Summary
Due to the key role of post-translational modifications (PTMs) such as lysine acetylation in numerous signaling and regulatory pathways, the ability to identify novel PTMs and to quantify their abundances provides invaluable information for understanding these signaling networks. Currently, mass spectrometry (MS) arguably serves as the most high-throughput and unbiased platform for studying post-translational modifications. Here we detail experimental and analytical procedures for the characterization of lysine acetylation on proteins in general and on histones in particular, which are among the most highly modified proteins in eukaryotic cells.
Keywords: Mass spectrometry, lysine acetylation, post-translational modification, proteomics, histones
1. Introduction
The identification of post-translational modifications is often more technically challenging than the identification of the modified protein itself for many both biological and technical reasons. First, the complexity of modifications, ranging from small groups such as acetylation to highly branched moieties such as poly-ADP-ribosylation, vastly dwarves the sequence complexity found with the 20 canonical amino acids. Second, most modifications occur at sub-stoichiometric levels relative to the total protein pool, often rendering the detection of such modifications below the dynamic range of the method used. Third, most enzymatically catalyzed modifications are reversible, and thus are dynamic in nature and have limited transient lifetimes. Undeniably the most extensively studied class of modifications in molecular biology is serine/threonine/tyrosine phosphorylation, where such investigations are greatly facilitated by several methods including 32P-radiolabeling, phosphorylation-specific antibodies, phosphospecific gel staining and enrichement using various approaches such as immobilized metal affinity chromatography (1-4).
In contrast to phosphorylation, relatively fewer tools are available for the identification of lysine acetylation. Such methodological deficit is particularly disconcerting given recent investigations identifying approximately 3500 acetylated residues in mammalian cells (5,6, comparable to the approximately >10,000 phosphorylated residues identified also in mammalian cells (7). Additionally, the number of reports describing protein phosphorylation far surpasses those studies characterizing acetylated proteins. Mass spectrometry has emerged as the most rigorous platform for both the identification and quantification of lysine acetylation (8). Essentially using the same logic as Edman degradation using using phenylthiohydantoin or dansyl-chloride, MS sequencing of a putative acetylated peptide relies on the fragmentation of a peptide into smaller peptide fragments sharing a common N- or C-terminus. The difference in mass between peptide fragments sharing either a common N- or C-terminus essentially corresponds to the residue mass of a single additional amino acid between the fragments. When that difference cannot be accounted by the residue mass of lysine alone but rather by the mass of lysine plus a nominal mass shift of 42 Da (or more accurately a mass of 42.010 Da), the possibility of an acetylated rather than unmodified lysine becomes highly likely. In this chapter, we detail the latest advancements in the MS analysis of lysine acetylation for proteins in general and for histones in particular, which require a more specialized workflow.
2. Materials
All reagents and samples should be handled with no contact to bare skin in order to minimize keratin contamination or to any detergents such as those used in glassware cleaning. Unless otherwise noted, all reagents can be stored at room temperature
2.1 Preparation of protein samples for MS analysis
Sequencing grade trypsin, stored at −80°C (see Note 1).
100mM ammonium bicarbonate, pH 8, stored at 4°C (see Note 2).
10 mM dithiothreiotol in 100 mM ammonium bicarbonate, prepared fresh prior to use.
55 mM iodoacetamide in 100 mM ammonium bicarbonate, prepared fresh prior to use and stored in dark.
Acetonitrile (see Note 3).
Glacial acetic acid
Peptide desalting kits, such as ZipTip pipette tips (Millipore) or Proxeon StageTips C18 material (Thermo Scientific) (see Note 4).
Razor blades (only if proteins were resolved by SDS-PAGE)
2.2 Preparation of histone samples for MS analysis
3. Methods
SDS-PAGE of protein samples is best reserved for complex solutions containing approximately 10 or more expected proteins. Using SDS-PAGE for relatively simple solutions will reduce the digestion efficiency and increase the risk of introducing acrylamide adducts that will complicate peptide sequencing. Highly complex protein samples containing approximately 100 or more expected proteins should be fractionated, for instance via reversed phase liquid chromatography, prior to SDS-PAGE. Similar logic is applicable to histone preparations, depending on the efficiency of the purification method used. All steps, unless otherwise noted, occur in room temperature.
