Abstract
SOCS3 (Suppressor of Cytokine Signaling 3) inhibits the intracellular signaling cascade initiated by exposure of cells to cytokines. SOCS3 regulates signaling via two distinct mechanisms: directly inhibiting the catalytic activity of Janus Kinases (JAKs) that initiate the intracellular signaling cascade and catalysing the ubiquitination of signaling components by recruiting components of an E3 ubiquitin ligase complex. Here we investigate the latter mode-of-action biochemically by reconstructing a SOCS3-based E3 ubiquitin ligase complex in vitro using fully purified, recombinant components and examining its ability to promote the ubiquitination of molecules involved in the cytokine signaling cascade. We show that SOCS3 is an active substrate recruitment module for a Cullin5-based E3 ligase and have defined the core protein components required for ubiquitination. SOCS3-induced poly-ubiquitination was rapid and could proceed through a number of different ubiquitin lysines. SOCS3 catalysed the ubiquitination of both the IL-6 receptor common chain (gp130) and JAK2.
Keywords: Suppressor of cytokine signaling, Ubiquitination, Cullin5, E3 ubiquitin ligase, SOCS3, JAK, cytokine receptor, cytokine signaling
INTRODUCTION
Cytokine signaling must be tightly controlled in order to prevent excessive proliferation which could lead to inflammation and tumorigenesis. The major regulator of interleukin-6 (IL-6) family cytokines, as well as granulocyte colony stimulating factor (G-CSF) and leptin, is SOCS3 (Suppressor of Cytokine Signaling 3). SOCS3 forms the effector molecule of a negative feedback loop; it is expressed in response to cytokine stimulation and then feeds back to inhibit the cytokine-induced signaling cascade, allowing the cell to return to its basal state1; 2; 3.
The primary mechanism of SOCS3 action is via direct inhibition of the catalytic activity of JAKs (Janus Kinases)4; 5; 6. JAKs are bound to the cytoplasmic domains of cytokine receptors and initiate the intracellular component of the signaling cascade by phosphorylating the STAT (Signal Transducers and Activators of Transcription) family of transcription factors. SOCS3 is able to block JAK phosphorylation of STAT and thereby inhibit further signaling7; 8. However, SOCS3 is believed to act via a second mode-of-action in addition to direct JAK inhibition. This second mechanism is its ability to recruit an E3 ubiquitin ligase complex and thereby inducing the ubiquitination and subsequent degradation of signaling components9. This relies upon a conserved domain at the SOCS3 C-terminus termed the SOCS Box2. The SOCS Box is present in all eight SOCS family members (SOCS1-7 and CIS, Cytokine Inducible SH2 domain containing protein) and is a small (~40 amino acid) domain found in approximately 50 human proteins. Its function is to recruit elongins B and C2; 9; 10; 11 and Cullin510; 11. Cullin5 is the scaffold protein for a large class of E3 ubiquitin ligases, enzymes that directly catalyse the covalent addition of ubiquitin to lysines on target proteins12; 13.
Ubiquitination is catalyzed by an enzymatic cascade. This consists of activating (E1), conjugating (E2) and ligating (E3) enzymes13. The Cullin-Ring-Ligase (CRL) family of ubiquitin ligases 14 are the largest class of E3 ligases and consist of multiple subunits all built around a Cullin scaffold12. The C-terminal domain of this scaffold binds activated ubiquitin whilst the N-terminal domain binds substrate14 thus allowing ubiquitin to be transferred to a substrate lysine. An intriguing aspect of Cullin-based ligases is that they themselves are post-translationally modified by a ubiquitin-like protein, Nedd815. Neddylation occurs on a conserved lysine near the E2-ubiquitin docking site and enhances the ubiquitin ligase efficiency by inducing a substantial conformational change in the Cullin scaffold such that the E2 is released from the core of the complex, allowing it more efficient access to the substrate lysine side chain16.
The purpose of the E3 ligase is to bring together the activated ubiquitin and the substrate that is to be ubiquitinated. Both of these binding events are co-ordinated by Cullin5 but mediated by further sub-units. For example, the interaction between Cullin5 and activated ubiquitin is mediated by a RING domain protein, Rbx217; 18, whilst the interaction with substrate is mediated by a diverse array of substrate-recruiting proteins that contain SOCS box domains, including SOCS3. Whilst E3 ligase activity has been demonstrated for a number of SOCS box-containing proteins17, of the eight members of the SOCS family, only for SOCS1, which promotes the degradation of the TEL-JAK oncoprotein18, vav19, IRS-1 and IRS-220 has E3 ligase activity been directly demonstrated.
Apart from SOCS1, SOCS3 is the most potent inhibitor of cytokine signaling. SOCS3 regulates the response to IL-621, granulocyte colony stimulating factor22, leukemia inhibitory factor23 and leptin24 and binds to both the cytoplasmic domains of the receptors for those cytokines as well as to the receptor-bound JAKs. However, SOCS3 can inhibit cytokine signaling through ubiquitin-independent mechanisms7,8 and therefore it is unknown whether a SOCS3-based E3 ligase will promote ubiquitination of JAK or cytokine receptors or other, as yet unidentified, molecules.
In this study we have reconstructed a full E1–E2-E3 cascade in vitro using purified components in order to study the mechanism of SOCS3/Cullin5-based ubiquitination and determine substrates for SOCS3-mediated ubiquitination. We find that SOCS3/Cullin5 is active in the presence of either Rbx1 or Rbx2 and these can recruit any one of the closely related E2 enzymes: UbcH5a, UbcH5b or UbcH5c. Cullin5 itself is efficiently mono-ubiquitinated on K724, the site of neddylation in vivo. Whilst K724 ubiquitination may mimic neddylation, it is not required for E3 ligase function. A Cullin5/SOCS3 E3 ligase promotes both JAK2 and receptor poly-ubiquitination. However the rate of and extent of poly-ubiquitination of gp130 (the IL-6 family shared receptor) suggests that it is the preferred substrate rather than JAK.
