarose gel electrophoresis, bands greater than 1 kbp, and less than 5 kbp were purified using Zymoclean Gel DNA Recovery kit (Zymo Research). Size-selected DNA fragments were end-repaired using End-It DNA End-Repair kit according to manufacturer’s instructions (Epicentre). The pET26b(+) expression vector was prepared by restriction digest with XbaI and HindIII. Restricted DNA was purified by agarose gel purification, blunt-ended by DNA end-repair, and dephosphorylated by treatment with Antarctic Phosphatase. Ligation of the expression vector to the eDNA was performed by incubating 500 ng of blunt-ended, dephosphorylated pET26b(+) with a mixture of the size-selected native staphylococcal plasmid DNA (92 ng for each species), in 20 µL T4 ligase buffer with 600 U of T4 ligase at 16°C overnight. The reactions were dialyzed against water and electroporated into DH5a. A serial dilution of the transformation mixture was plated on LB agar + antibiotic to titer the library. A liquid outgrowth of the library was subjected to plasmid miniprep and the library was electroporated into the BL21(DE3) expression strain.
Microfluidic Compartmentalization
Frozen aliquots of the native Staphylococcus plasmid eDNA library (E. coli BL21(DE3)::pET26b–eDNA) were thawed and grown in baffled shake flasks overnight in LB + selective antibiotic at 23°C. The target pathogen S. aureus were grown overnight at 30°C in DGMC-URA media. On the day of the experiment, the E. coli was diluted 1:50, and induced with 100 µM IPTG for 60 min. Both bacterial strains were washed twice with sterile PBS and resuspended to an OD600nm of ^100 (S. aureus) or ~5 (BL21(DE3)::pET26b–eDNA). Low melt agarose was dissolved in LB at 1.5% by heating, and then cooled to 50°C for sterile filtration. Once the agarose was equilibrated to 42°C, bacteria and additives were added: 100 µM IPTG, S. aureus to a final concentration of OD600nm 1.0, and BL21(DE3) to a final concentration of OD600nm 0.075. The agarose-microbe mixture was added to a sterilized glass vial with a magnetic stir bar and a PTFE/silicon septa screw-top (Fisher: 03–391-22), and incubated at 42°C on a heated magnetic stir plate. PicoSurf1 (2% in Novec 7500, Dolomite), a proprietary biocompatible surfactant in a fluorocarbon carrier oil was transferred into a second sterilized glass vial and heated to 42°C. Using a pressurized nitrogen tank and pneumatic pressure controllers (Airtrol, New Berlin, WI) and gauges Winters Instruments (Buffalo, NY), a pulseless gas pressure system was used to deliver the fluids to a polydimethylsiloxane (PDMS) microfluidic dropsplitting device (Figure S1) (Abate and Weitz, 2011). By controlling the ratio of pressures applied to the headspace of the glass vials, the size of droplets and rate of production can be controlled. The PDMS chips were fabricated by the Stanford Microfluidic foundry using standard soft lithography techniques. Mold height was 35 mm with a rectangular cross-section, and the PDMS chips were plasma-bonded to glass microscope slides. To deliver fluids to the microfluidic device, PTFE tubing (Grace 20033, 1/16″ ID × 0.020″ OD) was inserted directly into fluids and chips. The microfluidic device was mounted on a standard dissecting microscope (AmScope, Irvine, CA) and kept heated at 42°C using a digital temperature controller (Digi-Sense, Cole Parmer, Vernon Hills, IL). Agarose hydrogel microdroplets were flowed off-chip directly into 0.5 mL centrifuge tubes, and stored in emulsion at 30°C for 5–6 h. Before microscopic or FACS analysis, emulsions were broken by mixing with 1H,1H,2H,2H–Perfluoro-1-octanol (Sigma, St. Louis, MO). First, the carrier oil was removed, and then the creamed emulsion was mixed with PBS + 15% 1H,1H,2H,2H–Perfluoro-1-octanol by gentle agitation. The hydrogel droplets partition to the aqueous phase, and were pipetted off. After washing once in PBS, the hydrogel droplets were stained with 500 nM SyTox Orange and analyzed by fluorescence microscopy or FACS.
FACS Sorting of Agarose Hydrogel Microdroplets
Agarose hydrogel microdroplets stained with SyTox Orange were analyzed by flow cytometry on an iCYT Synergy using sterile PBS as sheath fluid. The sorter was equipped with a 126 mm nozzle, laser lines of 488 and 561 nm were used for excitation of yEGFP and SyTox Orange, respectively. Forward and back scatter patterns were used to gate for hydrogel microdroplets (i.e., to exclude analysis of free bacteria or yeast), and band-pass filters of 525/50 nm and 585/40 nm were used to capture fluorescence emission from yEGFP and SyTox, respectively. Events were analyzed at 3,000 events/ second in order to maintain sort stream stability, and sort gates were set between 0.01% and 2% of total events based on SyTox Orange signal. When sorting GMDs loaded with S. cerevisiae, GMDs were sorted into liquid growth media supplemented with 10 µg/mL cefazolin, an antibiotic with selectivity for S. aureus. This was necessary to prevent S. aureus, which are loaded in vast excess compared to S. cerevisiae, from overgrowing the culture of sorted droplets. In the final round of sorting, agarose droplets were sorted into sterile PBS and then plated directly onto nutrient agar media supplemented with heat-killed S. aureus to identify active clones.
Results
Preparation of and Characterization of Hydrogel Microdroplets
IVC has been utilized as a high-throughput screening technology for a variety of applications, with particularly impressive successes in directed evolution of enzyme activity (Agresti et al., 2010; Griffiths and Tawfik, 2003; Mastrobattista et al., 2005) and screening of chemical libraries (Brouzes et al., 2009). We hoped to utilize this powerful technique as a tool for screening libraries for antibiotic activity. The method involves emulsification of an aqueous hydrogel phase in which bacterial pathogens are co-cultured with a recombinant host microorganism capable of secreting biocatalytic antibiotics and/or secondary metabolites (Fig. 1). Recombinant microorganisms can be transformed with plasmids that have been engineered to express DNA from a variety of sources, including single genes with antibiotic activity, combinatorial mutant libraries of such genes, or metagenomic DNA from environmental sources. GMD are incubated under conditions permissive to recombinant protein expression and secretion, and should the recombinant gene product possess bactericidal activity, it will affect killing of the co-cultured pathogen. The GMD emulsion may then be broken, and the hydrogel droplets stained with a membrane-impermeant, DNA-intercalating, “dead cell” fluorogenic dye. Stained GMD can be sorted at rates exceeding 3,000 events per second by FACS. Using this technique, we have achieved screening throughputs exceeding 5 million clones/day.
