Abstract
Posttranslational modifications of histones, alterations in the recruitment and functions of non-histone proteins, DNA methylation, and changes in expression of noncoding RNAs contribute to current models of epigenetic regulation. Nuclear receptors (NRs) are a group of transcription factors that, through ligand-binding, act as sensors to changes in nutritional, environmental, developmental, pathophysiologic, and endocrine conditions and drive adaptive responses via gene regulation. One mechanism through which NRs direct gene expression is the assembly of transcription complexes with cofactors and coregulators that possess chromatin-modifying properties. Chromatin modifications can be transient or become part of the cellular “memory” and contribute to genomic imprinting. Because many food components bind to NRs, they can ultimately influence transcription of genes associated with biologic processes, such as inflammation, proliferation, apoptosis, and hormonal response, and alter the susceptibility to chronic diseases (e.g., cancer, diabetes, obesity). The objective of this review is to highlight how NRs influence epigenetic regulation and the relevance of dietary compound–NR interactions in human nutrition and for disease prevention and treatment. Identifying gene targets of unliganded and bound NRs may assist in the development of epigenetic maps for food components and dietary patterns. Progress in these areas may lead to the formulation of disease-prevention models based on epigenetic control by individual or associations of food ligands of NRs.
Introduction
Nuclear receptors (NRs)6 are a group of transcription factors that, through ligand binding, drive adaptive gene responses to changes in nutritional, environmental, developmental, pathophysiologic, and endocrine conditions (1). The NR superfamily includes endocrine, adopted orphan, orphan with evidence of natural and/or synthetic ligand, and orphan with no known ligand NR subclasses. Endocrine NRs bind the ovarian hormone receptors estradiol (ER) and progesterone, androgen receptor (AR), vitamin D receptor (VDR), retinoic acid receptor (RARα, RARβ, RARγ), retinoid acid receptor [retinoid X (RXRα, RXRβ, RXRγ)], glucocorticoid receptor (GR), mineralcorticoid receptor, and thyroid hormone receptor. The adopted orphan NRs are mediators of metabolic pathways and bind lipids [PPARα, PPARβ/δ, PPARγ liver X receptor (LXRα, LXRβ)] and bile acids [BAs; farnesoid X receptor (FXR)]. The orphan NR subclass, for which there is evidence of natural and/or synthetic ligands, includes the small heterodimer partner (SHP) and aromatic hydrocarbon receptor (AhR). The SHP binds retinoid-related molecules (2, 3) and regulates BAs and lipid homeostasis (4). The AhR binds dietary (e.g., resveratrol, indol-3-carbinol), endogenous (e.g., PGs), and synthetic polycyclic aromatic hydrocarbon (PAH), dioxin (e.g., 2,3,7,8-tetrachlorodibenzo-p-dioxin), and polychlorinated biphenyl compounds (5). Orphan NRs with no known ligands include the dosage-sensitive sex reversal-adrenal hypoplasia critical region on chromosome X, gene 1 (DAX1), whose mutation predisposes to X-linked adrenal hypoplasia congenital and hypogonadotropic hypogonadism (6). The DAX1 protein is a corepressor of FXR. Overexpression of DAX1 downregulates the expression of FXR target genes and interferes with BAs, TGs, and glucose metabolism (7).
It is well accepted that epigenetic mechanisms, i.e., changes in gene expression without modification in nt sequence, such as DNA methylation, histone modifications, and noncoding mRNAs, establish areas of active or repressed chromatin, which mediate adaptive responses to dietary, environmental, and developmental signals (8). Research evidence suggests that dietary compounds contribute to epigenetic regulation and phenotypic plasticity through both NRs and nongenomic pathways. For example, the soy compound genistein stimulates gene expression through induction of phosphoinositide 3-kinase/AKT, which in turn phosphorylates and represses the histone methyltransferase (HMT) enhancer of zeste homolog 2 (EZH2), thus reducing the amounts of the histone 3 trimethylated at lysine 27 (H3K27me3) repressive mark (9). In parallel, genistein induces transcription of estrogen-responsive genes by inducing the recruitment of ERα and cofactors with histone-modifying properties (e.g., p300) (10). Similarly, by engaging both cytosolic (i.e., extracellular signal-regulated kinase 2) and nuclear (i.e., RXR) mediators, retinoic acid modifies the epigenetic landscape and transcriptional regulation of genes involved in differentiation and embryonic development (11, 12). Moreover, a large number of dietary ligands influence the epigenetic machinery by acting as methyl donors, cofactors, coenzymes, and regulators of chromatin-modifying proteins (13). Therefore, the full impact of diet–NR interactions on health and disease can only be appreciated by developing models that integrate all the pathways that impinge on epigenetic regulation (14). This review will focus on how the interplay between food compounds and NRs influences epigenetic regulation and the relevance of these interactions for disease prevention and treatment.
NR Structure and Regulation
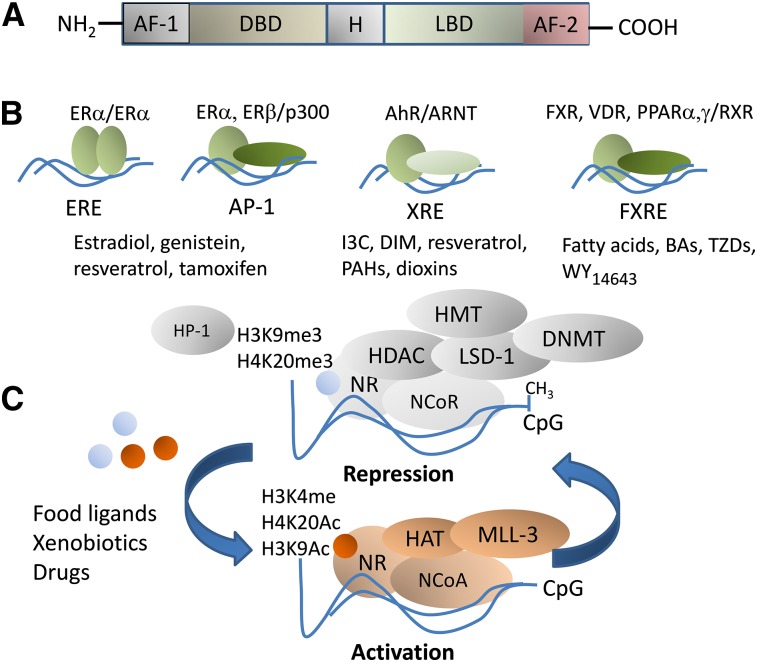
The general structure of NRs comprises an amino-terminal activation function domain-1 (AF-1), a DNA-binding domain (DBD), a hinge region, a ligand-binding domain, and a C-terminal activation function domain-2 (AF-2) (Fig. 1A). The ligand-binding domain comprises coregulator interaction domains. Through the DBD, NRs can bind directly to DNA as monomers, homodimers, or heterodimers (reviewed in references 15 and 16). For example, the ERα forms homodimers at estrogen response elements, whereas the FXR can form heterodimers with the RXR at FXR response elements (FXRE). Certain NRs function as promiscuous partners for different receptors. This is the case for the RXR, which forms complexes at target promoters with the PPAR, VDR, RAR, and FXR. In addition, NRs can modulate gene expression through physical interactions with DNA-bound transcription factors. For example, the ERα and ERβ can enhance or repress, respectively, transcription of genes through interactions with DNA-bound activator protein-1 (AP-1) complexes (17) (Fig. 1B).
FIGURE 1.
Nuclear receptor structure and coregulation by food ligands and nutritionally related xenobiotics and drug compounds. NRs share a general DNA structure comprising an amino-terminal AF-1, an H, an LBD, and a C-terminal AF-2 domain (A). Examples of NR interactions within the coregulator interaction domains: the ERα forms homodimers at EREs; the ERα and ERβ can enhance or repress, respectively, transcription of genes containing AP-1 sites through interactions with DNA-bound Jun/Fos complexes; the AhR/ARNT heterocomplex binds DNA at XREs; the FXR forms heterodimers with the RXR at FXRE (B). Certain NRs function as promiscuous partners for various receptors. This is the case for the RXR that forms complexes at target promoters with FXR and PPAR, VDR, and retinoic acid receptor. Examples of food ligands are listed below each NR. The diagram depicts general models of repression and activation via CpG methylation and demethylation mediated by NRs (C). In the absence of ligands or when NRs are bound to antagonists, NRs are found in NCoR complexes with factors that possess DNMT and HDAC, HMT, and histone demethylase (e.g., LSD-1) properties. Certain covalent modifications placed by NR corepressors on histones are markers of silenced heterochromatin and include H3K9me3, histone 3 trimethylated at lysine 27, and H4K20me3. Chromatin is occupied by the repressive HP-1 factor. Binding of food ligands to NRs triggers the dismissal of NCoR complexes and the recruitment at the target promoter of NCoA complexes that comprise HMT (e.g., MLL-3) and HAT (e.g., cAMP-response element-binding protein/p300) factors. The latter possess enzymatic activities that place “active” histone acetylation marks on histones 3 and 4 (H3K9Ac, H4K20Ac). AF-1, activation function 1; AF-2, activation function 2; AhR, aromatic hydrocarbon receptor; AP-1, activator protein-1; ARNT, aromatic hydrocarbon receptor nuclear translocator; BA, bile acid; CpG, cytosine-phosphate-guanine; DBD, DNA binding domain; DIM, diindolylmethane; DNMT, DNA methyltransferase; ERE, estrogen response element; ERα, estrogen receptor-α FXR, farnesoid X receptor; FXRE, farnesoid X receptor response element; H, hinge region; HAT, histone acetyltransferase; HDAC, histone deacetylase; HMT, methyltransferase; HP-1, heterochromatin protein-1; H3K4me4, histone 3 methylated at lysine 4; H3K9me3, histone 3 trimethylated at lysine 9; H4K20me3, histone 4 trimethylated at lysine 20; 3C, indol-3-carbinol; LBD, ligand-binding domain; LSD-1, lysine-specific demethylase-1; MLL-3, mixed lineage leukemia-3; NCoA, nuclear receptor coactivator; NCoR, nuclear corepressor; NR, nuclear receptor; PAH, polycyclic aromatic hydrocarbon; TZD, thiazolidinedione; VDR, vitamin D receptor; WY14643, 2-[[4-chloro-6-[(2,3-dimethylphenyl)amino]-2-pyrimidinyl]thio]acetic acid; XRE, xenobiotic response element.
The physical interaction between NRs and cofactors influences the magnitude and direction of the transcriptional response (18, 19). For instance, transcription by the ER, RAR (20), and PPARγ (15) is potentiated by interactions with the cofactors p160/steroid receptor coactivator-1 (SRC-1) and cAMP-response element-binding protein (CBP)/p300 through LXXLL-binding motifs (in which L indicates leucine and X indicates any amino acid) (20). Moreover, coregulator exchange contributes to gene-, cell-, and tissue-specific transcriptional regulation by NRs. For example, LXRα-regulated lipogenesis and cholesterol/BA homeostasis in the liver requires binding of LXRα through LXXLL motifs with the NR coactivator 6. However, NR coactivator 6 is not required for regulation of transcription by the ERα in the mammary gland (21). Conversely, repression of transcription by unliganded NRs is achieved through interactions with LXX I/H I XXX I/L motifs on nuclear corepressor (NCoR) and silencing-mediator for retinoic and thyroid (SMRT) factors (18).