EXPERIMENTAL PROCEDURES
Cloning and expression of the SOCS/elonginBC/cullin5/Rbx2 E3 ligase complex
Murine SOCS3/elonginsBC complex was cloned and expressed as previously described 10. Mouse Cullin5 was co-expressed as two domains, the N-terminal domain (1–384) and C-terminal domain (385–780) analagous to that described previously for Cullin114. The C-terminal domain of Cullin5 was cloned into the second multi-cloning site (MCS) of pACYC-DUET (Novagen) whilst mouse Rbx2 was cloned into the first MCS resulting in a HIS6 tag at its N-terminus. The N-terminal domain of Cullin5 was cloned as a GST-fusion protein into pGEX-4T1 and the two vectors were co-expressed in BL21(DE3) cells to yield a ternary GST-Cul5(NTD)/HIS6-Rbx2/Cul(CTD) complex. Expression was performed in LB broth at 18°C overnight following induction using 1mM IPTG when the O.D.600 was 0.7. The cells were harvested by centrifugation and then lysed using lysozyme and sonication. The complex was bound to Ni-NTA resin to capture Rbx2 and any excess Cullin5 removed by extensive washing. SDS-PAGE analysis of the Cul5/Rbx2 complex was used to verify the presence of both proteins. A ten-fold excess of SOCS3/elonginBC ternary complex was passed over the column to form the 5-protein E3 ligase complex. The complex was eluted in buffer containing 250mM imidazole and the eluate bound to Glutathione Sepharose (GE Healthcare) and washed thoroughly in PBS. The complex was then eluted from the resin by thrombin proteolysis of the GST fusion tag and purified by size exclusion chromatography using a Superdex 200 16/60 column (GE Healthcare). Finally the purified E3 ligase was concentrated to 2 mg/mL.
Cloning and expression of substrates: JAK2, gp130
The JH1 domain of JAK2, residues 836–1132 (Genbank: protein, AAH54807; cDNA, BC054807) fused to GST, was cloned into pFastBac HTb (Life Technologies), and the resulting bacmid was used to transfect Sf21 cells. High-titer baculovirus was used to infect 1–5 litres of Sf21 cells grown to a density of 2 × 106 ml−1. Cells were collected 72 h after infection and snap frozen. Cells were lysed by sonication and GST-JAK2 purified using glutathione sepharose (GE Healthcare) using standard procedures.
The cytoplasmic domain of the mouse IL-6 receptor (gp130 chain, residues 641–917) fused to GST was cloned into pGEX-4T. Expression was performed at 18°C for 6 hours in LB broth following induction with 1mM IPTG at a cell density of O.D600 = 1.0. The cells were harvested by centrifugation and lysed using lysozyme and sonication. The protein was bound to Glutathione Sepharose (GE Healthcare) and washed with 50 column volumes of PBS containing 2mM DTT. gp130 was eluted from the resin by thrombin proteolysis of the GST fusion tag, purified by size exclusion chromatography using a Superdex 200 26/60 column (GE Healthcare) and concentrated to 1 mg/mL. Two shorter fragments of gp130cyt (gp130cytNTD, residues 641–813 and gp130cytCTD, residues 720–917) were cloned and purified under identical conditions.
Cloning and expression of murine UbcH5a
Murine UbcH5a (E2) was expressed as a GST-fusion protein by cloning into pGEX-4T and expressed in BL21(DE3) cells at 37°C for two hours post IPTG induction. Following cell lysis, the protein was purified using Gluathione Sepharose and eluted by thrombin digestion. Size exclusion chromatography using a Superdex 75 16/60 column run in Tris-buffered saline pH7.5 was performed as the final step in purification.
Ubiquitin cascade components
Human E1 (His6 tagged), methylated-ubiquitin and a panel of human E2 enzymes (His6 tagged) were purchased from Biomol International. Bovine ubiquitin was purchased from Sigma-Aldrich.
Ubiquitination Assays
Ubiquitination assays were performed in 20 μl of 20mM Tris-HCl, 50mM NaCl, 5mM MgCl2, 2.5mM ATP, 0.1mM DTT. Reactions were stopped by the addition of 2x SDS PAGE loading buffer and heating at 95°C for 5 minutes. Typically reactions contained 0.1 μM E1, 2.5 μM E2, 2.5 μM E3, 50 μM ubiquitin and 5 μM substrate and were incubated for 30 minutes or as indicated. Reaction products were then detected by SDS-PAGE and Coomassie staining. Tryptic digestion followed by MS/MS analysis was used to identify protein species excised from SDS-polyacrylamide gels and was performed by the Joint Proteomics Service Facility (Parkville, Australia).