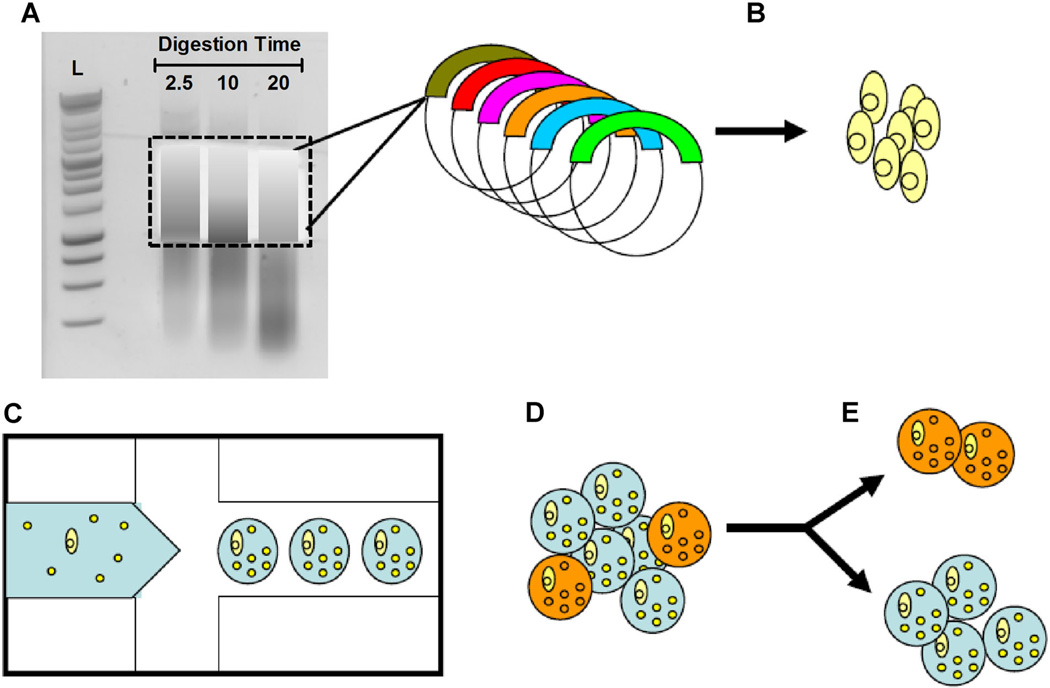
Figure 1.
Schematic of ultra-high-throughput screen for antibiotic drug discovery. A: Environmental DNA was subjected to a limited DNasel digestion and size selected for fragments between 1,000 and 5,000 bp. Digestion time in minutes indicated. B: Metagenomic DNA was cloned into an expression vector by blunt end ligation and then transformed into E. coli bacterial hosts. C: A microfluidic device is used to generate agarose-in-oil micro-emulsions in which individual recombinant E. coli are co-encapsulated with live S. aureus bacterial targets (small yellow spheres). D: GMDs in which the recombinant clone secretes an enzyme that is lytic towards S. aureus are labeled with the SyTox Orange viability probe, which selectively stains only dead bacteria (orange circles). E: Mixed GMD populations are sorted by FACS to isolate E. coli secreting bactericidal natural products. Genes from the sorted GMDs can be cloned, recombined, or rescreened iteratively to facilitate antibiotic drug discovery.
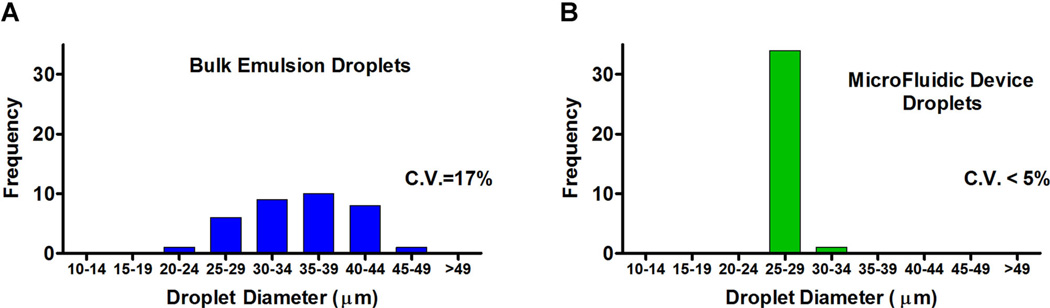
We demonstrate the utility of this screening strategy using two different emulsification techniques and two different recombinant host organisms. In early proof-of-concept experiments agarose hydrogel-in-oil emulsions were synthesized using a simple bulk emulsification method resulting in highly polydisperse emulsions. Microbes were added to a low melt agarose solution at specified cell densities such that, on average, each appropriately-sized droplet would contain < 1 antibiotic-producing S. cerevisiae and ~20–30 S. aureus. Agarose cell suspensions were then emulsified in mineral oil/surfactant solution by rapid mixing in a heated water bath using a standard magnetic stir plate. Various control experiments demonstrated that neither bacterial targets nor yeast hosts were subject to heat-mediated killing during GMD production (data not shown). The emulsion was cooled to solidify the agarose hydrogel, broken by iterative low-speed centrifugation and resuspension in surfactant-free mineral oil, and finally resuspended in aqueous growth media. Bulk emulsification generates a preponderance of GMD < 10 µm, but the low-speed centrifugation steps enrich for larger GMD (average diameter 24 µm, CV 31%). The suspension was first sieved over a 30 µm nylon mesh filter and the retained GMD were then sieved through a 40 µm nylon mesh filter. The resultant GMD are, on average, 35 µm in diameter, but the population remains relatively polydispersed (CV: 17%) (Fig. 2A). The size selected GMD are sufficiently large to support yeast and bacterial cell growth through several divisions, yet they are sufficiently small to be compatible with analysis by flow cytometry.
Figure 2.
Size distribution of agarose GMD. Polydisperse emulsions of agarose are made by homogenization of prewarmed agarose in oil-surfactant mixtures. Droplets are transferred to a continuous aqueous phase and sieved over a 30 µm nylon mesh filter, and then through a 40 µm nylon mesh filter. Micrographs of agarose droplets were analyzed using digital processing software (Axiovision, Zeiss, Thornwood, NY) to measure the diameter of 35 GMDs. The data were binned into groups of 5 µm, and represented as histograms. A: While the initial population trends toward small aGMDs, successive filtering through nylon mesh filters with pore sizes of 30 and 40 µm centers the population in the desired size range, but with considerable polydispersity. B: Monodisperse hydrogel microdroplets were produced with a drop-splitting microfluidic device, and droplet sizes were measured without any filtration.