NRs can also modify transcriptional regulation through interactions with cis-acting transcription factors. For example, the looping of the cytochrome P450, family 1, subfamily A, polypeptide 1 (CYP1A1) promoter region around histones facilitates interactions of DNA-bound AhR/AhR nuclear translocator (ARNT) complexes with the specificity protein 1 transcription factor recruited at adjacent sites and enhances transcription of the CYP1A1 gene in response to AhR ligands (22, 23). Examples of cooperativity between NRs and cis-acting transcription factors include those between the DNA-bound GR and the accessory nuclear factor-1 on the 11β-hydroxysteroid dehydrogenase (24) and phosphoenol-pyruvate-carboxykinase (PEPCK) genes (25). An example of negative cis-interaction is the recruitment of the activated AhR to a xenobiotic response element [XRE (5′-GCGTG-3′)] harbored in the breast cancer-1 (BRCA-1) gene, which displaces an ERα/p300 complex from an adjacent AP-1 site and represses estrogen-dependent activation of BRCA-1 expression (26). NRs may also repress transcription without direct binding to DNA through competition for ubiquitous cofactors as in the case of the trans-RA/RAR/RXR heterocomplex, which interferes with AP-1-dependent trans-activation by excluding the cofactor CBP (27). Similarly, the SHP orphan NR, which lacks a DBD, represses transcription through direct interaction with AF-2 coactivator domains on NRs or by stimulating the recruitment of NCoRs (4). Moreover, signaling pathways impart on NRs and cofactors posttranslational modifications (e.g., phosphorylation, methylation, and acetylation) that influence transcriptional activity. For instance, phosphorylation of p160 potentiates its coactivator functions on the AR and ER (28). Similarly, methylation and p300-dependent acetylation of the ERα protein potentiate its transcriptional activation of estrogen-responsive genes (29).
The cyclical recruitment of NRs and cofactors at target promoters contributes to transcriptional control (30). This is the case for the VDR on the 25-hydroxy vitamin D3 1α-hydroxylase (25OH-D3-1α) (31), the AR on the prostate specific antigen (32), the PPARδ on the pyruvate dehydrogenase kinase 4 (PDK4) (33), and the AhR on the CYP1A1 (34) and cyclooxygenase-2 (COX-2) (35) genes. Similarly, ligands of the AhR direct the cyclical recruitment of p300 to the COX-2 gene and its association with acetylated (Ac) histone 4 (H4) (35). Fluctuations in NR and cofactor recruitment fine-tune the duration of activation (or repression) and transcriptional rates of target genes. Thus, modeling the impact of food ligands for NRs on epigenetic regulation of target genes would require knowledge of the following: 1) the presence and spatial arrangement of core binding motifs for NRs and cofactors; 2) kinetics of recruitment of NR complexes; and 3) the effects on expression and posttranslational modification of NRs and related cofactors.
Mechanisms of Epigenetic Regulation by NRs
Epigenetic repression
In the absence of ligands or when NRs are bound to antagonists, NRs are found in corepressor complexes with factors that possess histone deacetylase (HDAC), HMT, histone demethylase (HDM), and phosphatase enzymatic activities (36) (Fig. 1C). The NR cofactors Sin3 transcription regulator family member A (Sin3A), NCoR, and SMRT lock target genes in a repressed state by forming bridges with HDACs. NR corepressor complexes include the ATP-dependent switch/sucrose non-fermentable (SWI/SNF) nucleosome remodeling factors. Certain covalent modifications placed by NR corepressors on histones are markers of silenced heterochromatin and include histone 3 trimethylated at lysine 9 (H3K9me3), H3K27me3, and H4K20me3. The binding of heterochromatin protein-1 to H3K9me3 locks heterochromatin in a transcriptionally silent state (37). The transrepressive functions of certain NCoR complexes (e.g., tailless-like X receptor) require the presence of the HDM lysine-specific demethylase-1, which demethylates H3K4me2 and H3K4me (38).
Competition among NRs for coregulators with histone-modifying properties (e.g., p300, SRC-1) is a mechanism that contributes to epigenetic repression. For example, by “squelching” p300 from ERα complexes, the agonist-bound AhR represses transcription of DNA repair (e.g., BRCA-1) (26) and other estrogen-inducible genes (39). Similarly, the binding of vitamin D to VDR causes the dissociation of p300 and the recruitment of HDAC corepressor complexes on the 25OH-D3-1α gene, hampering expression of 25OH-D3-1α and its participation in the de novo synthesis of vitamin D (40). Ubiquitin-mediated degradation of NCoRs is another mechanism that contributes to epigenetic repression by NRs (41). For example, agonist binding increases the affinity of PPARγ for NCoR/HDAC-3 repressor complexes and prevents ubiquitin-dependent removal of NCoR and transcriptional activation of the NF-κB-inducible proinflammatory COX-2 gene (42).
Epigenetic activation
For many genes, binding of agonists to NRs triggers the dismissal of NCoRs and the recruitment to the target promoter of coactivator complexes containing CBP and p300. The latter possess histone acetyltransferase (HAT) activities that place acetylation marks on H3 and H4 (H3K9Ac, H4K20Ac) (Fig. 1C). For example, transcriptional activation of the type 1 keratin KA11 (KA11) gene by IL-1β is accompanied by dismissal of NCoR from the AR bound to the KA11 promoter (43) Similarly, the estradiol-bound ERα induces transcription of the human pS2 (pS2) gene via dismissal of NCoRs and the ordered recruitment of activator complexes (44) comprising SRC-1 and HATs. These factors facilitate the acetylation and dimethylation of H3K14 and histone-4 at arginine-3 (H4R3) and the recruitment of transcription binding proteins and polymerase II, leading to activation of the pS2 gene.
Histone modifications associated with transcriptional activation by NR include methylation of H3K4 (H3K4me) by the H3K4-methyltransferase mixed lineage leukemia-3 (38, 45) and phosphorylation of H3 at serine 10. The later interferes with the binding of heterochromatin protein-1 to H3K27me3 and promotes nucleosomal opening and transcriptional activation (46). For example, elevation of phosphorylation of H3 at serine 10 and H3K9Ac are observed during early phases of ERα-induced transcription initiation (47).
The binding affinity of ligands toward NRs influences the dynamics of chromatin modifications at target genes. A study that examined the modulation of PPARγ transcriptional activity by insulin-sensitizing thiazolidinedione (TZD) compounds reported that pioglitazone and rosiglitazone were more effective than troglitazone in inducing the recruitment of PPARγ complexes to the PEPCK and PDK4 promoters and acetylation of PEPCK- and PDK4-associated histones (e.g., AcH4) (48). The stronger transcriptional effects of pioglitazone and rosiglitazone were mainly attributed to their higher binding affinity for PPARγ. Conversely, the preventive effects of resveratrol against transcriptional repression by the bound AhR of estrogen-inducible genes were related to formation of resveratrol/AhR heterocomplexes with lower affinity for XRE (49) and the agonist actions of resveratrol on the ERα (50) and related cofactors (e.g., p300) (51). Although the promiscuity of NRs for ligands with diverse binding affinity offers great variability and adaptability to environmental challenges, it also greatly expands the opportunities for the development of NR-based therapeutic strategies with food ligands (Table 1).
TABLE 1.
Food ligands of nuclear receptors and influence on epigenetic regulation1
| Food compound | Metabolic/epigenetic effects |
| ER | |
| Genistein, daidzein, equol | Potent activators of ERβ and ERα promoters through regulation of DNMT expression and enhanced HAT activity (122) |
| Resveratrol | Increases ER promoter methylation and inhibits HDAC (150) |
| EGCG | Induces chromatin remodeling by altering histone acetylation and methylation status leading to ER reactivation (47) |
| I3C | Disrupts interaction between ERα and Sp1 on promoter of the hTERT gene (151) |
| AR | |
| EGCG | Lowers AR acetylation and activation by inhibiting HAT activity (152) |
| DIM | Reverses epigenetic silencing of miR-34a in prostate cancer, inducing inactivation of the AR (153) |
| AhR | |
| Resveratrol | Prevents BRCA-1 promoter hypermethylation and silencing (49, 126) |
| EGCG | Inhibits binding of AhR to XRE in endothelial cells (154); directly inhibits DNMT by binding to catalytic subunit (122) |
| DIM, I3C | Abrogate dioxin-induced recruitment of AhR and AcH4 to the COX-2 promoter (35) |
| Quercetin, kaempferol, genistein, daidzein, apigenin | Inhibit AhR binding to XRE of target genes, such as CYP1A1 (123) |
| PPARα/γ | |
| Resveratrol | Activates PPARs in vitro and in vivo leading to reduced COX-2 expression (155) |
| PPARα | |
| Eicosanoids | Regulate β-oxidation genes in liver; inhibit NF-κB transcription in heart (156) |
| PPARγ | |
| Arachidonic acid | Increases adipocyte differentiation in brown adipose tissue (156) |
| Genistein | Promotes adipogenesis at concentrations >1 μmol/L through mechanisms involving downregulation of ER-mediated transcription (156) |
| Daidzein and equol | Activate PPARγ at lower concentrations than genistein (157) |
| CLA | Inhibits adipogenesis and inflammation, promotes osteoblastogenesis (158) |
| PPARβ/δ | |
| PUFAs | Increase FA oxidation (156) |
| PGs | Lower TGs and free FA amounts in adipose tissue (156) |
| RXR | |
| 9-cis RA | Binding to an NR partner (e.g., FXR, LXR, PPAR, RAR, TR, or VDR) promotes recruitment of cofactors and HATs that lead to transcriptional activation (159) |
| Lithocholic acid | |
| Phytanic acid | |
| VDR | |
| Vitamin D | Binding to VDR influences recruitment of coactivators or repressors and transcription of target genes (160–162) |
| Curcumin | Activates heterodimer formation with RXR and recruitment of coactivator SRC-1 (163) |
| FXR | |
| BAs | Increase interaction with NCoA6 (LXXLL motif) (126) |
| EGCG | Inhibits recruitment of coactivator SRC-2 to FXR and transcription of target genes in the intestine (7) |
| LXR | |
| Resveratrol | Increases RNA polymerase recruitment to the LXR promoter (164) |
| EGCG | Reduces expression of LXR in 3T3-LI liver cells (165) |
| Genistein | Increases promoter activity in rat liver, leading to insulin sensitization and improved lipid homeostasis (166) |
AcH4, acetylated histone-4; AhR, aromatic hydrocarbon receptor; AR, androgen receptor; BA, bile acid; BRCA-1, breast cancer-1; COX-2, cyclooxygenase-2; CYP1A1, cytochrome P450, family 1, subfamily A, polypeptide 1; DIM, diindolylmethane; DNMT, DNA methyltransferase; EGCG, epigallocatechin 3-O-gallate; ER, estrogen receptor; FXR, farnesoid X receptor; HAT, histone acetyltransferase; HDAC, histone deacetylase; hTERT, human telomerase reverse transcriptase; I3C, indole-3-carbinol; LXR, liver X receptor; LXXLL, L indicates leucine and X indicates any amino acid; miR-34a, microRNA-34a; NCoA6, nuclear receptor coactivator 6; NR, nuclear receptor; RA, retinoic acid; RAR, retinoic acid receptor; RXR, retinoid X receptor; Sp1, specificity protein 1; SRC-1, steroid receptor coactivator-1; SRC-2, SRC-1, steroid receptor coactivator-2; TR, thyroid receptor; VDR, vitamin D receptor; XRE, xenobiotic response element.