RESULTS
In vitro reconstitution of a SOCS3-based E3 ligase
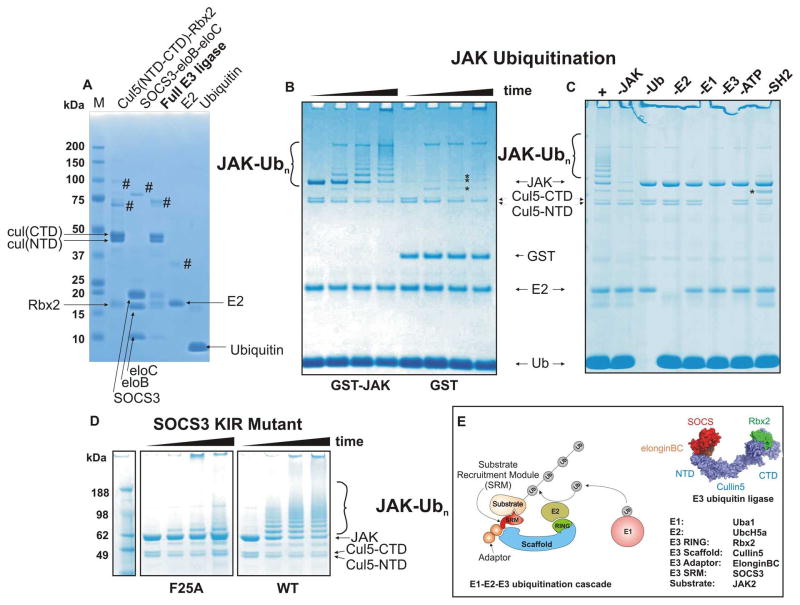
In order to investigate SOCS3-mediated ubiquitination we developed an in vitro ubiquitination assay consisting of fully purified, recombinant components. In particular we wished to construct a system that would allow direct visualisation of results (rather than visualisation by western blot) so that the ubiquitination state and stoichiometry of each component could be readily tracked. All subunits of the E3 ligase were expressed in E. coli using standard protocols. Cullin5, co-expressed with Rbx1 or Rbx2, was produced as two separate domains as first demonstrated for Cullin1 by Zheng, Schulman and colleagues 14; these associate with each other and with Rbx2, during expression and could be isolated as a ternary (Cul5(NTD):Cul5(CTD):Rbx2) complex (Figure 1A, lane 1). SOCS3/elonginsBC were also co-expressed and purified as a ternary complex as previously described (Figure 1A, lane 2). The SOCS3 construct lacked the first 21 residues and had the PEST motif replaced by a short Gly-Serx4 linker 25; 26 as these modifications enhance solubility but are not required for function. The two purified ternary complexes were then mixed and the correct stoichiometry of the full E3 ligase ensured by a combination of affinity tags and a final gel filtration step (Figure 1A, lane 3). This E3 complex could be prepared in large amounts and was highly soluble. All components were verified by mass-spectrometry. Recombinant human E1, E2 (UbcH5a) and purified bovine ubiquitin were then added to complete the ubiquitination assay system (Figure 1A, lanes 4,5).
Figure 1. A SOCS3/Cullin5 based E3 ligase catalyses JAK2JH1 ubiquitination.
(A) SDS-PAGE analysis of in vitro ubiquitination system components. Cullin5-Rbx2 and SOCS3-elonginBC are produced separately as ternary complexes (lanes 1,2) and then mixed and further purified to achieve the final E3 ubiquitin ligase. Cullin5 is produced as two domains (NTD and CTD) and these associate non-covalently to form the functional scaffold. Recombinant E2 (UbcH5a) and purified bovine ubiquitin are shown in lanes 4,5 respectively. # marks indicate impurities (B) GST-JAK2 (JH1 domain) is a substrate for polyubiquitination but GST alone is not. SOCS3 based E3 ligase (2.5uM) was incubated with ubiquitin (50uM), human E1 (100nM), purified recombinant E2 (UbcH5a, 2.5uM) and either GST-JAK2 (JH1 domain, 5uM, lanes 1–4) or GST alone (lanes 5–8) in the presence of 2.5mM Mg/ATP at 37°C for 0, 5, 30, 60 minutes. Note that the cullin C-terminal domain was polyubiquitinated in each case (asterisks). (C) GST-JAK polyubiquitination was dependent upon the presence of E1, E2, ATP, ubiquitin and the KIR/SH2 domain of SOCS3. Each reaction component was absent as indicated above the gel. -SH2 indicates that the SH2 domain of SOCS3 was deleted resulting in a SOCS box only:elonginBC ternary complex. (D) Mutating the JAK binding site (F25A) in SOCS3 abrogates its ability to promote JAK ubiquitination The reactions were incubated for 0, 10, 20 and 60 minutes (left to right in each case). All results are visualised by Coomassie staining following SDS-PAGE. (E). Schematic diagram illustrating the components of the Cullin5 in vitro ubiquitination system. A model of the E3 ubiquitin ligase is shown on the right, adapted from PDB 4JGH.
A Cullin5/Rbx2/SOCS3 based E3 ligase catalyses Jak2 (JH1) polyubiquitination
SOCS3 is known to bind directly to both JAKs and cytokine receptors7,8. We initially chose to investigate whether JAKs, rather than cytokine receptors, are substrates for ubiquitination as the SOCS:JAK interaction is not dependent upon post-translational modification. Although we were unable to produce full length JAK2, the kinase domain alone was expressed and purified as a GST fusion protein as previously described 27. As shown in Figure 1B, when added to our in vitro ubiquitination system, GST-JAK2 was rapidly ubiquitinated whereas GST alone was not. Ubiquitination was confirmed by excising the higher molecular weight products from the gel, digesting them using trypsin and performing mass-spectrometry. MS analyses detected multiple peptides from JAK and ubiquitin (>70% and >80% coverage respectively). Ubiquitination was E1-, E2-, E3- and ATP-dependent (Figure 1C). In addition, mutation of the kinase inhibitory region of SOCS3 (known to be involved in JAK binding8; 28) disrupted its ability to ubiquitinate JAK2 (Figure 1D). As highlighted by asterisks in Figure 1B, C Cullin5, the scaffold of the E3 complex, was also ubiquitinated in this system and this was investigated further.