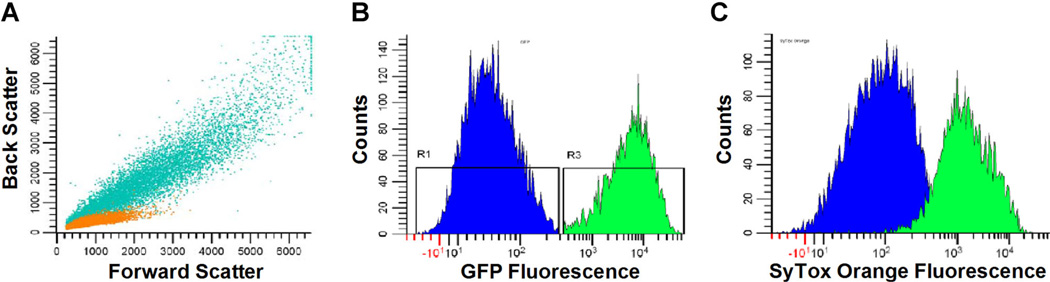
GMD were analyzed on an iCyt Synergy flow sorter equipped with a 126 mm nozzle. Forward scatter and backscatter dot plots show the GMD generate a distinct pattern compared to free yeast, although there is some small overlap (Fig. 3A). Visualization of these patterns is instructive in drawing gates that focus the analysis on GMD and exclude free yeast, which can escape the GMD during growth.
Figure 3.
FACS analysis of GMD populations. A: An overlay of flow cytometric scans of free S. cerevisiae cells (orange) and a population of GMD (teal). The two species have distinct sizes and morphologies that are easily distinguished by forward and back scatter analysis. Recombinant S. cerevisiae expressing either LYZ or yEGFP::ssLST were co-encapsulated with S. aureus, incubated for 16 h at30°C, stained with SyTox Orange, and analyzed using flow cytometry. The GFPwas used as an orthogonal fluorescent marker for the positive control cells. B: GFP signal of GMD deliminatestwo populations: negative LYZ secreting yeast (gated in blue: R1) and positive yEGFP::ssLST secreting yeast (gated in green: R3). C: FACS analysis of the SyTox Orange signal demonstrates >10-fold shift in the mean fluorescence intensity of the population secreting a lytic enzyme (yEGFP::ssLST) relative to the negative control population (LYZ, blue). SyTox Orange signal correlates with S. aureus killing.
Analysis of Hydrogel Droplets for Antibiotic Activity
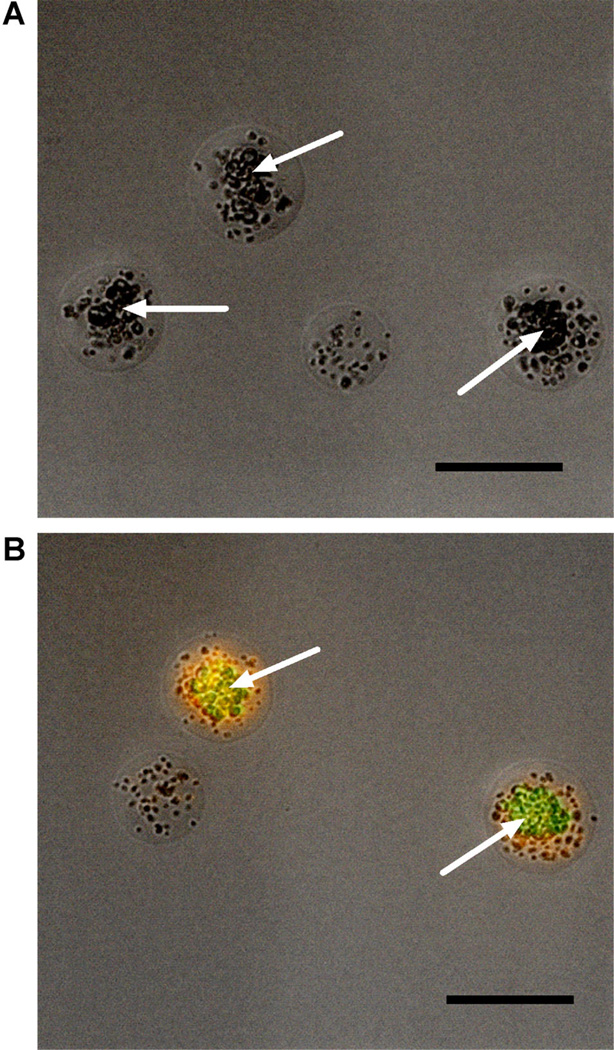
To conduct initial validation of our GMD-FACS screen, we chose to construct a model system wherein positive control BJ5464 S. cerevisiae constitutively express yeast enhanced GFP (yEGFP) (Sheff and Thorn, 2004) and inducibly secrete the well-studied bacteriolytic enzyme lysostaphin (ssLST, EC 3.4.24.75), which specifically kills the human pathogen S. aureus (Recsei et al., 1987). In parallel, non-fluorescent yeast were transformed with an expression plasmid for the galactose-inducible secretion of LYZ (Scanlon et al., 2010), a lytic hydrolase that is ineffective against S. aureus (Bera et al., 2005). The positive and negative control yeast strains were each mixed with S. aureus and emulsified using homogenization as described above. Following protein induction, the GMD populations were stained with the SyTox Orange viability dye and analyzed by flow cytometry (Fig. 3B and C) and fluorescence microscopy (Fig. 4A and B). Several interesting features of the experimental system can be visualized by fluorescence microscopy (Fig. 4A and B). Recombinant yeast remain viable within the GMD and continue to divide, forming clonal clusters during the course of incubation (30°C, 16–18 h). The S. aureus also remain viable and continue to divide, although at much slower rates than in shake flask cultures. The flow cytometry scans and micrographs clearly demonstrate that LYZ negative control yeast fail to elicit a SyTox Orange signal (Figs. 3C and 4A). In contrast, yeast secreting ssLST generate a robust SyTox signal indicating killing and lysis of S. aureus bacteria (Figs. 3C and 4B).
Figure 4.
Microscopic analysis of GMD fluorescence properties. Recombinant S. cerevisiae secreting various lytic hydrolase enzymes were co-encapsulated with S. aureus USA400, incubated for 16 h at 30°C, and stained with SyTox Orange viability stain. A: When co-encapsulated with recombinant yeast secreting LYZ, S. aureus retain viability and exhibit negligible SyTox Orange staining. B: When co-encapsulated with yeast that express ssLST (also expressing the yEGFP marker), S. aureus are killed and exhibit marked SyTox Orange staining. In each micrograph, yeast colonies are indicated by white arrows and scale bars are 50 µm. Images are from different fields of view from the same sample.