Cytosine-phosphate-guanine (CpG) methylation and demethylation at target promoters
Hypermethylation of cytosines comprised in CpG islands and its propagation through DNA replication have been related to gene silencing, whereas demethylation of CpGs has been linked to gene activation (52). Enzymes implicated in maintenance and de novo DNA hypermethylation are, respectively, DNA methyltransferase (DNMT)-1, and DNMT-3a and DNMT-3b, respectively (53). DNMT-1, but not DNMT-3a and DNMT-3b, and methylated cytosine binding protein-2 (MeCP2) recruit the biotin protein ligase holocarboxylase synthetase (HLCS) (54), leading to the assembly of a multiprotein gene repression complex comprising HDAC-1, NCoR, and the eukaryotic histone methyl transferase-1 (EHMT-1). The binding between HLCS and EHMT-1 is facilitated by the HLCS-dependent biotinylation of K161 in EHMT-1 (55). The close proximity between HLCS and histones facilitates the HLCS-dependent biotinylation of histone H3 and H4 (56). Nevertheless, biotinylation of histones is a rare event (≤0.001% of histone H3 and H4) and does not account for the repressive role of biotin on gene expression (57–59), in particular of repeats, which appears to be caused primarily by the HLCS-containing multiprotein complex (54). Repression of repeats is necessary for maintenance of genome stability, whereas chromosomal abnormalities have been reported in biotin-depleted human cell cultures (60).
DNMTs physically bind to HMTs and HDACs, providing a mechanistic link between transcriptional repression via DNA hypermethylation and histone methylation and deacetylation. In the case of the ERα-inducible pS2 gene, cycles of CpG methylation alternate with cycles of CpG demethylation at the pS2 promoter. The hypermethylation of CpGs occurs at the end of each productive transcription cycle and corresponds to the corecruitment of MeCP2, SWI/SNF, DNMT-1, and DNMT-3a/3b. Conversely, transcriptional activation by the ERα, GR, and VDR has been linked to stimulation of CpG demethylation at the pS2 (61, 62), tyrosine aminotransferase (63), and cytochrome P450, family 7, subfamily B, polypeptide 1 (64) promoter, respectively.
Whereas the mechanisms of DNA methylation have been established clearly, the processes that lead to active DNA demethylation remain an area of active investigation (53). Previous models of demethylation included the following: 1) the enzymatic removal of the methyl group from 5-methylcytosine; 2) nt excision repair involving the growth arrest and DNA damage 45 (GADD45) protein and XPG, a 3′ endonuclease; 3) direct base excision repair (BER) of 5-methylcytosine by DNA glycosylases [i.e., thymine DNA glycosylase (TDG)] and methyl-CpG binding domain-containing protein 4 (MBD4); 4) and deamination and repair of 5-methylcytosine via TDG and BER to replace the mismatched T with a C (53, 65–68). Evidence that GADD45 family members interact with many food-related NRs, such as RXRα, RARα, ERα, PPARα, PPARβ, and PPARγ, suggest that nt excision repair-based CpG demethylation plays an important role in epigenetic regulation by common dietary compounds. However, the role of GADD45 in demethylation is still debated, and mounting evidence now offers a model for a complete cycle of methylation and demethylation that involves 4 steps: 1) CpG methylation by DNMTs; 2) iterative oxidation of methylated cytosine by 10-11 translocation enzymes; 3) excision of methylated cytosine by TDG to generate an abasic site; and 4) placement of unmodified cytosine by BER (69–71). Evidence that TDG interacts with RAR, RXR, and ERα (72–74) further highlights the role of food-related NRs in epigenetic control via CpG demethylation.
Epigenetic Modifications Induced by Dietary and Nutrition-Related Ligands of NRs in Disease and Cell Fate Determination
PPAR
PPARα, PPARβ, and PPARγ play a key role in FA and glucose metabolism. PPARα is a binding target for several FAs, including palmitic acid, oleic acid, linoleic acid, CLA, arachidonic acid, EPA, DHA, and the PG metabolite 15D-J2 (75) (Table 1). PPARα is expressed primarily in liver, heart, and skeletal muscle. Gene targets for PPARα encode enzymes that contribute to FA oxidation: 1) acyl-CoA synthetase; 2) carnitine palmitoyl-transferase-1; 3) acyl-CoA oxidase; 4) very-long-chain and medium-chain acyl-CoA dehydrogenase; and 5) FA transport protein. Also, PPARα activates its own expression through binding to a hepatocyte nuclear factor-4α-binding element harbored in the PPARα promoter (76).
PPARα.
Gestational exposure to a protein-restricted diet elevates PPARα expression by lowering PPARα promoter CpG methylation in hepatic (77) and heart (78) tissue of rat offspring. Conversely, maternal supplementation with folic acid induces hypermethylation of the PPARα gene in liver tissue of offspring (79). These observations suggest that maternal nutrition influences epigenetic regulation of PPAR-regulated pathways in offspring.
Nonalcoholic fatty liver disease is a condition characterized by increased lipid influx into the liver and de novo hepatic lipogenesis. The hepatic accumulation of TGs has been linked to increased association of the repressive histone mark H3K9me3 on the PPARα gene and lowering of PPARα expression (80). Methyl-donor deficiency is a condition that has been related to impaired FA oxidation and decreased expression of PPARα. For example, administration during pregnancy and lactation of a methionine- and choline-deficient diet led to hypomethylation of the PPAR coactivator 1α (PGC-1α) protein and its reduced stimulation of PPARα expression in liver tissue of rat offspring (81). Thus, dietary disturbances (e.g., methyl-donor deficiency) may contribute to the pathogenesis of fatty liver by reducing expression of PPARα through epigenetic mechanisms. Conversely, targeting of PPARα with the synthetic agonist 2-[[4-chloro-6-[(2,3-dimethylphenyl)amino]-2-pyrimidinyl]thio]acetic acid (WY14,643) has been proposed as an option for the management of hypertriglyceridemia. The treatment with WY14,643 ameliorated liver insulin resistance, exerted anti-inflammatory effects, and activated β-oxidation enzymes in a rodent model of hepatic and muscle steatosis (82). Unfortunately, the benefits of lipid-lowering interventions based on WY14,643 and other agonists of PPARα could be offset by increased cancer risk. For example, increased PPARα expression was correlated with tumor development in humans, raising some concerns that constitutive hyperactivation of PPARα may exert tumor-promoting effects (83). In a rodent model (SV129 mice), the long-term exposure (5 mo) to WY14,463 increased liver cell proliferation and global hypomethylation but decreased the amounts of the repressive H4K20me3 mark. The latter changes were not observed in SV129/PPARα−/− mice. These data suggested a positive role for PPARα-mediated epigenetic regulation in tumorigenesis (84). As a result, more experimental evidence is needed to establish whether or not therapies based on dietary or synthetic agonists of PPARα pose increased cancer risks to humans through the induction of undesirable epigenetic changes (85).
PPARγ.
In mammals, white adipose tissue (WAT) plays a prominent role in storing excess energy in the form of TGs but also plays essential roles as an endocrine organ through secretion of leptin and adiponectin and metabolism of sex steroids and glucocorticoids (86). Resident macrophages in WAT are a meaningful source of inflammatory cytokines, such as TNF-α and IL-6. An increase in circulating amounts of these macrophage-derived factors in obesity leads to a chronic low-grade inflammatory state that has been linked to the development of insulin resistance and diabetes (87).
There are 2 known PPARγ splice variants, PPARγ1 and PPARγ2. Expression of the PPARγ1 isoform is higher in WAT, intestine and spleen. PPARγ2 is expressed preferentially in WAT and brown adipose tissue (BAT). The activation of PPARγ with the synthetic agonist rosiglitazone shifts the fate of pluripotent adipose-derived stem cells toward BAT (88). In addition, manipulation of PPARγ signaling in WAT produces gene expression representative of BAT and improves glucose tolerance in murine cell cultures and in mice, respectively (89).
Ligands of PPARγ comprise common dietary FAs (linoleic acid, arachidonic acid, EPA, DHA), metabolites (15d-PGJ2), synthetic TZDs, and nonsteroidal anti-inflammatory drugs (90, 91). Genes induced by PPARγ include those that regulate FA (e.g., FA-binding protein and lipoprotein lipase) and carbohydrate (PEPCK) metabolism (92). Chromatin-modifying cofactors that colocalize with PPARγ comprise p300/CBP, SRC-1, and jumonji histone demethylase 2A (JHDM2A), an HDM. Histone marks associated with activation of transcription by PPARγ are H3K4me2, H3K4me3, H4K20me1, acH3K9ac, and acH3K27. Conversely, the association of PPARγ with HDACs (HDAC-1 and HDAC-3), nuclear corepressors (NCoR-1, NCoR-2, and SMRT), and HMTs [SET domain, bifurcated 1 (SETDB1) and histone-lysine-N-methyltransferase (SUV39H1)] has been linked to transcriptional repression. Repressive histone marks associated with promoter recruitment of PPARγ include H3K9me2, H3K9me3, H3K27me2, and H3K27me3 (93).
The epigenetic silencing of the PPARγ gene by MeCP2- and EZH2-containing repressive complexes is a biomarker of colorectal cancer progression and adverse patient’s outcome (94). Hepatic reactivation of PPARγ expression via repression of EZH2 and MeCP2 functions has been documented for the dietary compounds rosmarinic acid and baicalin (95). Intrauterine growth restriction is a condition that, in neonatal rat lung, reduces expression of PPARγ by lowering the amounts of the HMT su(var)3-9, enhancer of zeste, trithorax, domain 8 (setd8) and H4K20me on the PPARγ gene. The latter effects are ameliorated in offspring by maternal supplementation with DHA (96).
A factor related to PPARγ that controls lipid metabolism is the PPARγ coactivator PGC-1α. In skeletal muscle, the acute exposure to a heavy lipid load (e.g., palmitate) reduces PGC-1α expression through DNMT-3b-dependent non-CpG methylation of the PGC-1α promoter and impairs FA oxidation (97). Conversely, exercise induces acute expression of PGC-1α, PDK4, and PPARδ in skeletal muscle via reduced CpG methylation of the respective promoters (98). This cumulative evidence illustrates the contribution of epigenetic modification through DNA methylation at the PGC-1α gene in the regulation of lipid metabolism by FAs and exercise in skeletal muscle.
TZDs are a group of insulin sensitizers, of which, troglitazone induces transcription of the PPARγ2 gene through placement of the active H4K20me1 mark on the PPARγ2 promoter (99). The induction of expression of PPARγ2 is linked to CpG demethylation of the PPARγ2 promoter in differentiated 3T3-Ll adipocytes (100). Similarly, the transcriptional activation of the glucose-dependent insulin-tropic polypeptide receptor and PEPCK genes with synthetic ligands of PPARγ is related to increased association of AcH3 and AcH4, respectively, with the glucose-dependent insulin-tropic polypeptide receptor and PEPCK promoters (48, 101). Because certain TZDs, such as rosiglitazone, induce severe health side effects, such as myocardial infarction (102, 103), future studies should examine whether or not feeding of certain food compounds alone or in combination with antidiabetic drugs offer safer alternatives for epigenetic activation of PPARγ networks and lowering of circulating glucose in patients with diabetes.