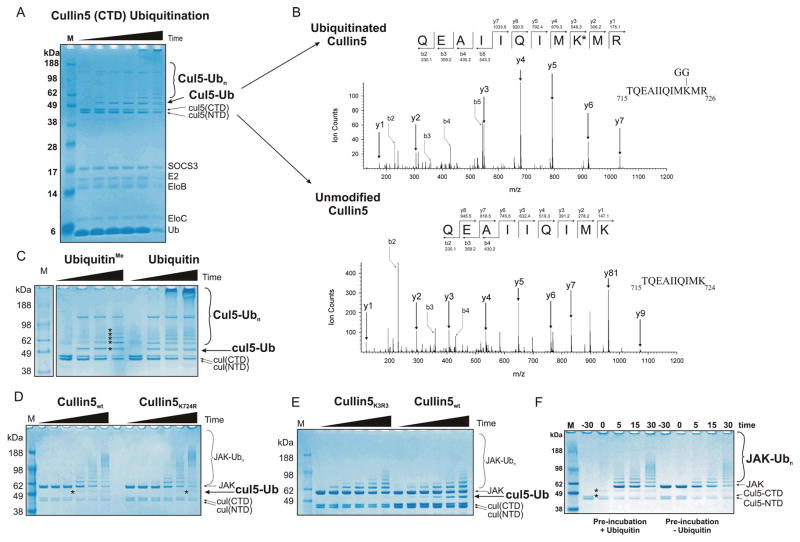
The Cullin5 C-terminal domain is ubiquitinated at K724, the Nedd8-attachment site but this does not affect substrate ubiquitination
As seen in Figure 1, in addition to JAK, another component of the reaction was consistently being polyubiquitinated in these assays. The protein bands were cut out of the SDS-PAGE gel and identified by tryptic digest MS/MS analysis as Cullin5. Tryptic digestion results in a gly-gly dipeptide remaining covalently bound to a lysine side chain if it had been ubiquitinated, resulting in an increased mass for that residue of 95 and this analysis identified the modified residue as K724 in the Cullin5 C-terminal domain (Figures 2A, B). All Cullin proteins are Neddylated in vivo at a conserved lysine in their C-terminal domain. In Cullin5, this conserved lysine is K724, therefore in vitro, Cullin5 was being ubiquitinated at its Neddylation site. No other ubiquitination site could be detected by mass-spectrometry; however experiments using lysine-methylated ubiquitin (which prevents polyubiquitination) showed there were at least four other sites of ubiquitination. (Figure 2C, asterisked). As K724 was the only lysine to be identified by mass-spectrometry, it seemed likely that it is the dominant site of ubiquitination. In order to investigate this further, a Cullin5 mutant (K724R) E3 ligase was constructed. Figure 2D shows that ubiquitination of Cullin5K724R was heavily delayed compared to wild-type. Mutating a further two nearby lysines (Cul5K724,727,728R) resulted in a further reduction in Cullin5 ubiquitination such that it was barely detectable (Figure 2E, lanes 1–6).
Figure 2. Cullin is modified by ubiquitin on K724.
(A) Ubiquitination reactions were performed in the absence of JAK2 to highlight Cullin5 ubiquitination.. The reactions were incubated for 0, 5, 10, 30, 60 and 120 minutes (left to right, upper panel). (B) Tryptic digest MS/MS analyses was used to identify ubiquitinated lysines on Cullin5 from the monoubiquitinated Cullin5 band shown in the left panel. (C) The use of methylated ubiquitin shows that Cullin5 is ubiquitinated on multiple lysines (asterisks). The reactions were incubated for 0, 5, 10, 30 minutes (left to right in each case). (D) Cullin K724R is self-ubiquitinated less efficiently than wild-type Cullin5 and is observable only after an extended incubation (60 minutes compared to 10 minutes, asterisks). The reactions were incubated for 0, 2, 5, 10, 30 and 60 minutes (left to right in each case). This mutant E3 ligase (right) was able to ubiquitinate JAK substrate as efficiently as wild-type. (E) Mutation of lysines 724, 727 and 728 to arginine abrogates Cullin auto-ubiquitination but has no effect on substrate ubiquitination. (F) Cullin ubiquitination does not impair E3 ligase function in vitro. The Cullin5 E3 ligase was auto-ubiquitinated prior to the addition of substrate (left) or incubated with buffer as a control (right). After 30 minutes, substrate (JAK2) and excess ubiquitin (50 μM final concentration) were added and substrate ubiquitination examined after the indicated times. Pre-incubation of the E3 ligase in this fashion resulted in at least 50% of the Cullin5 CTD becoming ubiquitinated (compare the intensity of the two asterisked bands, the lower band is unmodified Cul5(CTD) whilst the upper band is mono-ubiquitinated Cul5(CTD)))before the addition of substrate however no subsequent decrease in the rate of substrate ubiquitination was observed (compare the JAK-Ubn bands in lanes 2–5 with lanes 7–10). All results are visualised by Coomassie staining following SDS-PAGE. The ubiquitinated reaction products indicated using larger font are those referred to in the main text.