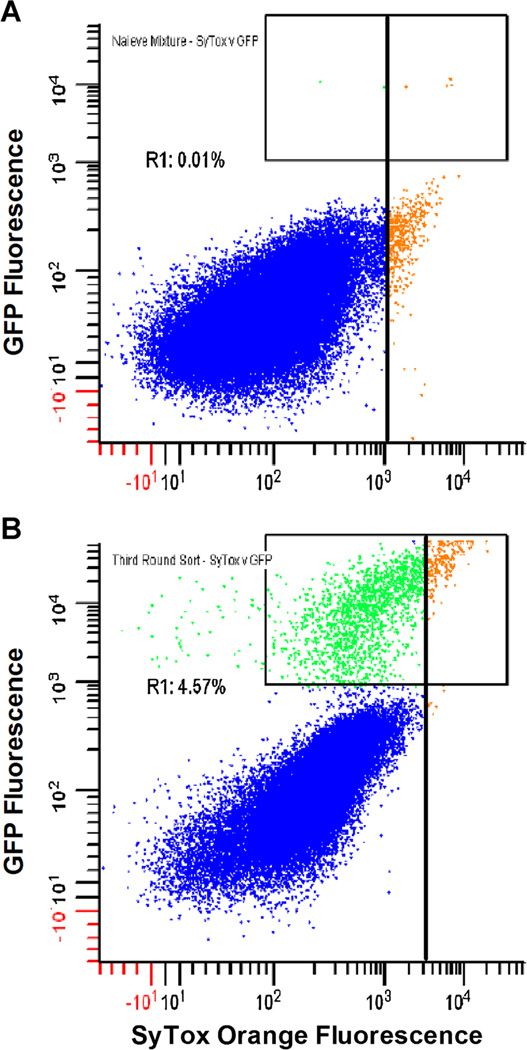
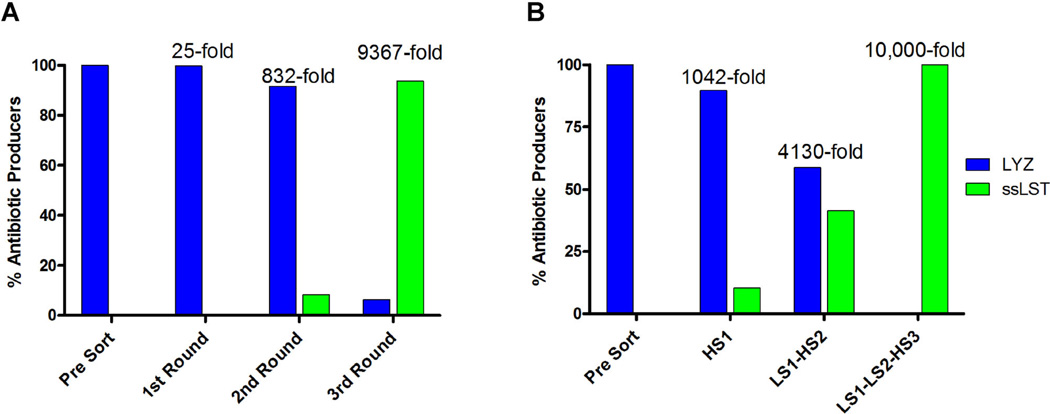
The method’s capacity to enrich rare positive clones was quantified by implementing a mock library screen. Negative control LYZ yeast were mixed at a 10,000:1 excess with positive control yEGFP::ssLST yeast, and the mixture was co-encapsulated with S. aureus in agarose GMD. Following a 17-h incubation in yeast induction media, the colloidal suspension of droplets was stained with SyTox Orange. Flow cytometric scans of the presort population are shown (Fig. 5A). Analysis of the yEGFP signal (γ-axis), used to quantify yEGFP::ssLST GMDs in the mixture, confirms the expected 1:10,000 input ratio of positive to negative clones. In the first round of sorting, a “low stringency” sort gate was set for the top 1% of SyTox Orange events (Fig. 5A, vertical bar). Yeast from sorted GMD were recovered by overnight outgrowth in selective media, archived as glycerol stocks, and the process of co-emulsification, screening and outgrowth was repeated iteratively. A representative FACS scan of the GMD population prior to the third round sort is shown (Fig. 5B). The third round sort gate stringency was increased to 0.5% of SyTox Orange events (Fig. 5B, vertical bar). GMD falling in this gate were almost entirely yEGFP::ssLST. To quantify yEGFP::ssLST enrichment during the mock library screen, outgrown yeast cultures from each round of sorting were analyzed for yEGFP fluorescence by flow cytometry. Starting at a frequency of 0.01%, the yEGFP::ssLST was enriched 25-fold, 33-fold, and 11-fold in successive rounds of low stringency sorting (Fig. 6A). Ultimately, 93.7% of yeast from the third round sort were yEGFP::ssLST, yielding a total enrichment factor of 9,370-fold relative to the initial mixture.
Figure 5.
Enrichment of S. aureus lytic enzymes by GMD-FACS. Recombinant S. cerevisiae expressing eitheryEGFP::ssLST or LYZ were mixed at a ratio of 1:10,000 and then co-encapsulated with S. aureus. After 16h at 30°C, the colloidal mixture of agarose droplets was stained with SyTox Orange and analyzed by flow cytometry. In both panels, unsorted LYZ GMD are colored blue, unsorted yEGFP::ssLST GMD are green, and GMD falling inside experimental sort gates are orange. A: FACS dot plot of SyTox Orange versus GFP of the presort mixture. Sort gates (vertical bars) were set on the top 1% of SyTox Orange signal. Analysis of GFP signal (region R1) confirms the 10,000:1 input ratio of LYZ yeast to yEGFP::ssLST yeast. Yeast were recovered from sorted GMD, and the process of co-encapsulation, incubation and screening was repeated two more times. B: FACS dot plot of SyTox Orange vs. GFP during the third round sort. By the third round, the GFP positive population has been enriched to 4.57% (region R1). The vast majority of the GMDs falling in the third round SyTox Orange sort gate are yEGFP::ssLST clones (93.7%, as determined by FACS analysis of yeast outgrowth).
Figure 6.
Enrichment of antibiotic producing yeast by GMD-FACS screen. GMD containing yeast expressing either a negative control (LYZ), or a positive control (yEGFP::ssLST) for antibiotic production were mixed at a ratio of 10,000:1, and submitted to three rounds of GMD encapsulation, SyTox Orange staining, and FACS screening. Numerical fold enrichment statistics relative to the starting mixture are provided above each experiment. The maximum theoretical enrichment is 10,000-fold. A: Sort gates were set at low stringency (top 0.5–1% of the population) based on SyTox Orange staining, and the sorted yeast populations were outgrown and analyzed by FACS to determine the percentage of yEGFP::ssLST. Starting at a frequency of 0.01%, the yEGFP::ssLST was enriched to 0.25%, 8.32%, and 93.7% in successive rounds of sorting. B: Increasing the stringency of the SyTox Orange sort gates resulted in increased enrichment rates. A single high stringency sort (HS1, top 0.05%) resulted in 1,000-fold enrichment for antibiotic producers in a single round. Combining an initial low stringency sort with a subsequent high stringency sort (LS1-HS2) resulted in 4,130-fold enrichment in only two rounds. Combining two low stringency sorts with a final high stringency sort enriched the yEGFP::ssLST population to completion (LS1-LS2-HS3).