ER
The ER is expressed in 2 isoforms, α and β. The ERα stimulates proliferation in reproductive organs (104). The estradiol-bound ERα recruits cofactors that possess HAT (p300, p160s), HMT (e.g., protein arginine methyltransferase 1), and HDM (e.g., lysine-specific demethylase-1) enzymatic activities. Conversely, ERα bound to antagonists (e.g., tamoxifen) associates with NCoR-1 and SMRT. The latter recruit nucleosome remodeling and deacetylase complexes that repress transcription. Amounts of NCoR-1 are usually lower in invasive breast cancers, implying that changes in epigenetic control linked to NCoR-1/HDACs may contribute to transition to more invasive phenotypes (105).
The activation of ERβ leads to transcriptional repression of ERα target genes through either the sequestration of cofactors for the ERα or formation at target genes of complexes that repress ERα-dependent transcription. For example, the genistein-bound ERβ triggers the recruitment of the cofactor GR interacting protein-1, which antagonizes the activation of ERα responsive promoters (106). The contribution of ERβ to epigenetic regulation appears to be gene and tissue specific. In differentiated mouse embryonic fibroblasts, ERβ induces hypomethylation of the glucose transporter 4 (GLUT4) promoter and restores GLUT4 gene transcription (107). Conversely, in prostate cancer cells, ERβ stimulates the recruitment of corepressor complexes containing NCoR, mSin3A, and HDAC-1 and the CpG methylation of the glutathione S-transferase P1-1 promoter (108).
Isoflavones are a class of phytoestrogens with higher (∼10-fold to 30-fold) binding affinity for ERβ compared with ERα (e.g., genistein > biochanin A > daidzein). The phytoalexin resveratrol also binds to the ERα and ERβ but with ∼7000-fold lower affinities compared with estradiol (109). Through epigenetic mechanisms, isoflavones exert tumor-protective or -promoting effects in ER-positive mammary tissue depending on the timing of exposure. In rodent models, the prenatal and neonatal exposure to genistein increased the incidence of carcinogen-induced mammary tumors (110) and uterine adenocarcinoma (111), respectively. The tumor-promoting effects of neonatal exposure to genistein in uterine tissue have been related to constitutive stimulation of hypomethylation and expression of the nucleosomal binding protein-1 (NSBP-1) gene (112). Notably, the NSBP-1 gene encodes for a member of the high-mobility group nucleosome-binding proteins that reduce compaction of chromatin and enhance transcription. The NSBP-1 protein is expressed at high amounts in cancer cells, whereas the knockdown of the NSBP1 gene inhibits tumor growth in nude mice but induces G2/M cell cycle arrest and apoptosis (113).
In adult life, genistein may increase breast cancer risk through epigenetic silencing of RARβ2. The latter mediates the anticancer effects of retinoic acid. DNA hypermethylation of the RARβ2 gene was observed in nipple aspirates obtained from premenopausal women supplemented for 1 menstrual cycle with phytoestrogens (90.6 mg/d genistein, 36.4 mg/d daidzein, and 1.8 mg/d glycitein) and plasma amounts of genistein >600 μg/L (114). The DNA methylation rate of RARβ2 is generally higher in breast tumors and precancerous breast lesions compared with normal breast tissue (115).
Some of the preventive effects of genistein and daidzein against ERα-positive breast cancer have been related to activation of BRCA-1 expression via reversal of CpG methylation at the BRCA-1 gene (116). Genistein mimics the stimulatory effects of estradiol on BRCA-1 (26, 117) and triggers the association of p300, SRC-1, and AcH4 with the BRCA-1 promoter (118, 119). Moreover, genistein may exert anticancer effects in endocrine tissue through epigenetic reactivation of ERα (120) and repression of DNMT-3b expression (121).
The antidiabetic effects of daidzein and its metabolite equol have been attributed to the induction of GLUT4 expression by ERβ through reduced CpG methylation at the GLUT4 promoter (107). Similarly, studies that focused on the anticancer effects of the phytoalexin resveratrol reported that it reduced promoter methylation of the phosphatase and tensin homolog gene in ERα-positive MCF-7 breast cancer cells (122, 123). Overall, these examples suggest a role for polyphenols, such as genistein, resveratrol, and daidzein, as modifiers of cancer and diabetes risk through epigenetic regulation of genes targeted by the ER.
AhR
Ligands of the AhR include environmental dioxins and PAHs, a multitude of dietary compounds (e.g., resveratrol, kaempferol, and indol-3-carbinol), and metabolites (i.e., PGs) (reviewed in references 5, 123). In the absence of ligands, the AhR is found in the cytoplasm in association with the chaperone proteins heat shock protein 90, HBV X-associated protein 2 (XAP2), and p23. The binding of ligands to the AhR displaces XAP2 and favors migration of the bound AhR to the nucleus in which it interacts with ARNT, of which 3 different isoforms (ARNT-1, ARNT-2, and ARNT-3) have been identified. After nuclear release of heat shock protein 90, the AhR/ARNT heterocomplex binds to core XREs and activates the expression of phase I and phase II enzymes. Phase I enzymes comprise CYP1A1, cytochrome P450, family 1, subfamily B, polypeptide 1, and cytochrome P450, family 1, subfamily A, polypeptide 2, which produce chemically reactive species. The latter are substrates for detoxification by phase II enzymes, such as NAD(P)H:quinine oxidoreductase and UDP-glucuronosyltransferase-1A6 (123). The role of CpG methylation in differential regulation of CYP1A1 and cytochrome P450, family 1, subfamily B, polypeptide 1 expression by the AhR has been documented clearly in breast and hepatic cells (124).
In addition to its putative role in detoxification, the AhR regulates transcription of estrogen-responsive genes (125). For example, in the absence of agonists, the AhR coactivates the ERα and is necessary for estrogen-dependent activation of BRCA-1 transcription (26). Conversely, in the presence of agonists (i.e., PAHs, dioxins), the bound AhR is recruited to XREs harbored in the BRCA-1 promoter and antagonizes ERα-dependent stimulation of BRCA-1 expression. The latter effect is paralleled by increased CpG methylation of the BRCA-1 promoter and association of HDAC-1, DNMTs (DNMT-1, DNMT-3a, and DNMT-3b), and H2K9me3 with the BRCA-1 gene (49). Among many dietary ligands of the AhR, resveratrol is a prototype food compound that overrides the placement of repressive marks by AhR agonists on the BRCA-1 gene (126).
VDR
Through epigenetic mechanisms, the VDR directs the expression of genes involved in vitamin D and bone tissue remodeling. The cholecalciferol, 1,25-dihydroxycholecalciferol, [1,25(OH)2-D3]-bound VDR heterodimerizes with RXR at VDR response elements [(A/G)G(G/T)TCA] and recruits SWI/SNF chromatin remodeling (e.g., BRG-1) and HAT factors (e.g., CBP/p300, SRC-1) that activate transcription of the 25-hydroxy vitamin D3-24-hydroxylase (cytochrome P450, family 24, subfamily A, polypeptide 1) and osteopontin genes (31). The cytochrome P450, family 24, subfamily A, polypeptide 1 enzyme and osteopontin maintain vitamin D and bone tissue homeostasis, respectively, through the conversion of 25OH-D3 and 1,25(OH)2-D3 into hydroxylated degradation products and stimulation of bone cell survival. Conversely, the recruitment of the RXR/VDR–1,25(OH)2-D3 heterocomplex to a VDR-interacting repressor bound to an E-box (CANNTG) harbored in the cytochrome P450, family 27, subfamily B, polypeptide 1 (CYP27B1) promoter represses CYP27B1 transcription (40). The CYP27B1 enzyme catalyzes the 1α-hydroxylation of vitamin D3. The transrepression of CYP27B1 transcription by the VDR/VDR-interacting repressor heterocomplex involves the recruitment of HDACs, Sin3A, NCoR, DNMTs, and MBDs on the CYP27B1 gene. The CpG demethylation and derepression of the CYP27B1 promoter is induced by the parathyroid hormone through PKC-dependent phosphorylation of the DNA glycosylase MBD4 and BER mechanisms (127, 128). Therefore, dietary ligands of VDR coordinate calcium and bone tissue metabolism by altering the epigenetic code of genes involved in vitamin D metabolism.
FXR
Endogenous ligands of FXR are the primary BAs chenodeoxycholic and cholic acid and the secondary metabolites deoxycholic acid and lithocholic acid. The chenodeoxycholic acid is the BA with the highest binding affinity for FXR (EC50 of 10–50 μmol/L). Conversely, the absence of a 7α-hydroxy group in deoxycholic acid and lithocholic acid or placement of a 12α-hydroxy group in CA reduces the binding affinity of these compounds for FXR, which directs transcription as a monomer, homodimer, or through heterodimerization with RXR. The activated FXR represses transcription of genes involved in BA synthesis (cytochrome P450, family 7, subfamily A, polypeptide 1) and gluconeogenesis (glucose-6-phosphatase). Conversely, it enhances the transcription of genes involved in BA shuttling [ileal bile acid-binding protein and metabolism (SHP)] (reviewed in reference 129).
Reduced FXR expression is found in intestinal epithelial cells with activated Wnt (wingless-type MMTV integration site family) signaling (130), human colon carcinoma (131), rodent colorectal cancers carrying a mutated adenomatous polyposis coli gene, and humans with familial adenomatous polyposis. FXR induces the expression of tumor suppressor (p21) and proapoptotic (FA synthase) but represses expression of antiapoptotic (B-cell CLL/lymphoma 2) and TNFα, genes. Also, FXR tethers the transcription factor p65, thus hampering the ability of NF-κB to activate proinflammatory (e.g., COX-2 and inducible nitric oxide synthase) genes (132).
The SHP and the bile salt export pump genes are targets for epigenetic regulation by the FXR. Under conditions of low exposure to BAs, the FXR is deacetylated by sirtuin 1, and transcription of SHP is repressed. Conversely, the activation of FXR by BAs induces the coordinate recruitment of p300 to the FXR/RXR complex, release of sirtuin 1, and association of AcH3K9 and AcH3K14 with the SHP promoter (133, 134). Similarly, FXR activation with the synthetic compound 3-[2-[2-chloro-4-[[3-(2,6-dichlorophenyl)-5-(1-methylethyl)-4-isoxazolyl]methoxy]phenyl]ethenyl]benzoic acid (GW4064) increases the association of AcH3K9 and AcH3K14 with the SHP promoter (135). FXR-dependent transactivation of the bile salt export pump gene requires histone methylation by the H3K mixed lineage leukemia-3 (136) and H4R3 protein arginine methyltransferase 1 (137) methylases. A posttranslational modification of FXR that increases binding of the FXR/RXR complex to FXRE and expression of FXR target genes is methylation of lysine 26 by the methylase histone-lysine N-methyltransferase SET domain 7/9 (SET7/9) (138). The latter enzyme also methylates the ERα at lysine 302 (139). These data clearly corroborate the role of epigenetic mechanisms in FRX-dependent transcriptional regulation of genes involved in maintenance of BA homeostasis.