3.1 In-gel digestion of protein bands and peptide extraction from gel bands
-
1
Excise out desired protein bands (see Note 6) with clean razor blades, and dice into approximately 1 mm3 pieces. Place gel pieces within a 1.5 ml tube. Ensure razor blades are wiped clean with methanol prior to cutting another band to minimize cross contamination between samples.
-
2
If bands were stained with coomassie blue, destain the band for at least 1 hour to reduce the amount of residual coomassie that eventually could be introduced into the mass spectrometer. If bands are irreversibly stained, proceed to next step.
-
3
Replace destain solution with 100 mM ammonium bicarbonate and shake or rotate vigorously for 5 minutes. Remove as much ammonium bicarbonate as possible and dehydrate gel slices using a vacuum centrifuge (see Note 7).
-
5
Add a sufficient volume of 10 mM dithiothreitol to cover gel and reduce samples for 1 hour at 51°C. Afterwards, allow tubes to cool to room temperature and replace dithiothreitol with equal volume of 55 mM iodoacetamide. Incubate for 1 hour in the dark.
-
6
Remove iodoacetamide and wash gel pieces with 100 mM ammonium bicarbonate by shaking/rotating for 5 minutes. After the wash, replace ammonium bicarbonate with acetonitrile to dehydrate the gel band and shake/rotate for 5 minutes. Remove acetonitrile and dry gel pieces completely.
-
7
Swell gels with minimal volume (i.e. enough to submerge the gel pieces) of 12.5 ng/μl sequencing grade trypsin in 100 mM ammonium bicarbonate for approximately 30 minutes on ice. After gel pieces are fully swollen, incubate samples at room temperature for 6-12 hours (see Note 8).
-
8
Collect solution that was not absorbed within gel into another tube, and dehydrate with minimal volume of 75% acetonitrile/5% acetic acid while shaking/rotating for 5-10 minutes. Afterwards, pool solution with the previously collected digest solution in the new tube and rehydrate gel pieces with minimal volume of 100 mM ammonium bicarbonate while shaking/rotating for 5-10 minutes. Pool this solution into the same new tube, and repeat the dehydration/rehydration step 1 additional time.
-
9
After pooling the 100 mM ammonium bicarbonate, wash for the second time, dehydrate the gel with 100% acetonitrile while shaking/rotating for 5-10 minutes. Without collecting the acetonitrile, add 100 mM ammonium bicarbonate at 3× excess volume of the acetonitrile solution while shaking/rotating for 5-10 minutes. Pool the acetonitrile/ammonium bicarbonate solution into the same new tube as before. Repeat the dehydration/rehydration step 1 additional time. Remove the acetonitrile content in the pooled solution using a vacuum centrifuge prior to desalting using commercially available kits.
3.2 In-solution digestion of protein samples
Adjust pH of the solution to pH 7.5-9, and determine protein concentration using the most reliable assay available. Add dithiothreitol to final concentration of 10 mM and reduce for 1 hour at 51°C. Add excess volume of iodoacetamide for final concentration of approximately 55 mM and alkylate for 45 minutes in dark.
Dry the solution to near completeness, with residual volume < 5 μl using a vacuum centrifuge, and add sequencing grade trypsin at a 1:10-1:20 enzyme:substrate concentration. Allow to digest for approximately 5-6 hours at 37°C. Quench reaction with addition of glacial acetic acid to approximately pH = 4. Samples are now ready for desalting using commercially available kits. (see Note 4).
3.3 In-solution derivatization of histone samples (9)
Adjust pH of histone samples to approximately pH 8. Determine histone concentration using the most convenient assay available. If histones are dried, solubilize the proteins with 100 mM ammonium bicarbonate. Prepare a 3:1 isopropanol:propionic anhydride reagent by volume and add reagent to histone sample at approximately half sample volume for derivatization.