Using the same 1:10,000 presort mixture of yEGFP::ssLST and LYZ yeast, we also evaluated enrichment efficiencies using high stringency sort gates. Screening the initial mixed population with a strict 0.05% gate on SyTox Orange fluorescence yielded a single round enrichment of 1,042-fold (Fig. 6B, HS1). By combining an initial low stringency SyTox Orange sort (top 1%) with a subsequent high stringency sort (top 0.05%), an enrichment of 4,100-fold could be achieved in two rounds (Fig. 6B, LS1-HS2). Finally, when two 1% low stringency sorts were followed with a single 0.05% high stringency sort, complete enrichment (i.e., 10,000-fold) was achieved (Fig. 6B, LS1-LS2-HS3).
Construction and Screening of a Metagenomic Library for Discovery of Antibiotic Molecules
Building on the successful enrichments in our model system, we sought to implement the GMD-FACS screen with an experimental metagenomic library. Additionally, we used this opportunity to demonstrate the versatility of the GMD-FACS screen. With respect to GMD production, we supplanted the bulk homogenization method with a microfluidics platform capable of high-throughput, monodispersed droplet generation. Second, we replaced our eukaryotic yeast expression host with the common Gram-negative bacterial host E. coli.
A metagenomic library was constructed from the native plasmids of Staphylococcus simulans, Staphylococcus arlettae, and Staphylococcus equorum, each a natural environmental competitor of S. aureus. Plasmids were subjected to a limited DNaseI digestion, and fragments between 1,000 and 5,000 bp were isolated (Fig. 1). To facilitate high level protein expression, these fragments were cloned into the pET26b(+) expression vector, which drives transcription from the powerful T7 promoter. Transformation into an E. coli host yielded ~2 million clones.
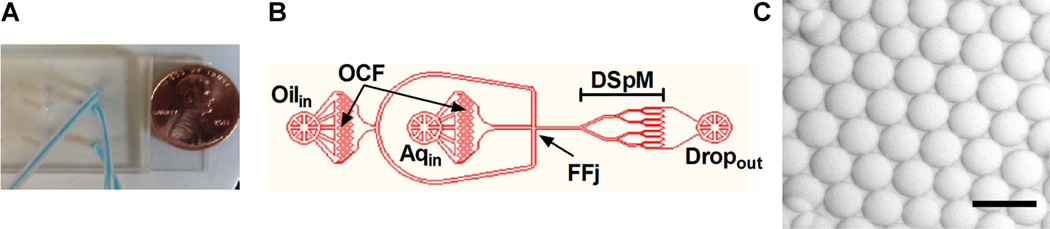
For screening, an overnight culture of the native Staphylococcus plasmid library was grown to exponential phase, treated with IPTG to induce expression, and mixed with a clinical isolate of S. aureus at a ratio that ensures, on average, <1 E. coli expression host and ~25 S. aureus per 25 pL droplet. A gas pressure system was used to deliver the molten agarose cell mixture to a PDMS microfluidic drop splitting device (Abate and Weitz, 2011) designed with a large diameter flow focusing arrangement that produces large agarose hydrogel droplets at a high volumetric rate. These droplets are subsequently fed to a drop-splitting array that ultimately divides the initial large droplet into eight equal portions (Fig. 7A–C). The size of droplets and rate of production are readily controlled by varying the oil verses the agarose feed pressures. Optimal drop productivity was achieved with a pressure ratio of 1.3:1 agarose:oil. Droplet size at this pressure ratio is ~27 µm in diameter, with <5% C.V. (Fig. 2B). Droplets were collected off-chip in 0.5 mL microfuge tubes, and microscopic analysis demonstrated that the liquid agarose droplets solidified rapidly when stored off-chip. The inherent stability of inverted emulsions in the fluorocarbon carrier oil prevented detectable droplet coalescence during incubations of 5+ days at 30°C (data not shown).
Figure 7.
High-throughput production of monodisperse GMD with microfluidic device. A: Drop-splitting microfluidic device mounted on glass microscope slide. B: A CAD schematic of a single device. Gas pressure pumps deliver PicoSurf oil through the oil inlet (Oilin), and molten agarose through the aqueous inlet (Aqin). The two immiscible fluids flow through on-chip filters (OCF) before meeting at a flow-focusing X-junction (FFj). Here, large droplets are formed in a jetting paradigm due to the Rayleigh–Plateau instability. Droplets then flow through a drop-splitting manifold (DSpM), comprised of a series of drop-splitting junctions. Finally, the surfactant-stabilized droplets exit the device through the outlet (Dropout). An AutoCAD file of the design can be downloaded from the supplementary online material. C: Micrograph of emulsified droplets generated with the microfluidic droplet splitter at a rate of ~10 million drops/h. Scale bar is 50 µm.
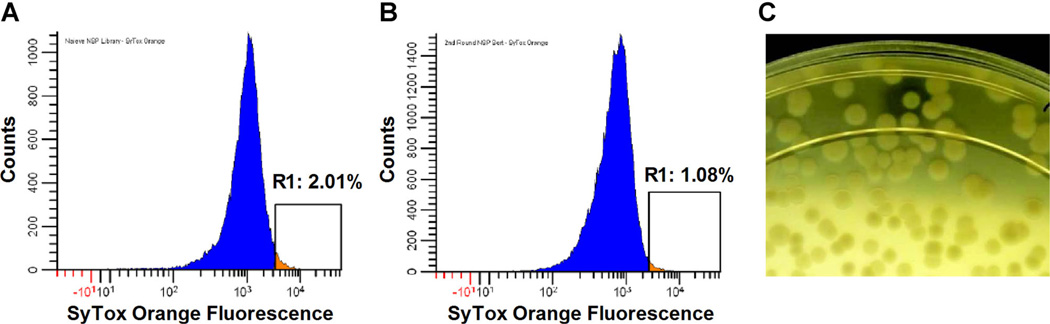
Emulsions were incubated at 30°C for 5–6 h to allow expression of metagenomic DNA and S. aureus killing. Following induction and incubation, the inverted emulsion was broken with 1H,1H,2H,2H–Perfluoro-1-octanol and stained with 500 nM SyTox Orange. The naïve library was screened by FACS using a sort gate set to the highest 2% of SyTox Orange events (Fig. 8A). Approximately 2 million clones were interrogated in “recovery” mode (~66% coverage of the library (Patrick et al., 2003)), and 55,000 events were collected. Sorted E. coli were outgrown in selective media and subjected to a second round of emulsification and FACS screening. A total of 455,000 clones were interrogated during the second round of screening (comprehensive coverage of the 55,000 member population) and 5,500 events were sorted in “recovery” mode using a 1% SyTox Orange gate (Fig. 8B). The second round-sorted events were not outgrown in liquid media, but were instead plated directly onto radial diffusion agar containing embedded S. aureus bacteria (Fig. 8C). Six clones that generated visible zones of clearance during 1 day of incubation at 30° C were restreaked to verify the S. aureus lytic phenotype and then archived. Sequencing their respective metagenomic inserts revealed that all six clones encoded the lysostaphin gene, which was derived from plasmid pACK1 of S. simulans.