Significance and Future Perspectives
The field of epigenetics finds its roots in the pioneering theories of Conrad Hal Waddington (140) and others (141), who developed the concept that the epigenotype is the sum of the mechanisms that bridges the gap between genotype and phenotype (142). Research findings discussed in this review highlighted the notion that NRs influence the epigenotype, and thus the phenotypic response, by directing gene expression through the recruitment to target promoters of factors that possess chromatin, histone, and DNA-modifying properties. The ability of NRs to affect gene expression via epigenetic mechanisms would be sequence specific, i.e., via direct binding to core elements (i.e., XRE, VDR response element, FXRE, etc,) or to factors (i.e., AP-1, specificity protein 1) bound to DNA motifs. The precise sequence of the core motifs and their spatial arrangement would determine the specificity, strength, and impact of NRs–response element interactions on gene expression (143). Therefore, the identification and cataloging of binding sequences would greatly improve our understanding of how unliganded and food-bound NRs contribute to the epigenotype associated with chronic diseases. Notably, NRs interact promiscuously with many food ligands (Table 1). Therefore, future research should examine how synergies and antagonisms between food components and drugs for NRs influence the epigenotype and response to disease therapy. For example, studies reported that the efficacy of breast cancer therapies based on the ERα antagonist tamoxifen was augmented by the food isoflavone and ERα ligand genistein (144). Conversely, the treatment with the PPARα agonist fenofibrate prevented glucocorticoid-induced hyperinsulinemia of mice fed a high-fat diet but potentiated the anti-inflammatory effects of the GR on NF-κB. These examples provide the rationale for the development of combination therapies for the management of chronic disorders, such as breast cancer and chronic inflammation (145). Hence, future research should help clarify why diseased cells/tissues that share the same genotypes with healthy ones have different epigenotypes and predict their response to therapies based on food ligands of NRs.
Finally, it is relevant to highlight that epigenetic events, such as DNA methylation, may be imprinted through somatic transmission or even exert transgenerational effects if they affect the germ line (146). Nevertheless, unlike genetic changes (i.e., mutations), DNA methylation is reversible. For example, reversal of DNA hypermethylation has been documented for genistein on the RARβ (147) and ERα (148) and for epigallocatechin 3-O-gallate on the RARβ (149) genes. Whereas the anticancer effects of genistein and epigallocatechin 3-O-gallate may be due to combinatorial actions on NRs and other factors that influence epigenetic control, we suggest that vast opportunities exist for nutritional therapy of disease-related epigenotypes using food ligands of NRs.
Acknowledgments
The authors read and approved the final manuscript.
Footnotes
Abbreviations used: Ac, acetylated; AF-1, activation function domain-1; AF-2, activation function domain-2; AhR, aromatic hydrocarbon receptor; AP-1, activator protein-1; AR, androgen receptor; ARNT, AhR nuclear translocator; BA, bile acid; BAT, brown adipose tissue; BER, base excision repair; BRCA-1, breast cancer-1; CBP, cAMP-response element-binding protein; COX-2, cyclooxygenase-2; CpG, cytosine-phosphate-guanine; CYP1A1, cytochrome P450, family 1, subfamily A, polypeptide 1; CYP27B1, cytochrome P450, family 27, subfamily B, polypeptide 1; DAX1, dosage-sensitive sex reversal-adrenal hypoplasia critical region on chromosome X, gene 1; DBD, DNA binding domain; DNMT, DNA methyltransferase; EHMT-1, eukaryotic histone methyl transferase-1; ER, estrogen receptor; EZH2, enhancer of zeste homolog 2; FXR, farnesoid X receptor; FXRE, FXR response elements; GADD45, growth arrest and DNA damage 45; GLUT4, glucose transporter 4; GR, glucocorticoid receptor; HAT, histone acetyltransferase; HDAC, histone deacetylase; HDM, histone demethylase; HLCS, holocarboxylase synthetase; HMT, histone methyltransferase; H3K4, histone 3 at lysine 4; H3K9, histone 3 at lysine 9; H3K14, histone 3 at lysine 14; H3K27, histone 3 at lysine 27; H4K20, histone 4 at lysine 20; LXR, liver X receptor; MBD4, methyl-CpG binding domain-containing protein 4; me, methylated; me2, dimethylated; me3, trimethylated; MeCP2, methylated cytosine binding protein-2; NCoR, nuclear corepressor; NR, nuclear receptor; NSBP-1, nucleosomal binding protein-1; PAH, polycyclic aromatic hydrocarbon; PDK4, pyruvate dehydrogenase kinase 4; PEPCK, phosphoenol-pyruvate-carboxykinase; PGC-1α, PPAR coactivator 1α pS2, human pS2; RAR, retinoic acid receptor; RXR, retinoid acid receptor; SHP, small heterodimer partner; Sin3A, Sin3 transcription regulator family member A; SMRT, silencing-mediator for retinoic and thyroid; SRC-1, steroid receptor coactivator-1; SWI/SNF, switch/sucrose non-fermentable; TDG, thymine DNA glycosylase; TZD, thiazolidinedione; VDR, vitamin D receptor; WAT, white adipose tissue; WY14,643, 2-[[4-chloro-6-[(2,3-dimethylphenyl)amino]-2-pyrimidinyl]thio]acetic acid; XRE, xenobiotic response element; 1,25(OH)2-D3, cholecalciferol, 1,25-dihydroxycholecalciferol, 25-hydroxycholecalciferol; 25OH-D3-1α, 25-hydroxy vitamin D3-1α.
References
- 1.Sladek FM. What are nuclear receptor ligands? Mol Cell Endocrinol. 2011;334:3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miao J, Choi SE, Seok SM, Yang L, Zuercher WJ, Xu Y, Willson TM, Xu HE, Kemper JK. Ligand-dependent regulation of the activity of the orphan nuclear receptor, small heterodimer partner (SHP), in the repression of bile acid biosynthetic CYP7A1 and CYP8B1 genes. Mol Endocrinol 2011;25:1159–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farhana L, Dawson MI, Leid M, Wang L, Moore DD, Liu G, Xia Z, Fontana JA. Adamantyl-substituted retinoid-related molecules bind small heterodimer partner and modulate the Sin3A repressor. Cancer Res 2007;67:318–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garruti G, Wang HH, Bonfrate L, de Bari O, Wang DQ, Portincasa P. A pleiotropic role for the orphan nuclear receptor small heterodimer partner in lipid homeostasis and metabolic pathways. J Lipids 2012;2012:304292. [DOI] [PMC free article] [PubMed]
- 5.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol 2003;43:309–34 [DOI] [PubMed] [Google Scholar]
- 6.Muscatelli F, Strom TM, Walker AP, Zanaria E, Récan D, Meindl A, Bardoni B, Guioli S, Zehetner G, Rabl W, et al. Mutations in the DAX-1 gene give rise to both X-linked adrenal hypoplasia congenita and hypogonadotropic hypogonadism. Nature 1994;372:672–6 [DOI] [PubMed] [Google Scholar]
- 7.Li J, Lu Y, Liu R, Xiong X, Zhang Z, Zhang X, Ning G, Li X. DAX1 suppresses FXR transactivity as a novel co-repressor. Biochem Biophys Res Commun 2011;412:660–6 [DOI] [PubMed] [Google Scholar]
- 8.Baylin SB, Jones PA. A decade of exploring the cancer epigenome – biological and translational implications. Nat Rev Cancer 2011;11:726–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greathouse KL, Bredfeldt T, Everitt JI, Lin K, Berry T, Kannan K, Mittelstadt ML, Ho SM, Walker CL. Environmental estrogens differentially engage the histone methyltransferase EZH2 to increase risk of uterine tumorigenesis. Mol Cancer Res 2012;10:546–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong T, Nakagawa T, Pan W, Kim MY, Kraus WL, Ikehara T, Yasui K, Aihara H, Takebe M, Muramatsu M, et al. Isoflavones stimulate estrogen receptor-mediated core histone acetylation. Biochem Biophys Res Commun 2004;317:259–64 [DOI] [PubMed] [Google Scholar]
- 11.Wei LN. Non-canonical activity of retinoic acid in epigenetic control of embryonic stem cell. Transcription 2013;June 14 (Epub ahead of print; DOI:10.4161/trns.25395) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kashyap V, Gudas LJ. Epigenetic regulatory mechanisms distinguish retinoic acid-mediated transcriptional responses in stem cells and fibroblasts. J Biol Chem 2010;285:14534–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zempleni J, Cordonier EL, Baier SR, Xue J. Vitamins, bioactive food compounds, and histone modifications. Handbook of vitamins. 5th ed. Zempleni J, Suttie JW, Gregory JF III, Stover PJ, eds. Boca Raton, FL: CRC Press, Taylor and Francis Group; 2013:551–64.