Quickly adjust pH of samples with addition of ammonium hydroxide to return to pH 8. If samples become too basic at pH ≥ 10, adjust by addition of isopropanol:propionic anhydride reagent. Once pH is adjusted, incubate samples at 37°C for 15 minutes.
Reduce sample to original volume using a vacuum centrifuge, and repeat the derivatization 1 additional time. Afterwards, add 100 mM ammonium bicarbonate at 3-4× excess volume of the sample and digest with sequencing grade trypsin at 1:20 enzyme:substrate concentration at 37°C for 5-6 hours. Quench digestion with addition of acetic acid to pH 4.
Reduce sample volume, and after readjusting to pH 8, repeat derivatization method 2 times. After the second derivatization of the tryptic peptides, histone peptides are now ready for desalting using commercially available kits.
3.4. LC-MS operation and analysis of MS data
Depending on the instrument available, there are various setups for peptide sequencing. This chapter assumes upfront sample separation with an online HPLC followed either by a linear ion trap quadrupole mass spectrometer or a hybrid linear ion trap-orbitrap mass spectrometer.
The optimal HPLC gradient should be determined for each protein sample. Gradient parameters include the buffer composition, rate of change of the buffers, and gradient duration. Generally, reversed-phase chromatography provides a versatile gradient for resolving most tryptic peptides.
For non-histone proteins, a typical instrument method would comprise the acquisition of 1 full mass spectrum (MS) followed by 5-10 data-dependent tandem mass spectra (MS/MS) for the 5-10 most abundant peptide ions detected from the mass spectrum. Collisional induced dissociation (CID) of the precursor ion should be appropriate for most tryptic peptides. (see Note 9)
After the MS run is completed, proceed to a bioinformatic search of putative acetylated peptides. Various software packages are available commercially, and other programs are developed in-house. Note that one needs to specify the addition of acetamide (+57.043 Da) on cysteines in the program used, the possible oxidation (+15.995 Da) of methionine, which can arise during sample preparation or electrospray ionization under high voltage, and the possible acetylation (+42.010 Da) on lysine. For propionylated histone samples, one should specify the addition of a propionyl group (+56.026 Da) on the N-terminus, on unmodified (+56.026 Da) and monomethylated lysines (+70.042 Da = 56.026 Da +14.016 Da) rather than the acetamide derivatization of cysteines. It is important to allow for several (e.g. 1-4) mis-cleavages by trypsin, since the protease will not cleave at an acetylated lysine.