Figure 8.
Selection of S. aureus lytic clones from a metagenomic library. Metagenomic DNA from environmental competitors of S. aureus was purified and cloned into an E. coli BL21(DE3) expression host. GMD emulsions were produced using a microfluidic device and then incubated for 5 h at 30°C. Droplets were stained with SyTox Orange and sorted using FACS. A: In the first round of sorting, >2 million GMD were interrogated in recovery mode. Using a 2% SyTox Orange sort gate, 55,000 GMD were sorted, and the isolated clones were amplified by overnight growth in selective media. B: In the second round of sorting, 455,000 GMD were interrogated and 5,500 GMD were sorted using a 1% SyTox Orange sort gate. C: Sorted clones were spread on radial diffusion agarose containing embedded USA400 S. aureus target cells. E. coli colonies producing zones of S. aureus clearance were picked and sequenced. The photograph has been uniformly contrast-enhanced to aid visualization of the positive clone (top center of plate).
Discussion
During the past decade, IVC techniques have found many applications in molecular engineering and the study of molecular evolution. IVC has been used for engineering catalytic activities (Agresti et al., 2010; Fallah-Araghi et al., 2012; Ghadessy et al., 2001, 2004; Lee et al., 2002; Paegel and Joyce, 2010), for discovery and engineering of binding proteins (Matochko et al., 2012; Sepp et al., 2002), and in screening chemical libraries (Brouzes et al., 2009). Non-biological compartmentalization of genotype and phenotype typically leverages water-in-oil emulsions in which a surfactant molecule forms a chemical partition between droplets. While these surfactant-stabilized droplets resist coalescence when incubated on the bench, the fact that they reside in a continuous oil phase precludes high-throughput screening techniques such as FACS. To enable FACS screening of IVC reactions, various bead capture techniques (Bertschinger and Neri, 2004; Griffiths and Tawfik, 2003; Levy et al., 2005) and product-gene linkage strategies (Doi and Yanagawa, 1999) have been devised, but these multistep techniques are labor intensive, time consuming, and not directly amenable to assaying for antibacterial activity against live pathogens. Direct FACS screening of live microbes within IVC has been accomplished previously using water-in-oil-in-water (w/o/w) double emulsions (Aharoni et al., 2005). In comparison, GMD are more amenable to various manipulations such as concentration by centrifugation, media or buffer exchange during the course of incubation, or timed addition of enzyme inhibitors, reactants, or molecular probes. Moreover, compared to double emulsions, GMD are more compatible with FACS analysis. It bears noting, however, that FACS screening of GMD requires an insoluble or at least diffusion limited substrate.
Recently, impressive developments in microfluidic technology have been used to make monodisperse emulsions using laminar flow paradigms. These approaches enable precise control over droplet size and production rate, and have opened the door to droplet splitting, merging, mixing, off-chip incubation and re-injection, and on-chip fluorescence-activated sorting at high speed (recently reviewed in Kintses et al. (2010)). While these technologies represent a paradigm shift relative to bulk-emulsion IVC techniques, the technology requires elaborate set-up and expertise in fluid dynamics and microelectronics, rendering the methods inaccessible to the average biochemistry lab.
We have developed a high-throughput screening methodology suitable for use in discovery of natural product antibiotics. The method employs an IVC scheme utilizing an agarose hydrogel as the aqueous phase, thereby creating hydrogel-in-oil emulsions in which a recombinant host microorganism can be co-cultured with a bacterial pathogen target. A key aspect of this screen is the use of a micron scale hydrogel to physically link the recombinant host and bacterial pathogen. This feature enables high speed quantitative screening and sorting by conventional flow cytometry, based on selective labeling with a fluorogenic viability probe.
We report the use of bulk emulsification as a technique to generate IVC material suitable for screening libraries for antibiotic activity. Emulsions are fabricated on heated stir plates by adding agarose drop-wise to solutions of stirring mineral oil/Span80. After sufficient time, the emulsion is cooled, solidifying the agarose. Emulsions are then broken, and colloidal suspensions of agarose microdroplets are suspended in nutrient growth media or assay buffer. Because agarose mineral oil and Span80 are inexpensive lab reagents, large quantities (25 mL) of highly polydisperse emulsion can be economically produced and then sieved with nylon mesh filters to focus the droplet size over a narrow range appropriate for FACS; 30–40 µm droplets were used in this study, but readily available nylon mesh filters having a wide range of pore diameters may be employed to isolate droplets of any size. Using this method, one can easily screen libraries exceeding 10 million unique clones in a single experiment. We implemented this technique to isolate recombinant yeast engineered to secrete the antibacterial biocatalyst ssLST from a mixture where it was present at an exceptionally low frequency (1:10,000). By setting high stringency sort gates, we were able to achieve enrichment factors exceeding 1,000-fold in a single round of sorting.
Using bulk emulsification, we rely on the extreme proximity of the antibiotic producing microbe and the target bacterium to facilitate interaction between bioactive molecules and target bacteria. While yielding tens of millions of size-appropriate GMD in minutes, bulk emulsification is limited by the fact that solidified GMD are suspended in aqueous growth media, and secreted biomolecules will therefore diffuse passively out of their source hydrogel. Bioactive molecules can then cross into other GMD, potentially confounding the genotype-phenotype linkage. This limitation is effectively circumvented by simply diluting the suspension into a large volume of aqueous media. Fifty millilitres of media was used for experiments in which >5 million GMD were screened at a “hit rate” of 0.01%. For larger library sizes or higher hit rates, the volume of aqueous media should be scaled appropriately. Because the GMD can be effectively pelleted by low speed centrifugation, recovery from large volumes of growth media is facile.
To concentrate secreted gene products within the GMD over time, we sought to incubate the microbe-containing hydrogel droplets in emulsion. Bulk emulsification of agarose-in-mineral oil/Span80 resulted in unstable emulsions that were not suitable for in emulsion incubation. As a result, we implemented a previously described drop-splitting microfluidic device for generation of monodisperse hydrogel-in-oil emulsions (Abate and Weitz, 2011). We used the commercially available oil/surfactant mix PicoSurf1, a fluorinated oil/ surfactant that is reported to be biocompatible and highly stable as the continuous phase. By concentrating the secreted bioactive molecules in emulsion, we expected an increase in the sensitivity of the assay compared to the bulk emulsification technique. Indeed, by performing in emulsion co-culture of S. cerevisiae secreting lysostaphin and S. aureus, we were able to reduce the incubation time necessary to generate a SyTox signal from 16 to 6h in emulsion (data not shown).