- 14.Zhang X, Ho SM. Epigenetics meets endocrinology. J Mol Endocrinol 2011;46:R11–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev 2001;81:1269–304 [DOI] [PubMed] [Google Scholar]
- 16.Gadaleta RM, Magnani L. Nuclear receptors and chromatin: an inducible couple. J Mol Endocrinol 2014:52:R137–49 [DOI] [PubMed] [Google Scholar]
- 17.Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov 2004;3:950–64 [DOI] [PubMed] [Google Scholar]
- 18.McKenna NJ, O’Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 2002;108:465–74 [DOI] [PubMed] [Google Scholar]
- 19.Jeyakumar M, Webb P, Baxter JD, Scanlan TS, Katzenellenbogen JA. Quantification of ligand-regulated nuclear receptor corepressor and coactivator binding, key interactions determining ligand potency and efficacy for the thyroid hormone receptor. Biochemistry 2008;47:7465–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McInerney EM, Rose DW, Flynn SE, Westin S, Mullen TM, Krones A, Inostroza J, Torchia J, Nolte RT, Assa-Munt N, et al. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev 1998;12:3357–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q, Chu MJ, Xu J. Tissue- and nuclear receptor-specific function of the C-terminal LXXLL motif of coactivator NCoA6/AIB3 in mice. Mol Cell Biol 2007;27:8073–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlberg C, Seuter S. Dynamics of nuclear receptor target gene regulation. Chromosoma 2010;119:479–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F, Hoivik D, Pollenz R, Safe S. Functional and physical interactions between the estrogen receptor Sp1 and nuclear aryl hydrocarbon receptor complexes. Nucleic Acids Res 1998;26:3044–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hebbar PB, Archer TK. Chromatin-dependent cooperativity between site-specific transcription factors in vivo. J Biol Chem 2007;282:8284–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stafford JM, Wilkinson JC, Beechem JM, Granner DK. Residues 88–109 of factor IXa are important for assembly of the factor X activating complex. J Biol Chem 2001;276:39885–9111518712 [Google Scholar]
- 26.Hockings JK, Thorne PA, Kemp MQ, Morgan SS, Selmin O, Romagnolo DF. The ligand status of the aromatic hydrocarbon receptor modulates transcriptional activation of BRCA-1 promoter by estrogen. Cancer Res 2006;66:2224–32 [DOI] [PubMed] [Google Scholar]
- 27.Benkoussa M, Brand C, Delmotte MH, Formstecher P, Lefebvre P. Retinoic acid receptors inhibit AP1 activation by regulating extracellular signal-regulated kinase and CBP recruitment to an AP1-responsive promoter. Mol Cell Biol 2002;22:4522–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Q, Burghardt R, Safe S. Vitamin D-interacting protein 205 (DRIP205) coactivation of estrogen receptor alpha (ERalpha) involves multiple domains of both proteins. J Biol Chem 2004;279:53602–12 [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Tanaka K, Yan J, Li J, Peng D, Jiang Y, Yang Z, Barton MC, Wen H, Shi X. Regulation of estrogen receptor α by histone methyltransferase SMYD2-mediated protein methylation. Proc Natl Acad Sci USA 2013;110:17284–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenuwein T, Allis CD. Translating the histone code. Science 2001;293:1074–80 [DOI] [PubMed] [Google Scholar]
- 31.Kim S, Shevde NK, Pike JW. 1, 25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J Bone Miner Res 2005;20:305–17 [DOI] [PubMed] [Google Scholar]
- 32.Kang Z, Pirskanen A, Jänne OA, Palvimo JJ. Involvement of proteasome in the dynamic assembly of the androgen receptor transcription complex. J Biol Chem 2002;277:48366–71 [DOI] [PubMed] [Google Scholar]
- 33.Degenhardt T, Rybakova KN, Tomaszewska A, Mone MJ, Westerhoff HV, Bruggeman FJ, Carlberg C. Population-level transcription cycles derive from stochastic timing of single-cell transcription. Cell 2010;143:651 Retraction of: Degenhardt T, Rybakova KN, Tomaszewska A, Mone MJ, Westerhoff HV, Bruggeman FJ, Carlberg C. Cell. 2009;138:489–501 [DOI] [PubMed] [Google Scholar]
- 34.Hestermann EV, Brown M. Agonist and chemopreventative ligands induce differential transcriptional cofactor recruitment by aryl hydrocarbon receptor. Mol Cell Biol 2003;23:7920–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Degner SC, Papoutsis AJ, Selmin O, Romagnolo DF. Targeting of aryl hydrocarbon receptor-mediated activation of cyclooxygenase-2 expression by theindole-3-carbinol metabolite 3,3′-diindolylmethane in breast cancer cells. J Nutr 2009;139:26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev 2006;20:1405–28 [DOI] [PubMed] [Google Scholar]
- 37.Hiragami-Hamada K, Shinmyozu K, Hamada D, Tatsu Y, Uegaki K, Fujiwara S, Nakayama J. N-terminal phosphorylation of HP1{alpha} promotes its chromatin binding. Mol Cell Biol 2001;31:1186–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yokoyama A, Takezawa S, Schüle R, Kitagawa H, Kato S. Transrepressive function of TLX requires the histone demethylase LSD1. Mol Cell Biol 2008;28:3995–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohtake F, Takeyama K, Matsumoto T, Kitagawa H, Yamamoto Y, Nohara K, Tohyama C, Krust A, Mimura J, Chambon P, et al. Modulation of oestrogen receptor signalling by association with the activated dioxin receptor. Nature 2003;423:545–50 [DOI] [PubMed] [Google Scholar]
- 40.Murayama A, Kim MS, Yanagisawa J, Takeyama K, Kato S. Transrepression by a liganded nuclear receptor via a bHLH activator through co-regulator switching. EMBO J 2004;23:1598–608 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol Cell 2003;12:723–34 [DOI] [PubMed] [Google Scholar]
- 42.Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG, et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell 2005;122:707–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell 2002;110:55–67 [DOI] [PubMed] [Google Scholar]
- 44.Métivier R, Penot G, Hübner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 2003;115:751–63 [DOI] [PubMed] [Google Scholar]
- 45.Lee S, Roeder RG, Lee JW. Roles of histone H3-lysine 4 methyltransferase complexes in NR-mediated gene transcription. Prog Mol Biol Transl Sci 2009;87:343–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dormann HL, Tseng BS, Allis CD, Funabiki H, Fischle W. Dynamic regulation of effector protein binding to histone modifications: the biology of HP1 switching. Cell Cycle 2006;5:2842–51 [DOI] [PubMed] [Google Scholar]
- 47.Li Y, Sun L, Zhang Y, Wang D, Wang F, Liang J, Gui B, Shang Y. The histone modifications governing TFF1 transcription mediated by estrogen receptor. J Biol Chem 2011;286:13925–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sears DD, Hsiao A, Ofrecio JM, Chapman J, He W, Olefsky JM. Selective modulation of promoter recruitment and transcriptional activity of PPARgamma. Biochem Biophys Res Commun 2007;364:515–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papoutsis AJ, Lamore SD, Wondrak GT, Selmin OI, Romagnolo DF. Resveratrol prevents epigenetic silencing of BRCA-1 by the aromatic hydrocarbon receptor in human breast cancer cells. J Nutr 2010;140:1607–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gehm BD, McAndrews JM, Chien PY, Jameson JL. Resveratrol, a polyphenolic compound found in grapes and wine, is an agonist for the estrogen receptor. Proc Natl Acad Sci USA 1997;94:14138–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hardy TM, Tollefsbol TO. Epigenetic diet: impact on the epigenome and cancer. Epigenomics 2011;3:503–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol 2007;213:384–90 [DOI] [PubMed] [Google Scholar]
- 53.Niehrs C. Active DNA demethylation and DNA repair. Differentiation 2009;77:1–11 [DOI] [PubMed] [Google Scholar]
- 54.Xue J, Wijeratne SS, Zempleni J. Holocarboxylase synthetase synergizes with methyl CpG binding protein 2 and DNA methyltransferase 1 in the transcriptional repression of long-terminal repeats. Epigenetics 2013;8:504–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, Hassan YI, Moriyama H, Zempleni J. doi: 10.1016/j.jnutbio.2012.12.003. Holocarboxylase synthetase interacts physically with euchromatic histone-lysine N-methyltransferase, linking histone biotinylation with methylation events. J Nutr Biochem. 2013;24:1446–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bao B, Pestinger V, Hassan YI, Borgstahl GE, Kolar C, Zempleni J. Holocarboxylase synthetase is a chromatin protein and interacts directly with histone H3 to mediate biotinylation of K9 and K18. J Nutr Biochem 2011;22:470–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stanley JS, Griffin JB, Zempleni J. Biotinylation of histones in human cells. Effects of cell proliferation. Eur J Biochem 2001;268:5424–9 [DOI] [PubMed] [Google Scholar]
- 58.Bailey LM, Ivanov RA, Wallace JC, Polyak SW. Artifactual detection of biotin on histones by streptavidin. Anal Biochem 2008;373:71–7 [DOI] [PubMed] [Google Scholar]
- 59.Kuroishi T, Rios-Avila L, Pestinger V, Wijeratne SS, Zempleni J. Biotinylation is a natural, albeit rare, modification of human histones. Mol Genet Metab 2011;104:537–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chew YC, West JT, Kratzer SJ, Ilvarsonn AM, Eissenberg JC, Dave BJ, Klinkebiel D, Christman JK, Zempleni J. Biotinylation of histones represses transposable elements in human and mouse cells and cell lines and in Drosophila melanogaster. J Nutr 2008;138:2316–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Métivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature 2008;452:45–50 [DOI] [PubMed] [Google Scholar]
- 62.Wilks AF, Cozens PJ, Mattaj IW, Jost JP. Estrogen induces a demethylation at the 5′ end region of the chicken vitellogenin gene. Proc Natl Acad Sci USA 1982;79:4252–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomassin H, Flavin M, Espinás ML, Grange T. Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J 2001;20:1974–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kouzmenko A, Ohtake F, Fujiki R, Kato S. Hormonal gene regulation through DNA methylation and demethylation. Epigenomics 2010;2:765–74 [DOI] [PubMed] [Google Scholar]
- 65.Kress C, Thomassin H, Grange T. Active cytosine demethylation triggered by a nuclear receptor involves DNA strand breaks. Proc Natl Acad Sci USA 2006;103:11112–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barreto G, Schäfer A, Marhold J, Stach D, Swaminathan SK, Handa V, Döderlein G, Maltry N, Wu W, Lyko F, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 2007;445:671–5 [DOI] [PubMed] [Google Scholar]
- 67.Dalton SR, Bellacosa A. DNA demethylation by TDG. Epigenomics 2012;4:459–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 2011;146:67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet 2009;43:143–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 2013;502:472–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 2014;156:45–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen D, Lucey MJ, Phoenix F, Lopez-Garcia J, Hart SM, Losson R, Buluwela L, Coombes RC, Chambon P, Schär P, et al. G mismatch-specific thymine-DNA glycosylase potentiates transcription of estrogen-regulated genes through direct interaction with estrogen receptor alpha. J Biol Chem 2003;278:38586–92 [DOI] [PubMed] [Google Scholar]
- 73.Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol Cell 2002;9:265–77 [DOI] [PubMed] [Google Scholar]