3.5. Validation of putative acetylated lysines on histone and non-histone proteins
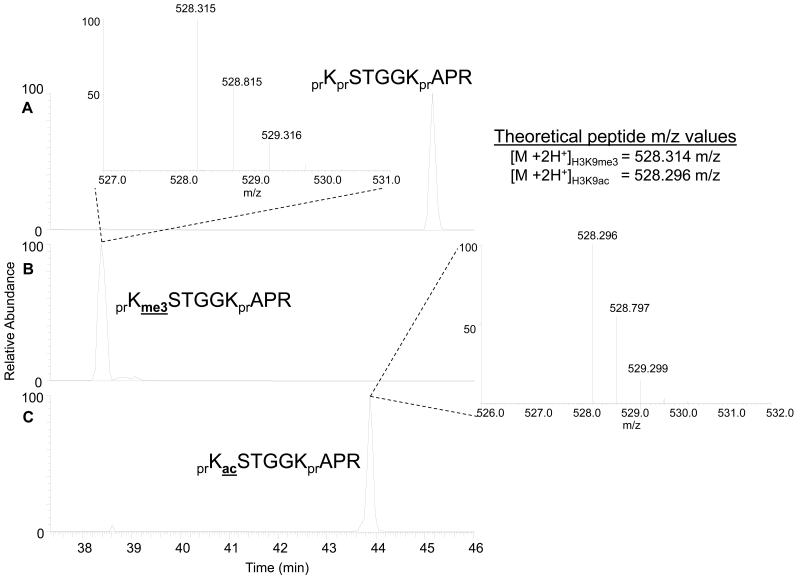
Confirm whether the precursor mass found in the MS is consistent with an acetyl moiety (42.010 Da) or the nearly isobaric trimethyl moiety (42.046 Da) if the samples were analyzed on a high-mass accuracy mass spectrometer, such as the hybrid linear ion trap-Orbitrap this should be possible (see Note 10). Shown in Figure 1 are the total ion chromatograms for the doubly charged ions from three histone peptides: the histone H3 unmodified 9-17 peptide prKprSTGGKprAPR (Fig.1A), the H3K9me3 9-17 peptide prKme3STGGKprAPR (Fig. 1B) and the H3K9ac 9-17 peptide prKacSTGGKprAPR (Fig. 1C). Note that pr = propionyl group added to unmodified lysines. Differentiation of the H3K9me3 and H3K9ac peptides based on accurate mass can be accomplished, as the mass of the peptide in Figure 1B is closer to the calculated mass of the H3K9me3 (1.89 ppm error for me3 compared to 35.9 ppm error for H3K9ac), while the mass of the peptide in Figure 1C is closer to the calculated mass of the H3K9ac (0 ppm error for me3 compared to 34 ppm error for H3K9ac).
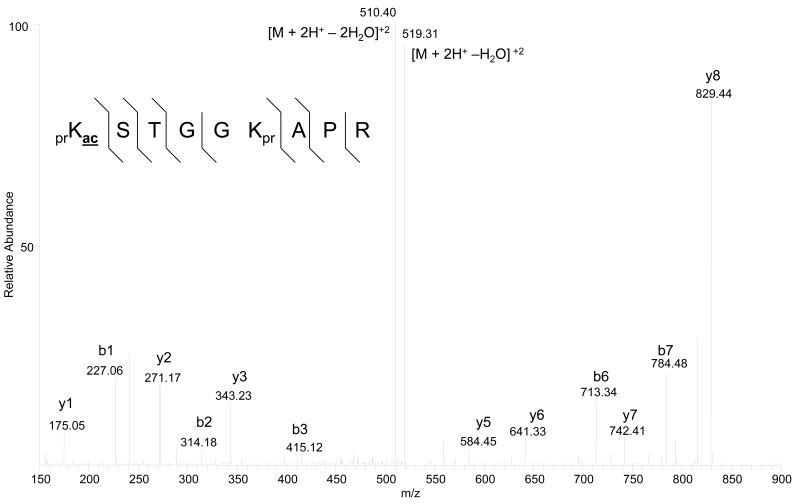
Inspect manually the MS/MS corresponding to the putative acetylated peptide. First confirm the presence of particular b- (n-terminal) and y- (c-terminal) ion fragments that arise only from the acetylated residue. For instance, in the 9-17 histone H3 peptide prKacSTGGKprAPR (pr = propionyl group added to unmodified lysines), the presence of the expected y4-y8 fragment ions is more critical than the presence of the expected y1-y3 fragment ions for localizing the acetyl moiety to K9 rather than K14 (Figure 2). Similar logic can be applied for finding the appropriate b-ion series. One should not only be concerned with the extent of b- and y-ion coverage, but also the overall noise of the tandem mass spectrum. Maximum b- and y-ion coverage is readily observed for a very noisy tandem mass spectrum. An ideal MS/MS should contain relatively few ions that cannot be accounted by the expected CID fragmentation pattern (see Note 11).
Confirm that the acetylated histone peptide elutes earlier than the respective unmodified histone peptide if a reversed phase gradient was used, due to the absence of a propionyl group on the acetylated peptide and thus a decrease in relative hydrophobicity. Furthermore, a peptide containing an acetylated lysine should elute later than the same peptide containing a trimethylated lysine, due to charge stabilization on the side-chain amine from the three methyl groups compared to charge removal on the amine from the acetyl group (see Note 12).
Fig 1.