Using a gas pressure pump constructed from inexpensive materials, we fabricated a droplet maker with a productivity of ~10 million droplets/h. While this level of throughput was sufficient for the experiments described here, we note that, in contrast to common syringe pump set-ups, the gas pressure pump described here can be parallelized by simply adding inlet lines to the aqueous and oil phases. We currently use a PDMS device on which seven drop-splitting devices are printed on a standard glass microscope slide. Thus, without any modifications to the system or any additional equipment, the system can be scaled to 70 million droplets/h.
We used the in emulsion GMD-FACS assay to screen recombinant E. coli expressing large libraries of eDNA isolated from S. simulans, S. arlettae, and S. equorum, which are environmental competitors of S. aureus. Greater than 1 million clones were screened, and ssLST was efficiently isolated in less than 4 days (Fig. 8). It bears noting that ssLST cleaves pentaglycine crosslinks in peptidoglycan, and these crosslinks are absent in the cell wall of E. coli. We speculate that other antimicrobials encoded in our library were lost as a result of expression host toxicity, a possibility that underscores the importance of our highly adaptable platform, that is our system permits the use of any expression host capable of co-culture with the target pathogen.
The approach reported here is a natural extension of previous work in which metagenomic libraries were screened for antibiotic activity using standard agar overlay methodology and an E. coli expression host (Brady, 2007). Our partially automated workflow enables greater throughput in shorter timeframes, thereby raising the bar for antibiotic screens. We anticipate that GMD-FACS screening of larger more complex libraries will yield a commensurate increase in the number of bioactive antibiotics discovered.
Conclusion
In summary, we report the validation of a novel ultra-high-throughput screen for antibiotic drug discovery. Our platform benefits from miniaturization of reaction volumes by IVC and the unparalleled speed of FACS. We designed this methodology so as to provide screening throughput better matched to the enormous diversity of antimicrobial agents that is likely hidden behind the veil of unculturable microbes. Moreover, our platform will enable combinatorial protein engineering projects geared towards development of functionally enhanced antibiotics, efforts that are currently bottlenecked by screening limitations. Importantly, our approach is relatively low tech and therefore accessible to the average biochemistry lab, provided access to a flow cytometry core facility; our lab is not a traditional microfluidics group, and the entire microfluidics set-up (chips, vials, tubing, microscope, etc.) can be acquired for less than $2,000. We expect that the system and procedures described here will facilitate similar screening efforts in other laboratories.
Acknowledgments
This work was supported by the NIH National Institute of Allergy and Infectious Disease, grants 1R21AI094391 and 1R21AI098122 (awarded to K.E.G.). T.C.S. acknowledges the Carol Basbaum Memorial Postdoctoral Fellowship (SCANLO08F0) from the Cystic Fibrosis Foundation.
Abbreviations
- MRSA
methicillin-resistant Staphylococcus aureus
- GMD
gel microdroplet
- FACS
fluorescence-activated cell sorting
- DGMC-URA
dextrose growth media with casamino acids
- GIM-URA
galactose induction media
- PDMS
polydimethylsiloxane
- yEGFP
yeast-enhanced GFP
- ssLST
Staphylococcus simulans lysostaphin
- LYZ
human lysozyme
- IVC
in vitro compartmentalization
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
References
- Abate AR, Weitz DA. Faster multiple emulsification with drop splitting. Lab Chip. 2011;11(11):1911–1915. doi: 10.1039/c0lc00706d. [DOI] [PubMed] [Google Scholar]
- Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, Baret JC, Marquez M, Klibanov AM, Griffiths AD, Weitz DA. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc Natl Acad Sci USA. 2010;107(9):4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, Amitai G, Bernath K, Magdassi S, Tawfik DS. High-throughput screening of enzyme libraries: Thiolactonases evolved by fluorescence-activated sorting of single cells in emulsion compartments. Chem Biol. 2005;12(12):1281–1289. doi: 10.1016/j.chembiol.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Athanassa Z, Siempos II, Falagas ME. Impact of methicillin resistance on mortality in Staphylococcus aureus VAP:A systematic review. Eur Respir J. 2008;31(3):625–632. doi: 10.1183/09031936.00081007. [DOI] [PubMed] [Google Scholar]
- Bamberger DM, Boyd SE. Management of Staphylococcus aureus infections. Am Fam Physician. 2005;72(12):2474–2481. [PubMed] [Google Scholar]
- Banik JJ, Brady SF. Cloning and characterization of new glycopeptide gene clusters found in an environmental DNA megalibrary. Proc Natl Acad Sci USA. 2008;105(45):17273–17277. doi: 10.1073/pnas.0807564105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera A, Herbert S, Jakob A, Vollmer W, Gotz F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus . Mol Microbiol. 2005;55(3):778–787. doi: 10.1111/j.1365-2958.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- Bertschinger J, Neri D. Covalent DNA display as a novel tool for directed evolution of proteins in vitro. Protein Eng Des Sel. 2004;17(9):699–707. doi: 10.1093/protein/gzh082. [DOI] [PubMed] [Google Scholar]
- Brady SF. Construction of soil environmental DNA cosmid libraries and screening for clones that produce biologically active small molecules. Nat Protoc. 2007;2(5):1297–1305. doi: 10.1038/nprot.2007.195. [DOI] [PubMed] [Google Scholar]
- Brady SF, Chao CJ, Clardy J. Long-chain N-acyltyrosine synthases from environmental DNA. Appl Environ Microbiol. 2004;70(11):6865–6870. doi: 10.1128/AEM.70.11.6865-6870.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML. Droplet microfluidic technology for single-cell high-throughput screening. Proc Natl Acad Sci USA. 2009;106(34):14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci USA. 2002;99(16):10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel R. The soil metagenome—A rich resource for the discovery of novel natural products. Curr Opin Biotechnol. 2004;15(3):199–204. doi: 10.1016/j.copbio.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Doi N, Yanagawa H. STABLE: Protein-DNA fusion system for screening of combinatorial protein libraries in vitro. FEBS Lett. 1999;457(2):227–230. doi: 10.1016/s0014-5793(99)01041-8. [DOI] [PubMed] [Google Scholar]