Previous studies on Cullin1 have speculated that ubiquitination of K720 (the equivalent of K724 in Cullin5) may mimic neddylation of this lysine that occurs in vivo and accelerate the ubiquitination of substrates16. However mutating K724 to arginine, thereby blocking the ubiquitination of this residue, neither inhibited nor enhanced substrate ubiquitination in our system. In order to examine this more carefully, the E3 ligase was pre-incubated with ubiquitin, E1 and E2 before the addition of substrate and substrate ubiquitination was compared to that of reactions performed under identical conditions but without pre-ubiquitination (Figure 2F). Once again, there was no difference in the rate of substrate ubiquitination suggesting that ubiquitination of K724 has no effect on Cullin5 function in this system in vitro
JAK2 is polyubiquitinated on multiple lysines through multiple ubiquitin linkages
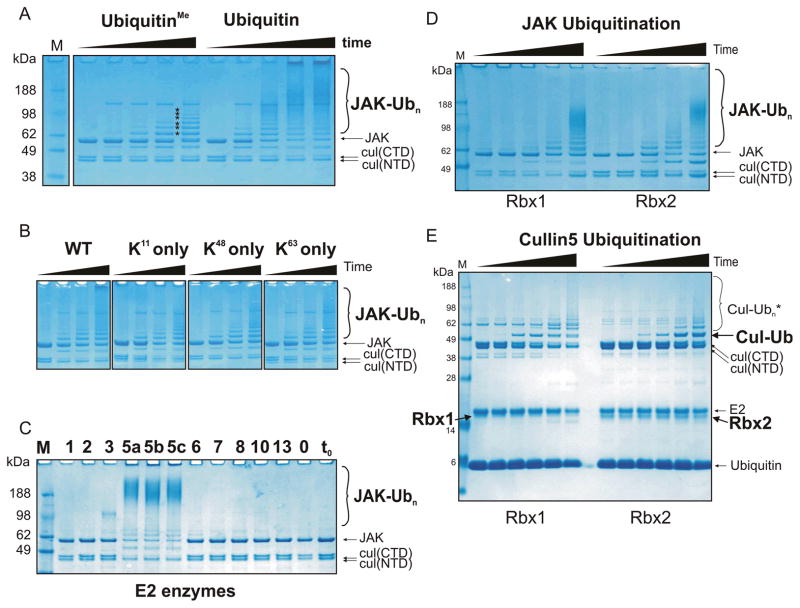
The ladder of ubiquitinated JAK produced in these experiments could result from a single poly-ubiquitinated lysine, multiple mono-ubiquitinated lysines or multiple polyubiquitinated lysines. In order to determine which of these were present, ubiquitin that was lysine-methylated was used, precluding poly-ubiquitination. Based on the number of bands in the ubiquitination ladder we consistently observed at least six sites of ubiquitination on GST-JAK2JH1 (Figure 3A). However ubiquitination experiments using methylated ubiquitin did not show the very high molecular weight products (>100 kDa) seen when using wild type ubiquitin. This suggests that JAK2 is polyubiquitinated.
Figure 3. Rbx1 and Rbx2 are active components of a SOCS3/Cullin5 based E3 ligase which catalyses poly-ubiquitination of multiple lysines in the presence of UbcH5a, UbcH5b or UbcH5c.
(A) Ubiquitination reactions were performed as described in Figure 1 using either methylated (left) or wild-type (right) ubiquitin. The use of methylated ubiquitin indicates there are at least six individual lysines on GST-JAK2 (JH1) that are ubiquitinated in this system (asterisks). The reactions were incubated for 0, 10, 30, 60 and 120 minutes (left to right in each case). (B) Polyubiquitination of JAK was observed when ubiquitin with only a single lysine (K11, K48 or K63) was used. (C) A panel of different E2 enzymes were tested for the ability to promote JAK2 ubiquitination. Each E2 enzyme was added to a standard ubiquitination assay (see Figure 1) at a concentration of 2.5 μM and the reaction performed for 30 minutes at 37°C. Only UbcH5a, UbcH5b and UbcH5c were able to promote JAK ubiquitination. The results were visualised by Coomassie staining following SDS-PAGE. (D) Rbx1 and Rbx2 were both active RING finger components of a Cullin5/SOCS3 based E3 ligase. Experiments were performed under identical conditions to those in (A). (E) In the presence of Rbx2, mono-ubiquitinated Cullin5 accumulates to a greater extent than seen in the presence of Rbx1 (indicated by the black label to the right of the gel). Experimental conditions were the same as (D) only JAK was omitted. All results are visualised by Coomassie staining following SDS-PAGE. The ubiquitinated reaction products indicated using larger font at the right of each gel are those referred to in the main text.
In order to determine whether polyubiquitination was of a particular topology, mutant ubiquitins which only contained a single lysine (K11 only, K48 only and K63 only) were tested in this system. These contain K>R mutations at all other lysine positions. As shown in Figure 3B, polyubiquitination, as evidenced by the smear >100 kDa, occurred using either K11 only, K48 only, K63 only or WT ubiquitin. There was no obvious difference in reaction rate using these three ubiquitin mutants. Again, although polyubiquitination was clearly occurring using these single lysine ubiquitins the very high molecular weight ubiquitinated products that do not migrate into the gel were less prevalent. These species are presumably due to the formation of branched ubiquitin chains, which cannot form when ubiquitin contains only one lysine, as noted previously 29.
UbcH5a,b,c are active E2 components of the SOCS3/Cullin5/Rbx2 E3 ligase
The human genome encodes >50 ubiquitin conjugating enzymes (E2s). In order to determine whether any E2 enzyme, apart from UbcH5a was active in a SOCS/Cullin5/Rbx2 system we tested eleven different E2 enzymes in our Cullin5-based ubiquitination assay. As shown in Figure 3C, UbcH5a, UbcH5b and UbcH5c were most active in catalysing ubiquitination of JAK2. Of the other E2s, only UbcH3 was active in catalysing JAK ubiquitination; however the rate of ubiquitination was significantly lower using this enzyme. Interestingly, we consistently observed a qualitative difference in the pattern of ubiquitination catalysed by UbcH3 (compared to UbcH5), in that the number of ubiquitins added seemed to be more tightly regulated; there was very little mono-ubiquitination and very little high molecular weight ubiquitination products observed.
Both Rbx1 and Rbx2 are active components of a Cullin5 E3 ligase
Although Rbx2 appears to be the preferential RING protein associated with Cullin5 in vivo, Rbx1 can bind Cullin5 and was active in a Cul5/Cul1 hybrid E3 ligase 16. We wished to examine whether there were any consequences to using Rbx2 as the E2 recruitment module, rather than Rbx1. A Cullin5/Rbx1 E3 ligase was therefore cloned, expressed and purified. As shown in Figure 3D, both Rbx1 and Rbx2 catalysed substrate ubiquitination in a Cullin5 based assay. Over multiple experiments there was no discernible difference in ubiquitination rate comparing Rbx1 with Rbx2. The same ubiquitin E2 enzymes were active in Cul5/Rbx1 and Cul5/Rbx2 complexes (data not shown). The only difference between Rbx1- and Rbx2-based E3 ligases was that predominantly mono-ubiquitinated Cullin5 accumulated in the presence of Rbx2, whereas when Rbx1 was the associated RING protein, Cullin5 tended to progress through to a polyubiquitinated state (Figure 3E).