Extracted ion chromatograms of the doubly charged 9-17 peptide (KSTGGKAPR) on histone H3 unmodified on K9 (A), trimethylated on K9 (B), and monoacetylated on K9 (C). Differences in retention time under reversed phase chromatography facilitate PTM assignment of propionylated histone peptides. In general, the acetylated peptide elutes earlier than the unmodified peptide, which elutes earlier than the monomethylated peptide. Inserts depict the MS of the respective trimethylated and monoacetylated histone peptides acquired in the Orbitrap. Note that the spacing of the isotopes (~0.5 m/z) indicates the histone peptide as doubly charged. Abbreviations: pr = propionyl, me3 = trimethyl, ac1 = acetyl.
Fig 2.
Tandem mass spectrum of the monoacetylated H3 histone 9-17 peptide, KacSTGGKAPR, with the expected and observed mass of the doubly charged molecular ion [M+2H+] provided. The MS/MS was acquired from CID fragmentation in the linear ion trap, while the precursor peptide mass [M+2H+] was determined from the orbitrap. Hatched lines above and below the peptide sequence correspond to b- and y-ions respectively that were positively annotated in the tandem mass spectrum. Abbreviation: [M+2H+ - H2O]+2 = doubly charged ion with loss of 1 water molecule, [M+2H+ - 2H2O]+2 = doubly charged ion with loss of 2 water molecules.
6. Acknowledgment
BMZ is supported by the NSF Graduate Research Fellowship Program and BAG gratefully acknowledges support from a National Science Foundation (NSF) Early Faculty CAREER award, an NIH Innovator award (DP2OD007447) from the Office Of The Director, NIH, and an American Society for Mass Spectrometry research award sponsored by the Waters Corporation. We also thank all members of the Garcia lab for helpful discussion during the composition of this chapter.
Footnotes
Despite the catalytic robustness of trypsin, the protease should not undergo multiple freeze-thaw cycles for maximal digestion efficiency. Do not substitute with tissue culture-grade trypsin, due to the increased likelihood of autolysis and reduced substrate specificity resulting from the digested trypsin. Furthermore, one may substitute the ammonium bicarbonate buffer with the accompanied buffer provided by the vendor if the mass spectrometer detects a high abundance of undigested intact proteins. The use of ammonium bicarbonate is simply for convenience with later preparation steps.
Over time, the ammonium bicarbonate buffer may lose its buffering capacity. Thus, one should confirm the buffer pH = 8 prior to use.
One does not have to use HPLC-grade acetonitrile for gel desiccation. Standard ACS grade acetonitrile reagent is sufficient.
Desalting helps prevent the reduction in ionization efficiency during electrospray ionization and to reduce the likelihood of salt adducts (such as Na+ or K+ or buffer salts) forming with the peptide ions. One should be mindful that the desalting resin used should be the same as or similar to the resin used for online LC separation of the peptides during MS analysis. Desalting should only be omitted if one knows a priori that the peptide(s) of interest are unusually hydrophilic (and thus will not be retained on the resin) or that the peptide(s) are of very low abundance (and thus more likely to suffer from sample loss during the desalting procedure).
To help maintain anhydrous conditions for isopropanol and propionic anhydride, one can purge the reagent containers with nitrogen (N2) or argon every time the container is opened, and securely seal the lid. Excess moisture in the reagents will reduce the efficiency of derivatization.
Be mindful that the actual and apparent molecular weights of proteins are not necessarily equal. The migration distance of proteins in SDS-PAGE, although determined mostly on the basis of size, can be influence by other parameters such as phosphorylation and overall charge state.
If one does not have access to a vacuum centrifuge, one could leave the samples to air-dry at room temperature for an extended period of time.
The optimal digestion temperature of sequencing-grade trypin is approximately 37°C. However, trypsin digestion can proceed in a relatively broad range of temperatures. If one knows that the ambient temperature in the laboratory has a relatively wide fluctuation, one should digest the samples in a 25°C to 37°C heat block or water bath.