- Eun YJ, Utada AS, Copeland MF, Takeuchi S, Weibel DB. Encapsulating bacteria in agarose microparticles using microfluidics for high-throughput cell analysis and isolation. ACS Chem Biol. 2010;6(3):260–266. doi: 10.1021/cb100336p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallah-Araghi A, Baret JC, Ryckelynck M, Griffiths AD. A completely in vitro ultrahigh-throughput droplet-based microfluidic screening system for protein engineering and directed evolution. Lab Chip. 2012;12(5):882–891. doi: 10.1039/c2lc21035e. [DOI] [PubMed] [Google Scholar]
- Garau J, Bouza E, Chastre J, Gudiol F, Harbarth S. Management of methicillin-resistant Staphylococcus aureus infections. Clin Microbiol Infect. 2009;15(2):125–136. doi: 10.1111/j.1469-0691.2009.02701.x. [DOI] [PubMed] [Google Scholar]
- Ghadessy FJ, Ong JL, Holliger P. Directed evolution of polymerase function by compartmentalized self-replication. Proc Natl Acad Sci USA. 2001;98(8):4552–4557. doi: 10.1073/pnas.071052198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadessy FJ, Ramsay N, Boudsocq F, Loakes D, Brown A, Iwai S, Vaisman A, Woodgate R, Holliger P. Generic expansion of the substrate spectrum of a DNA polymerase by directed evolution. Nat Biotechnol. 2004;22(6):755–759. doi: 10.1038/nbt974. [DOI] [PubMed] [Google Scholar]
- Gillespie DE, Brady SF, Bettermann AD, Cianciotto NP, Liles MR, Rondon MR, Clardy J, Goodman RM, Handelsman J. Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Appl Environ Microbiol. 2002;68(9):4301–4306. doi: 10.1128/AEM.68.9.4301-4306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths AD, Tawfik DS. Directed evolution of an extremely fast phosphotriesterase by in vitro compartmentalization. EMBO J. 2003;22(1):24–35. doi: 10.1093/emboj/cdg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem Biol. 1998;5(10):R245–R249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- Hassan M, Tuckman HP, Patrick RH, Kountz DS, Kohn JL. Cost of hospital-acquired infection. Hosp Top. 2010;88(3):82–89. doi: 10.1080/00185868.2010.507124. [DOI] [PubMed] [Google Scholar]
- Iqbal HA, Feng Z, Brady SF. Biocatalysts and small molecule products from metagenomic studies. Curr Opin Chem Biol. 2012;16(1-2):109–116. doi: 10.1016/j.cbpa.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EW. Tackling the protease problem in Saccharomyces cerevisiae . Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- Kintses B, van Vliet LD, Devenish SR, Hollfelder F. Microfluidic droplets: New integrated workflows for biological experiments. Curr Opin Chem Biol. 2010;14(5):548–555. doi: 10.1016/j.cbpa.2010.08.013. [DOI] [PubMed] [Google Scholar]
- Lee YF, Tawfik DS, Griffiths AD. Investigating the target recognition of DNA cytosine-5 methyltransferase HhaI by library selection using in vitro compartmentalisation. Nucleic Acids Res. 2002;30(22):4937–4944. doi: 10.1093/nar/gkf617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Griswold KE, Ellington AD. Direct selection of trans-acting ligase ribozymes by in vitro compartmentalization. RNA. 2005;11(10):1555–1562. doi: 10.1261/rna.2121705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrobattista E, Taly V, Chanudet E, Treacy P, Kelly BT, Griffiths AD. High-throughput screening of enzyme libraries: in vitro evolution of a beta-galactosidase by fluorescence-activated sorting of double emulsions. Chem Biol. 2005;12(12):1291–1300. doi: 10.1016/j.chembiol.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Matochko WL, Ng S, Jafari MR, Romaniuk J, Tang SK, Derda R. Uniform amplification of phage display libraries in monodisperse emulsions. Methods. 2012;58(1):18–27. doi: 10.1016/j.ymeth.2012.07.012. [DOI] [PubMed] [Google Scholar]
- Mazutis L, Gilbert J, Ung WL, Weitz DA, Griffiths AD, Heyman JA. Single-cell analysis and sorting using droplet-based microfluidics. Nat Protoc. 2013;8(5):870–891. doi: 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: Comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22(25):5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156(1):119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Narayanan J, Xiong J-Y, Liu X-Y. Determination of agarose gel pore size: Absorbance measurements vis a vis other techniques. J Phys: Conf Ser. 2006;28(1):83. [Google Scholar]
- Paegel BM, Joyce GF. Microfluidic compartmentalized directed evolution. Chem Biol. 2010;17(7):717–724. doi: 10.1016/j.chembiol.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick WM, Firth AE, Blackburn JM. User-friendly algorithms for estimating completeness and diversity in randomized protein-encoding libraries. Protein Eng. 2003;16(6):451–457. doi: 10.1093/protein/gzg057. [DOI] [PubMed] [Google Scholar]
- Recsei PA, Gruss AD, Novick RP. Cloning, sequence, and expression of the lysostaphin gene from Staphylococcus simulans . Proc Natl Acad Sci USA. 1987;84(5):1127–1131. doi: 10.1073/pnas.84.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon TC, Gray EC, Griswold KE. Quantifying and resolving multiple vector transformants in S. cerevisiae plasmid libraries. BMC Biotechnol. 2009;9:95. doi: 10.1186/1472-6750-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon TC, Teneback CC, Gill A, Bement JL, Weiner JA, Lamppa JW, Leclair LW, Griswold KE. Enhanced antimicrobial activity of engineered human lysozyme. ACS Chem Biol. 2010;5(9):809–818. doi: 10.1021/cb1001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepp A, Tawfik DS, Griffiths AD. Microbead display by in vitro compartmentalisation: Selection for binding using flow cytometry. FEBS Lett. 2002;532(3):455–458. doi: 10.1016/s0014-5793(02)03740-7. [DOI] [PubMed] [Google Scholar]
- Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae . Yeast. 2004;21(8):661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- Taubes G. The bacteria fight back. Science. 2008;321(5887):356–361. doi: 10.1126/science.321.5887.356. [DOI] [PubMed] [Google Scholar]
- Tumarkin E, Tzadu L, Csaszar E, Seo M, Zhang H, Lee A, Peerani R, Purpura K, Zandstra PW, Kumacheva E. High-throughput combinatorial cell co-culture using microfluidics. Integr Biol (Camb) 2011;3(6):653–662. doi: 10.1039/c1ib00002k. [DOI] [PubMed] [Google Scholar]
- Zhang H, Jenkins G, Zou Y, Zhu Z, Yang CJ. Massively parallel single-molecule and single-cell emulsion reverse transcription polymerase chain reaction using agarose droplet microfluidics. Anal Chem. 2012;84(8):3599–3606. doi: 10.1021/ac2033084. [DOI] [PubMed] [Google Scholar]
- Ziemert N, Ishida K, Weiz A, Hertweck C, Dittmann E. Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Appl Environ Microbiol. 2010;76(11):3568–3574. doi: 10.1128/AEM.02858-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotchev SB, Sekurova ON, Katz L. Genome-based bioprospecting of microbes for new therapeutics. Curr Opin Biotechnol. 2012;23(6):941–947. doi: 10.1016/j.copbio.2012.04.002. [DOI] [PubMed] [Google Scholar]