- 74.Um S, Harbers M, Benecke A, Pierrat B, Losson R, Chambon P. Retinoic acid receptors interact physically and functionally with the T:G mismatch-specific thymine-DNA glycosylase. J Biol Chem 1998;273:20728–36 [DOI] [PubMed] [Google Scholar]
- 75.Sampath H, Ntambi JM. Polyunsaturated fatty acid regulation of genes of lipid metabolism. Annu Rev Nutr 2005;25:317–40 [DOI] [PubMed] [Google Scholar]
- 76.Pineda Torra I, Jamshidi Y, Flavell DM, Fruchart JC, Staels B. Characterization of the human PPARalpha promoter: identification of a functional nuclear receptor response element. Mol Endocrinol 2002;16:1013–28 [DOI] [PubMed] [Google Scholar]
- 77.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr 2005;135:1382–6 [DOI] [PubMed] [Google Scholar]
- 78.Slater-Jefferies JL, Lillycrop KA, Townsend PA, Torrens C, Hoile SP, Hanson MA, Burdge GC. Feeding a protein-restricted diet during pregnancy induces altered epigenetic regulation of peroxisomal proliferator-activated receptor-α in the heart of the offspring. J Dev Orig Health Dis 2011;2:250–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoyo C, Murtha AP, Schildkraut JM, Forman MR, Calingaert B, Demark-Wahnefried W, Kurtzberg J, Jirtle RL, Murphy SK. Folic acid supplementation before and during pregnancy in the Newborn Epigenetics STudy (NEST). BMC Public Health 2011;11:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jun HJ, Kim J, Hoang MH, Lee SJ. Hepatic lipid accumulation alters global histone h3 lysine 9 and 4 trimethylation in the peroxisome proliferator-activated receptor alpha network. PLoS One 2012;7:e44345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pooya S, Blaise S, Moreno Garcia M, Giudicelli J, Alberto JM, Guéant-Rodriguez RM, Jeannesson E, Gueguen N, Bressenot A, Nicolas B, et al. Methyl donor deficiency impairs fatty acid oxidation through PGC-1α hypomethylation and decreased ER-α, ERR-α, and HNF-4α in the rat liver. J Hepatol 2012;57:344–51 [DOI] [PubMed] [Google Scholar]
- 82.Chou CJ, Haluzik M, Gregory C, Dietz KR, Vinson C, Gavrilova O, Reitman ML. WY14,643, a peroxisome proliferator-activated receptor alpha (PPARalpha) agonist, improves hepatic and muscle steatosis and reverses insulin resistance in lipoatrophic A-ZIP/F-1 mice. J Biol Chem 2002;277:24484–9 [DOI] [PubMed] [Google Scholar]
- 83.Knapp P, Chabowski A, Błachnio-Zabielska A, Jarząbek K, Wołczyński S. doi: 10.1155/2012/471524. Altered peroxisome-proliferator activated receptors expression in human endometrial cancer. PPAR Res. 2012;2012:471524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pogribny IP, Tryndyak VP, Woods CG, Witt SE, Rusyn I. Epigenetic effects of the continuous exposure to peroxisome proliferator WY-14,643 in mouse liver are dependent upon peroxisome proliferator activated receptor alpha. Mutat Res 2007;625:62–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guyton KZ, Chiu WA, Bateson TF, Jinot J, Scott CS, Brown RC, Caldwell JC. A reexamination of the PPAR-alpha activation mode of action as a basis for assessing human cancer risks of environmental contaminants. Environ Health Perspect 2009;117:1664–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kershaw EE, Flier JS. Adipose tissue as an endocrine organ. J Clin Endocrinol Metab 2004;89:2548–56 [DOI] [PubMed] [Google Scholar]
- 87.Galic S, Oakhill JS, Steinberg GR. Adipose tissue as an endocrine organ. Mol Cell Endocrinol 2010;316:129–39 [DOI] [PubMed] [Google Scholar]
- 88.Elabd C, Chiellini C, Carmona M, Galitzky J, Cochet O, Petersen R, Pénicaud L, Kristiansen K, Bouloumié A, Casteilla L, et al. Human multipotent adipose-derived stem cells differentiate into functional brown adipocytes. Stem Cells 2009;27:2753–60 [DOI] [PubMed] [Google Scholar]
- 89.Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, Rosenbaum M, Zhao Y, Gu W, Farmer SR, et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of PPARγ. Cell 2012;150:620–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Koble CS, Devchand P, Wahli W, Willson TM, Lenhard JM, et al. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc Natl Acad Sci USA 1997;94:4318–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Edwards IJ, O’Flaherty JT. doi: 10.1155/2008/358052. Omega-3 fatty acids and PPARgamma in cancer. PPAR Res. 2008;2008:358052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Francis GA, Fayard E, Picard F, Auwerx J. Nuclear receptors and the control of metabolism. Annu Rev Physiol 2003;65:261–311 [DOI] [PubMed] [Google Scholar]
- 93.Sugii S, Evans RM. Epigenetic codes of PPARγ in metabolic disease. FEBS Lett 2011;585:2121–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pancione M, Sabatino L, Fucci A, Carafa V, Nebbioso A, Forte N, Febbraro A, Parente D, Ambrosino C, Normanno N, et al. Epigenetic silencing of peroxisome proliferator-activated receptor γ is a biomarker for colorectal cancer progression and adverse patients’ outcome. PLoS One 2010;5:e14229. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 95.Yang MD, Chiang YM, Higashiyama R, Asahina K, Mann DA, Mann J, Wang CC, Tsukamoto H. Rosmarinic acid and baicalin epigenetically derepress peroxisomal proliferator-activated receptor γ in hepatic stellate cells for their antifibrotic effect. Hepatology 2012;55:1271–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Joss-Moore LA, Wang Y, Baack ML, Yao J, Norris AW, Yu X, Callaway CW, McKnight RA, Albertine KH, Lane RH. IUGR decreases PPARγ and SETD8 Expression in neonatal rat lung and these effects are ameliorated by maternal DHA supplementation. Early Hum Dev 2010;86:785–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Barrès R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, Krook A, Zierath JR. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab 2009;10:189–98 [DOI] [PubMed] [Google Scholar]
- 98.Barrès R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, Caidahl K, Krook A, O’Gorman DJ, Zierath JR. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab 2012;15:405–11 [DOI] [PubMed] [Google Scholar]
- 99.Wakabayashi K, Okamura M, Tsutsumi S, Nishikawa NS, Tanaka T, Sakakibara I, Kitakami J, Ihara S, Hashimoto Y, Hamakubo T, et al. The peroxisome proliferator-activated receptor gamma/retinoid X receptor alpha heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol Cell Biol 2009;29:3544–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fujiki K, Kano F, Shiota K, Murata M. Expression of the peroxisome proliferator activated receptor gamma gene is repressed by DNA methylation in visceral adipose tissue of mouse models of diabetes. BMC Biol 2009;7:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim SJ, Nian C, McIntosh CH. Adipocyte expression of the glucose-dependent insulinotropic polypeptide receptor involves gene regulation by PPARγ and histone acetylation. J Lipid Res 2011;52:759–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rosen CJ. Revisiting the rosiglitazone story–lessons learned. N Engl J Med 2010;363:803–6 [DOI] [PubMed] [Google Scholar]
- 103.Woodcock J, Sharfstein JM, Hamburg M. Regulatory action on rosiglitazone by the U.S. Food and Drug Administration. N Engl J Med 2010;363:1489–91 [DOI] [PubMed] [Google Scholar]
- 104.Oseni T, Patel R, Pyle J, Jordan VC. Selective estrogen receptor modulators and phytoestrogens. Planta Med 2008;74:1656–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kurebayashi J, Otsuki T, Kunisue H, Tanaka K, Yamamoto S, Sonoo H. Expression levels of estrogen receptor-alpha, estrogen receptor-beta, coactivators, and corepressors in breast cancer. Clin Cancer Res 2000;6:512–8 [PubMed] [Google Scholar]
- 106.An J, Tzagarakis-Foster C, Scharschmidt TC, Lomri N, Leitman DC. Estrogen receptor beta-selective transcriptional activity and recruitment of coregulators by phytoestrogens. J Biol Chem 2001;276:17808–14 [DOI] [PubMed] [Google Scholar]
- 107.Rüegg J, Cai W, Karimi M, Kiss NB, Swedenborg E, Larsson C, Ekström TJ, Pongratz I. Epigenetic regulation of glucose transporter 4 by estrogen receptor β. Mol Endocrinol 2011;25:2017–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Re A, Aiello A, Nanni S, Grasselli A, Benvenuti V, Pantisano V, Strigari L, Colussi C, Ciccone S, Mazzetti AP, et al. Silencing of GSTP1, a prostate cancer prognostic gene, by the estrogen receptor-β and endothelial nitric oxide synthase complex. Mol Endocrinol 2011;25:2003–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bowers JL, Tyulmenkov VV, Jernigan SC, Klinge CM. Resveratrol acts as a mixed agonist/antagonist for estrogen receptors alpha and beta. Endocrinology 2000;141:3657–67 [DOI] [PubMed] [Google Scholar]
- 110.Hilakivi-Clarke L, Cho E, Onojafe I, Raygada M, Clarke R. Maternal exposure to genistein during pregnancy increases carcinogen-induced mammary tumorigenesis in female rat offspring. Oncol Rep 1999;6:1089–95 [DOI] [PubMed] [Google Scholar]
- 111.Newbold RR, Banks EP, Bullock B, Jefferson WN. Uterine adenocarcinoma in mice treated neonatally with genistein. Cancer Res 2001;61:4325–8 [PubMed] [Google Scholar]
- 112.Tang WY, Newbold R, Mardilovich K, Jefferson W, Cheng RY, Medvedovic M, Ho SM. Persistent hypomethylation in the promoter of nucleosomal binding protein 1 (Nsbp1) correlates with overexpression of Nsbp1 in mouse uteri neonatally exposed to diethylstilbestrol or genistein. Endocrinology 2008;149:5922–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jiang N, Zhou LQ, Zhang XY. Downregulation of the nucleosome-binding protein 1 (NSBP1) gene can inhibit the in vitro and in vivo proliferation of prostate cancer cells. Asian J Androl 2010;12:709–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Qin W, Zhu W, Shi H, Hewett JE, Ruhlen RL, MacDonald RS, Rottinghaus GE, Chen YC, Sauter ER. Soy isoflavones have an antiestrogenic effect and alter mammary promoter hypermethylation in healthy premenopausal women. Nutr Cancer 2009;61:238–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sun J, Xu X, Liu J, Liu H, Fu L, Gu L. Epigenetic regulation of retinoic acid receptor β2 gene in the initiation of breast cancer. Med Oncol 2011;28:1311–8 [DOI] [PubMed] [Google Scholar]
- 116.Bosviel R, Dumollard E, Déchelotte P, Bignon YJ, Bernard-Gallon D. Can soy phytoestrogens decrease DNA methylation in BRCA1 and BRCA2 oncosuppressor genes in breast cancer? OMICS 2012;16:235–44 [DOI] [PubMed] [Google Scholar]
- 117.Gudas JM, Nguyen H, Li T, Cowan KH. Hormone-dependent regulation of BRCA1 in human breast cancer cells. Cancer Res 1995;55:4561–5 [PubMed] [Google Scholar]
- 118.Romagnolo D, Annab LA, Thompson TE, Risinger JI, Terry LA, Barrett JC, Afshari CA. Estrogen upregulation of BRCA1 expression with no effect on localization. Mol Carcinog 1998;22:102–9 [DOI] [PubMed] [Google Scholar]
- 119.Jeffy BD, Hockings JK, Kemp MQ, Morgan SS, Hager JA, Beliakoff J, Whitesell LJ, Bowden GT, Romagnolo DF. An estrogen receptor-alpha/p300 complex activates the BRCA-1 promoter at an AP-1 site that binds Jun/Fos transcription factors: repressive effects of p53 on BRCA-1 transcription. Neoplasia 2005;7:873–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li Y, Meeran SM, Patel SN, Chen H, Hardy TM, Tollefsbol TO. Epigenetic reactivation of estrogen receptor-α (ERα) by genistein enhances hormonal therapy sensitivity in ERα-negative breast cancer. Mol Cancer 2013;12:9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li H, Xu W, Huang Y, Huang X, Xu L, Lv Z. Genistein demethylates the promoter of CHD5 and inhibits neuroblastoma growth in vivo. Int J Mol Med 2012;30:1081–6 [DOI] [PubMed] [Google Scholar]