Cullin5/Rbx2/SOCS3 E3 ligase catalyses efficient gp130 polyubiquitination in a phosphorylation-dependent manner
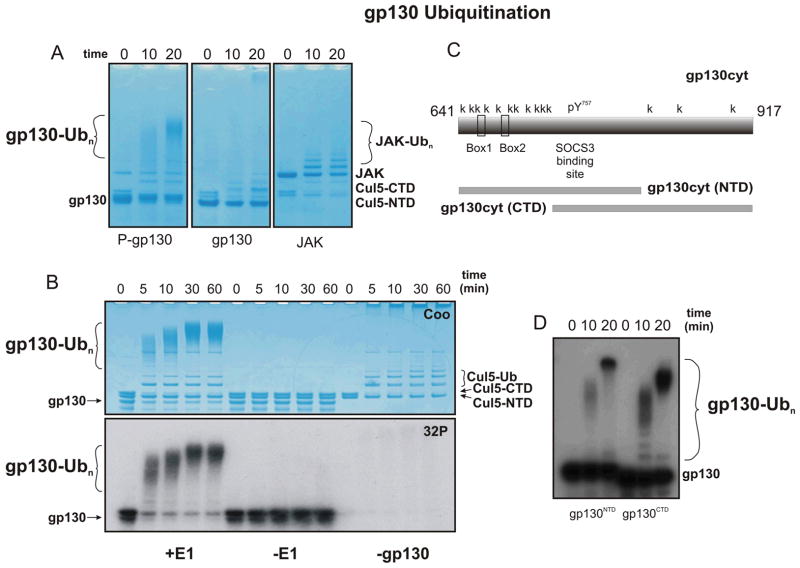
The SH2 domain of SOCS3 is known to bind to phosphotyrosine motifs in the intracellular domains of several cytokine receptors. The most well-characterised binding site is pY757 on the IL-6 receptor β-chain (gp130). In vivo Y757 is phosphorylated by JAK1 or JAK2. Therefore, in order to determine whether a SOCS3/Cullin5 based E3 ligase can catalyse the ubiquitination of cytokine receptors, we expressed and purified the cytoplasmic domain of gp130. This was incubated with catalytic amounts of recombinant JAK2 kinase domain to promote its tyrosine-phosphorylation and then tested as a substrate in our ubiquitination assay. As shown in Figure 4A, gp130 was very efficiently ubqiuitinated using this assay but only if it was phosphorylated. By radiolabelling gp130 using γ-32P-ATP, its ubiquitination could be clearly distinguished from that of any other reaction component (Figure 4B). The cytoplasmic domain of gp130 contains 14 lysine residues, eleven of these are upstream of the SOCS3 binding site (pY757) and three are downstream. We generated two deletion constructs of gp130cyt. The first fragment, termed gp130cyt(NTD), consisted of residues 641–813. This contains 10 lysines, all upstream of the SOCS3 binding site. The second fragment, termed gp130cyt(CTD), consisted of residues 720–917 and contains four lysines, all downstream of the SOCS3 binding site (Figure 4C). Both of these were rapidly polyubiquitinated in our assay system, indicating that there is little, if any, topological constraint upon ubiquitination of gp130 (Figure 4D).
Figure 4. Cullin5/Rbx2/SOCS3 based E3 ligase catalyses efficient gp130 polyubiquitination in a phosphorylation-dependent manner.
(A) SOCS3 catalyses the ubiquitination of the gp130 cytoplasmic domain (gp130cyt) when it is phosphorylated (left panel) but not in the absence of phosphorylation (center panel). Phosphorylation was achieved by including 500 nM JAK2JH1 in the reaction. Note that whereas ubiquitination of JAK proceeds through a series of mostly mono- di- and tri-ubiquitinated intermediates (right panel), ubiquitination of gp130 is qualitatively different in that the intermediates are all highly poly-ubiquitinated. (B) Pre-phosphorylation of gp130 shows that its SOCS3/Cullin5-catalysed ubiquitination is highly efficient and proceeds through poly-ubiquitinated intermediates. Results are visualized by Coomassie staining (upper) and autoradiography (lower). (C) Schematic showing the gp130 cytoplasmic domain and the two truncation mutants tested here. (D) Lysines both N- and C-terminal to the SOCS3 binding site on gp130 are ubiquitinated. The ubiquitinated reaction products referred to in the main text are indicated using larger font in each panel.
DISCUSSION
SOCS3 is a potent suppressor of signaling by IL-6 family cytokines, G-CSF and leptin. The main mechanism by which SOCS3 regulates these cytokines is via its ability to directly inhibit JAK1, JAK2 and TYK2, the kinases that initiate the intracellular part of the signaling cascade. However, genetic deletion of the SOCS3 SOCS box domain has shown that the SOCS box also contributes to control of cytokine signaling in vivo30. Bone marrow from SOCS box-deleted mice had an increased number of colony forming cells responsive to IL-6 and G-CSF and the mice were hyper-responsive to externally administered G-CSF30. Clearly then, the action of the SOCS box is important for correct functioning of SOCS3 in vivo. Presumably, the requirement for the SOCS box is due its ability to recruit components of an E3 ubiquitin ligase, even though biochemical studies have shown that the SOCS3 SOCS box, like that of SOCS1, interacts with the core E3 scaffold, Cullin5, with low affinity11.