If one digests the protein or histone samples with an alternative protease, such as Arg-C, a different fragmentation method may be more suitable for peptide sequencing. Proteases such as Arg-C have an increased likelihood to produce peptides of higher charge states than trypsin, due to the presence of internal lysines on the peptides. Higher charge states (z ≥ 3) lead to increased likelihood for internal CID fragmentation of the peptide, as opposed to fragmentation from the N- or C-terminus, and vastly complicates the sequencing process. Furthermore, because CID is known as a non-ergodic process, the efficiency of CID fragmentation decreases as peptide length increases. An alternative fragmentation scheme potentially more conducive to ArgC digests (or other similarly non-tryptic proteases) is electron transfer dissociation (ETD), which is less sensitive to longer peptide length and more conducive towards higher-charged peptides.
If one intends to use a hybrid linear ion trap-orbitrap mass spectrometer for peptide sequencing, one should perform external mass calibration prior to the experiments in order to take advantage of the mass accuracy of the precursor peptide ion. In addition, one can use internal ion standard(s) with known masses that allow for mass calibration while the data is being collected (10).
It is possible that the isolation width for acquiring the tandem mass spectrum is wide enough to pick up multiple different peptides, resulting in a mixed tandem mass spectrum of several ion species. One may solve this with a narrower isolation width or a different LC gradient for improved peptide ion resolution.
Generally, the presence of an acetylated lysine is accompanied by an unmodified lysine. As described for histones, one can rationalize whether the putative acetylated histone peptide has the expected chromatographic behavior relative to its respective unmodified peptide. For acetylated peptides in general, the unmodified peptide will be longer due to the miscleavage of the acetylated lysine and the peptide chromatographic behavior will be harder to predict. For instance, if one detects the acetylated peptide PEAKacATR, the unmodified peptides would be PEAKun and ATR. Thus, one should use an alternative protease such as Arg-C that will digest an unmodified and acetylated protein into the same peptide sequence. In this case unlike the histones, the acetylated peptide should elute later than the unmodified peptide.
Lysine acetylation is a reversible process mediated by acetyltransferases and deacetylases. If no acetylation is found, confirm that deacetylase inhibitors such as sodium butyrate were included in the protein isolation and purification steps. Conversely, the presence of protease inhibitor cocktails commonly used in protein isolation and purification will negatively affect the trypsin digestion step. Thus one should either omit protease inhibitors in the final step of purification prior to digestion or dialyze the proteins away from the inhibitors.
References
- 1.Tolkovsky AM, Wyttenbach A. Differential phosphoprotein labeling (DIPPL) using 32P and 33P. Methods in Molecular Biology. 2009;527:21–29. doi: 10.1007/978-1-60327-834-8_2. [DOI] [PubMed] [Google Scholar]
- 2.Archuleta AJ, Stutzke CA, Nixon KM, Browning MD. Optimized protocol to make phosphor-specific antibodies that work. Methods in Molecular Biology. 2011;717:69–88. doi: 10.1007/978-1-61779-024-9_4. [DOI] [PubMed] [Google Scholar]
- 3.Steinberg TH, Agnew BJ, Gee KR,, Leung WY, Goodman T, Schulenberg B, Hendrickson J, Beechem JM, Haugland RP, Patton WF. Global quantitative phosphoprotein analysis using Multiplexed Proteomics technology. Proteomics. 2003;3(7):1128–1144. doi: 10.1002/pmic.200300434. [DOI] [PubMed] [Google Scholar]
- 4.Villén J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nature Protocols. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norris KL, Lee J, Yao T. Acetylation goes global: the emergence of acetylation biology. Science Signaling. 2009;2:pe76. doi: 10.1126/scisignal.297pe76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 7.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nature Biotechnology. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 9.Zee BM, Levin RS, DiMaggio PA, Garcia BA. Global turnover of histone post-translational modifications and variants in human cells. Epigenetics & Chromatin. 2010;3:22. doi: 10.1186/1756-8935-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Parts per million mass accuracy on an orbitrap mass spectrometer via lock mass injection into a C-trap. Molecular & Cellular Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]