- 122.Stefanska B, Salamé P, Bednarek A, Fabianowska-Majewska K. Comparative effects of retinoic acid, vitamin D and resveratrol alone and in combination with adenosine analogues on methylation and expression of phosphatase and tensin homologue tumour suppressor gene in breast cancer cells. Br J Nutr 2012;107:781–90 [DOI] [PubMed] [Google Scholar]
- 123.Romagnolo DF, Degner SC, Selmin O. Dietary Compounds that target the AhR and Cancer Risk. In: Milner JA and Romagnolo DF, eds. Bioactive food components and cancer. NY: Humana Press/Springer. 2010;761–82 [Google Scholar]
- 124.Beedanagari SR, Taylor RT, Bui P, Wang F, Nickerson DW, Hankinson O. Role of epigenetic mechanisms in differential regulation of the dioxin-inducible human CYP1A1 and CYP1B1 genes. Mol Pharmacol 2010;78:608–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Safe S, Wang F, Porter W, Duan R, McDougal A. Ah receptor agonists as endocrine disruptors: antiestrogenic activity and mechanisms. Toxicol Lett 1998;102–103:343–7 [DOI] [PubMed] [Google Scholar]
- 126.Papoutsis AJ, Borg JL, Selmin OI, Romagnolo DF. BRCA-1 promoter hypermethylation and silencing induced by the aromatic hydrocarbon receptor-ligand TCDD are prevented by resveratrol in MCF-7 cells. J Nutr Biochem 2012;23:1324–32 [DOI] [PubMed] [Google Scholar]
- 127.Kim MS, Kondo T, Takada I, Youn MY, Yamamoto Y, Takahashi S, Matsumoto T, Fujiyama S, Shirode Y, Yamaoka I, et al. DNA demethylation in hormone-induced transcriptional derepression. Nature 2012;486:1280 Retraction of: Kim MS, Kondo T, Takada I, Youn MY, Yamamoto Y, Takahashi S, Matsumoto T, Fujiyama S, Shirode Y, Yamaoka I, et al. Nature. 2009;461:1007–12 [DOI] [PubMed] [Google Scholar]
- 128.Leuenberger N, Pradervand S, Wahli W. Sumoylated PPARalpha mediates sex-specific gene repression and protects the liver from estrogen-induced toxicity in mice. J Clin Invest 2009;119:3138–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Modica S, Gadaleta RM, Moschetta A. Deciphering the nuclear bile acid receptor FXR paradigm. Nucl Recept Signal 2010;8:e005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Modica S, Murzilli S, Salvatore L, Schmidt DR, Moschetta A. Nuclear bile acid receptor FXR protects against intestinal tumorigenesis. Cancer Res 2008;68:9589–94 [DOI] [PubMed] [Google Scholar]
- 131.Lax S, Schauer G, Prein K, Kapitan M, Silbert D, Berghold A, Berger A, Trauner M. Expression of the nuclear bile acid receptor/farnesoid X receptor is reduced in human colon carcinoma compared to nonneoplastic mucosa independent from site and may be associated with adverse prognosis. Int J Cancer 2012;130:2232–9 [DOI] [PubMed] [Google Scholar]
- 132.Wang YD, Chen WD, Wang M, Yu D, Forman BM, Huang W. Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology 2008;48:1632–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, Tsang S, Wu SY, Chiang CM, Veenstra TD. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab 2009;10:392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fang S, Tsang S, Jones R, Ponugoti B, Yoon H, Wu SY, Chiang CM, Willson TM, Kemper JK. The p300 acetylase is critical for ligand-activated farnesoid X receptor (FXR) induction of SHP. J Biol Chem 2008;283:35086–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lee J, Seok S, Yu P, Kim K, Smith Z, Rivas-Astroza M, Zhong S, Kemper JK. Genomic analysis of hepatic farnesoid X receptor binding sites reveals altered binding in obesity and direct gene repression by farnesoid X receptor in mice. Hepatology 2012;56:108–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ananthanarayanan M, Li Y, Surapureddi S, Balasubramaniyan N, Ahn J, Goldstein JA, Suchy FJ. Histone H3K4 trimethylation by MLL3 as part of ASCOM complex is critical for NR activation of bile acid transporter genes and is downregulated in cholestasis. Am J Physiol Gastrointest Liver Physiol 2011;300:G771–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rizzo G, Renga B, Antonelli E, Passeri D, Pellicciari R, Fiorucci S. The methyl transferase PRMT1 functions as co-activator of farnesoid X receptor (FXR)/9-cis retinoid X receptor and regulates transcription of FXR responsive genes. Mol Pharmacol 2005;68:551–8 [DOI] [PubMed] [Google Scholar]
- 138.Balasubramaniyan N, Ananthanarayanan M, Suchy FJ. Direct methylation of FXR by Set7/9, a lysine methyltransferase, regulates the expression of FXR target genes. Am J Physiol Gastrointest Liver Physiol 2012;302:G937–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, Peng J, Cheng X, Vertino PM. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol Cell 2008;30:336–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Waddington CH. The epigenotype. Endeavour 1942;1:18–20 [Google Scholar]
- 141.Holliday R. Epigenetics: a historial overview. Epigenetics 2006;1:76–80 [DOI] [PubMed] [Google Scholar]
- 142.Jamniczky HA, Boughner JC, Rolian C, Gonzalez PN, Powell CD, Schmidt EJ, Parsons TE, Bookstein FL, Hallgrímsson B. Rediscovering Waddington in the post-genomic age: operationalising Waddington’s epigenetics reveals new ways to investigate the generation and modulation of phenotypic variation. Bioessays 2010;32:553–8 [DOI] [PubMed] [Google Scholar]
- 143.Nakshatri H, Bhat-Nakshatri P. Multiple parameters determine the specificity of transcriptional response by nuclear receptors HNF-4, ARP-1, PPAR, RAR and RXR through common response elements. Nucleic Acids Res 1998;26:2491–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mai Z, Blackburn GL, Zhou JR. Genistein sensitizes inhibitory effect of tamoxifen on the growth of estrogen receptor-positive and HER2-overexpressing human breast cancer cells. Mol Carcinog 2007;46:534–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bougarne N, Paumelle R, Caron S, Hennuyer N, Mansouri R, Gervois P, Staels B, Haegeman G, De Bosscher K. PPARalpha blocks glucocorticoid receptor alpha-mediated transactivation but cooperates with the activated glucocorticoid receptor alpha for transrepression on NF-kappaB. Proc Natl Acad Sci USA 2009;106:7397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gapp K, von Ziegler L, Tweedie-Cullen RY, Mansuy IM. Early life epigenetic programming and transmission of stress-induced traits in mammals: how and when can environmental factors influence traits and their transgenerational inheritance? Bioessays 2014; Feb 28 (Epub ahead of print; DOI:) [DOI] [PubMed] [Google Scholar]
- 147.Fang MZ, Chen D, Sun Y, Jin Z, Christman JK, Yang CS. Reversal of hypermethylation and reactivation of p16INK4a, RARbeta, and MGMT genes by genistein and other isoflavones from soy. Clin Cancer Res 2005;11:7033–41 [DOI] [PubMed] [Google Scholar]
- 148.Li Y, Meeran SM, Patel SN, Chen H, Hardy TM, Tollefsbol TO. Epigenetic reactivation of estrogen receptor-α (ERα) by genistein enhances hormonal therapy sensitivity in ERα-negative breast cancer. Mol Cancer 2013;12:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang CS. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res 2003;63:7563–70 [PubMed] [Google Scholar]
- 150.Berner C, Aumüller E, Gnauck A, Nestelberger M, Just A, Haslberger AG. Epigenetic control of estrogen receptor expression and tumor suppressor genes is modulated by bioactive food compounds. Ann Nutr Metab 2010;57:183–9 [DOI] [PubMed] [Google Scholar]
- 151.Marconett CN, Sundar SN, Tseng M, Tin AS, Tran KQ, Mahuron KM, Bjeldanes LF, Firestone GL. Indole-3-carbinol downregulation of telomerase gene expression requires the inhibition of estrogen receptor-alpha and Sp1 transcription factor interactions within the hTERT promoter and mediates the G1 cell cycle arrest of human breast cancer cells. Carcinogenesis 2011;32:1315–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lee YH, Kwak J, Choi HK, Choi KC, Kim S, Lee J, Jun W, Park HJ, Yoon HG. EGCG suppresses prostate cancer cell growth modulating acetylation of androgen receptor by anti-histone acetyltransferase activity. Int J Mol Med 2012;30:69–74 [DOI] [PubMed] [Google Scholar]
- 153.Kong D, Heath E, Chen W, Cher M, Powell I, Heilbrun L, Li Y, Ali S, Sethi S, Hassan O, et al. Epigenetic silencing of miR-34a in human prostate cancer cells and tumor tissue specimens can be reversed by BR-DIM treatment. Am J Transl Res 2012;4:14–23 [PMC free article] [PubMed] [Google Scholar]
- 154.Gu JW, Makey KL, Tucker KB, Chinchar E, Mao X, Pei I, Thomas EY, Miele L. EGCG, a major green tea catechin suppresses breast tumor angiogenesis and growth via inhibiting the activation of HIF-1α and NFκB, and VEGF expression. Vasc Cell 2013;5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nakata R, Takahashi S, Inoue H. Recent advances in the study on resveratrol. Biol Pharm Bull 2012;35:273–9 [DOI] [PubMed] [Google Scholar]
- 156.Ricketts ML, Moore DD, Banz WJ, Mezei O, Shay NF. Molecular mechanisms of action of the soy isoflavones includes activation of promiscuous nuclear receptors. A review. J Nutr Biochem 2005;16:321–30 [DOI] [PubMed] [Google Scholar]
- 157.Penumetcha M, Santanam N. doi: 10.1155/2012/858352. Nutraceuticals as ligands of PPARγ. PPAR Res. 2012;2012:858352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ing SW, Belury MA. Impact of conjugated linoleic acid on bone physiology: proposed mechanism involving inhibition of adipogenesis. Nutr Rev 2011;69:123–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dawson MI, Xia Z. doi: 10.1016/j.bbalip.2011.09.014. The retinoid X receptors and their ligands. Biochim Biophys Acta. 2012;1821:21–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Haussler MR, Whitfield GK, Kaneko I, Haussler CA, Hsieh D, Hsieh JC, Jurutka PW. Molecular mechanisms of vitamin D action. Calcif Tissue Int 2013;92:77–98 [DOI] [PubMed] [Google Scholar]
- 161.Fleet JC, DeSmet M, Johnson R, Li Y. Vitamin D and cancer: a review of molecular mechanisms. Biochem J 2012;441:61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jiang Y, Fleet JC. Phorbol esters enhance 1α,25-dihydroxyvitamin D3-regulated 25-hydroxyvitamin D-24-hydroxylase (CYP24A1) gene expression through ERK-mediated phosphorylation of specific protein 3 (Sp3) in Caco-2 cells. Mol Cell Endocrinol 2012;361:31–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bartik L, Whitfield GK, Kaczmarska M, Lowmiller CL, Moffet EW, Furmick JK, Hernandez Z, Haussler CA, Haussler MR, Jurutka PW. Curcumin: a novel nutritionally derived ligand of the vitamin D receptor with implications for colon cancer chemoprevention. J Nutr Biochem 2010;21:1153–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sevov M, Elfineh L, Cavelier LB. Resveratrol regulates the expression of LXR-alpha in human macrophages. Biochem Biophys Res Commun 2006;348:1047–54 [DOI] [PubMed] [Google Scholar]
- 165.Moon HS, Chung CS, Lee HG, Kim TG, Choi YJ, Cho CS. Inhibitory effect of (−)-epigallocatechin-3-gallate on lipid accumulation of 3T3–L1 cells. Obesity (Silver Spring) 2007;15:2571–82 [DOI] [PubMed] [Google Scholar]
- 166.Ronis MJ, Chen Y, Badeaux J, Badger TM. Dietary soy protein isolate attenuates metabolic syndrome in rats via effects on PPAR, LXR, and SREBP signaling. J Nutr 2009;139:1431–8 [DOI] [PubMed] [Google Scholar]