Whilst the kinase inhibitory activity of SOCS3 has been well studied biochemically, its ability to function as an E3 ligase has not. In part, this is due to the difficulty of studying ubiquitination in defined in vitro systems. SOCS3, like all SOCS box-containing proteins, interacts with Cullin531 and to date a Cullin5-based ubiquitination system has not been published. We were able to construct an in vitro ubiquitination assay system consisting of fully purified, recombinant proteins and used this to determine whether SOCS3 could function as an efficient E3 ligase. In addition, we used our in vitro ubiquitination assay to address questions regarding the mechanism of ubiquitination, including which E2 and RING domain-containing proteins were active, whether the E3 ligase catalysed mono- or poly-ubiquitination and (if the latter) by which lysine linkage. Whilst such defined systems are inevitably limited by the fact that they are ex vivo, they are invaluable in answering questions regarding mechanism, and in their ability to identify the essential components of molecular machines such as the E2–E3 ubiquitin transfer complex investigated here.
Using our assay we showed that SOCS3 formed an active Substrate Recruitment Module (SRM) for a Cullin5-based E3 ligase. SOCS3 formed a stable 1:1:1:1:1, SOCS3:elonginB:elonginC:Cullin5:Rbx E3 ligase complex that could be isolated as a single species via gel filtration. We tested two individual E3 ligase complexes, the first contained Rbx1 as the RING domain-containing subunit and the second contained Rbx2. Both of these were shown to be active. There are approximately fifty E2 enzymes encoded in the human genome32; 33. We tested a panel of 13 of these and discovered four that were active. Three of these E2 enzymes, the related UbcH5a, UbcH5b and UbcH5c, gave the most robust ubiquitination (with both Rbx1 and Rbx2), as seen previously for a number of different E3 ligase systems34; 35. Two known SOCS3-binding targets, JAK2 and gp130, were each good substrates for ubiquitination. Ubiquitination relied upon a robust SOCS3-substrate interaction, as mutating the JAK2-binding site in SOCS3, or using unphosphorylated gp130 (which does not bind to the SOCS3 SH2 domain36), resulted in complete loss of ubiquitination.
The type of ubiquitination catalysed by this system was manifold. We observed K11, K48 and K63-linked polyubiquitin chains. K48- and K11- mediated poly-ubiquitination is generally thought to lead to proteasome-mediated degradation of the target protein whilst K63-mediated ubiquitination is more commonly involved in intracellular trafficking13. In vivo the type of poly-ubiquitination will depend in part upon which E2 enzyme is most active / available. Whilst the UbcH5 family were most active in our assays it may be that others, for example UbcH3 will be more relevant in certain cell-types. UbcH3 (cdc34 in yeast) is highly evolutionarily conserved, whereas UbcH5 is not34. Whilst ubiquitination of JAK2 using UbcH3 occurred at a much slower rate compared to UbcH5, there was a qualitative difference in the pattern of ubiquitination:. There was relatively little mono- or di-ubiquitinated species visible when UbcH3 was included in the reaction; instead there was a slow accumulation of higher-order ubiquitin chains. It is thought that a chain-length of >4 is required for proteasome recognition and that, given the preponderance of de-ubiquitinases in the cell, this may have to occur within the lifetime of the E3-substrate complex association34. Therefore, in vivo UbcH3 may be more efficient at producing the level of poly-ubiquitination required for proteasomal degradation compared to UbcH5 which, although it gave a faster rate of poly-ubiquitination, tended to progress through mono- di- and tri- ubiquitinated species.
We observed that the E3 scaffold protein, Cullin5, was itself ubiquitinated in our system. In particular, Cullin5 was ubiquitinated at K724. In vivo, this lysine is modified by Nedd8. Ubiquitination of this site in other Cullins has been observed in numerous studies16. It has even been suggested that ubiquitination of this lysine (by mimicking the neddylation that occurs in vivo) may be necessary in order for the E3 to be fully active16. Neddylation unhinges the Rbx protein from the core of the E3 complex, allowing it the conformational flexibility to position the E2 at the target lysine16. However we observed no change at all in the rate of substrate ubiquitination when this lysine (or two other nearby lysines) were mutated to arginine, suggesting that neddylation may not be required for Cullin5 activity in this system.
There were qualitative and quantitative differences in the ubiquitination of JAK2 compared to gp130. Ubiquitination of gp130 was more rapid than JAK2. In addition, gp130 was more highly poly-ubiquitinated. Poly-ubiquitination of gp130 occurred through the accumulation of higher-order ubiquitin chains whereas poly-ubiquitination of JAK2 occurred via mono- and di-ubiqutinated species. This is likely due to the highly flexible nature of the gp130 structure. Unlike the extracellular domain, the cytoplasmic domain of gp130 is disordered and therefore there will be relatively few spatial restraints in accessing the E2 active site for linking the first, and subsequent, ubiquitin moieties. Models of SOCS-based E3 ligases suggest that there is > 80 Å between the substrate binding site and the reactive cysteine of the E2 enzyme when it is docked onto the SOCS-elonginBC-Cullin5-Rbx2 scaffold31; 37; 38. The cytoplasmic domain of gp130 is 276 residues in length and, given that it is unstructured, would be predicted to easily span this distance. In fact we observed ubiquitnation of lysines both N- and C-terminal to the SOCS3-binding site (which is located roughly centrally) indicated a high level of flexibility. As the major targets of other SOCS family members are other cytokine receptors, all of which are predicted to contain unstructured cytoplasmic domains, this may be a common theme. It is tempting to conclude that gp130 is a preferred substrate for SOCS3-induced ubiquitination compared to JAK2. However, we must note that we could only test the kinase domain of JAK2 (rather than the full length protein). The full length protein may span the gap between E2 and SOCS3 more effectively, allowing more efficient ubiquitination. To date, successful purification of full-length JAKs in quantities required for these assays has not been reported and as such we cannot test this hypothesis. Nevertheless, given the conservation within the SOCS family of specifically targeting cytokine receptors and given the robust ubiquitination of gp130 seen in our assays, we speculate that gp130 (along with potentially the G-CSF and Leptin receptors) is a true target of SOCS3-based ubiquitination.
Given that SOCS3 is a potent inhibitor of cytokine signaling, purely by virtue of its ability to inhibit JAK1, JAK2 and TYK2, why is its ability to induce ubiquitination required? One possible answer is that upon JAK or receptor degradation, signaling can only be re-initiated following synthesis of new JAK and receptor chains, which must then be trafficked to the cell membrane. This type of inhibition is presumably more long-lasting than simple kinase inhibition (whose duration is limited by the off-rate of SOCS3 from the JAK:Receptor complex) and may be required to reset the cell back to its basal, unstimulated, state following cytokine exposure.
In summary, we have produced a fully functional, purified recombinant Cullin5-based E3 ligase and shown that the SOCS box-containing protein SOCS3 can recruit its binding partners JAK2 and phospho-gp130 for ubiquitination. The SOCS box, which binds to the ElonginB/ElonginC/Cullin5 complex, is present in all eight SOCS family proteins as well as a wide variety of other molecules. We believe that our defined assay system will be of great use in determining and verifying other Cullin5 substrates. This includes substrates recruited by the other seven SOCS family members as well as those recruited by the wider class of SOCS box containing proteins, including viral proteins such as HIV Vif (Viral Infectivity Factor) which co-opts Cullin5 to induce degradation of the host anti-viral cytidine deaminase APOBEC3G39.
Acknowledgments
We thank Tracy Willson for the gift of plasmids. This work was supported by the National Health and Medical Research Council of Australia (program grant nos. 461219 and 487922, 1011804), the U.S. National Institutes of Health (grant no. CA22556), the Victorian State Government Operational Infrastructure Support Grant, and the NHMRC Independent Research Institutes Infrastructure Support Scheme (361646). N.A.N. acknowledges fellowship support from the National Health and Medical Research Council and J.J.B. from the Australian Research Council.
Abbreviations used
- DTT
dithiothreitol
- GdnHCl
guanidine hydrochloride
- IPTG
isopropyl-β-D thiogalactopyranoside
- JAK
Janus kinase
- KIR
kinase inhibitory region
- PBS
phosphate-buffered saline
- SOCS
suppressor of cytokine signaling
- STAT
signal transduction and activator of transcription. IL-6, interleukin 6
- G-CSF
granulocyte colony stimulating factor
Footnotes
Declaration of interests
NAN is a patent holder on SOCS3 and its activities. This work was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS. This research was supported by an NHMRC Program Grant (461219), NIH Grant (CA022556), ARC Future Fellowship (FT110100169) and NHMRC Project Grant (1011804). The authors alone are responsible for the content and writing of the paper.
References
- 1.Hilton DJ. Negative regulators of cytokine signal transduction. Cell Mol Life Sci. 1999;55:1568–77. doi: 10.1007/s000180050396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hilton DJ, Richardson RT, Alexander WS, Viney EM, Willson TA, Sprigg NS, Starr R, Nicholson SE, Metcalf D, Nicola NA. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:114–119. doi: 10.1073/pnas.95.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babon JJ, Nicola NA. The biology and mechanism of action of suppressor of cytokine signaling 3. Growth Factors. 2012;30:207–19. doi: 10.3109/08977194.2012.687375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sasaki A, Yasukawa H, Suzuki A, Kamizono S, Syoda T, Kinjyo I, Sasaki M, Johnston JA, Yoshimura A. Cytokine-inducible SH2 protein-3 (CIS3/SOCS3) inhibits Janus tyrosine kinase by binding through the N-terminal kinase inhibitory region as well as SH2 domain. Genes to Cells. 1999;4:339–351. doi: 10.1046/j.1365-2443.1999.00263.x. [DOI] [PubMed] [Google Scholar]
- 5.Yasukawa H, Misawa H, Sakamoto H, Masuhara M, Sasaki A, Wakioka T, Ohtsuka S, Imaizumi T, Matsuda T, Ihle JN, Yoshimura A. The JAK-binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. Embo J. 1999;18:1309–20. doi: 10.1093/emboj/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki A, Yasukawa H, Shouda T, Kitamura T, Dikic I, Yoshimura A. CIS3/SOCS-3 suppresses erythropoietin (EPO) signaling by binding the EPO receptor and JAK2. Journal of Biological Chemistry. 2000;275:29338–29347. doi: 10.1074/jbc.M003456200. [DOI] [PubMed] [Google Scholar]
- 7.Kershaw NJ, Murphy JM, Liau NP, Varghese LN, Laktyushin A, Whitlock EL, Lucet IS, Nicola NA, Babon JJ. SOCS3 binds specific receptor-JAK complexes to control cytokine signaling by direct kinase inhibition. Nat Struct Mol Biol. 2013 doi: 10.1038/nsmb.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babon JJ, Kershaw NJ, Murphy JM, Varghese LN, Laktyushin A, Young SN, Lucet IS, Norton RS, Nicola NA. Suppression of cytokine signaling by SOCS3: characterization of the mode of inhibition and the basis of its specificity. Immunity. 2012;36:239–50. doi: 10.1016/j.immuni.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang JG, Farley A, Nicholson SE, Willson TA, Zugaro LM, Simpson RJ, Moritz RL, Cary D, Richardson R, Hausmann G, Kile BJ, Kent SB, Alexander WS, Metcalf D, Hilton DJ, Nicola NA, Baca M. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc Natl Acad Sci U S A. 1999;96:2071–6. doi: 10.1073/pnas.96.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babon JJ, Sabo JK, Soetopo A, Yao S, Bailey MF, Zhang JG, Nicola NA, Norton RS. The SOCS box domain of SOCS3: structure and interaction with the elonginBC-cullin5 ubiquitin ligase. J Mol Biol. 2008;381:928–40. doi: 10.1016/j.jmb.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babon JJ, Sabo JK, Zhang JG, Nicola NA, Norton RS. The SOCS Box Encodes a Hierarchy of Affinities for Cullin5: Implications for Ubiquitin Ligase Formation and Cytokine Signalling Suppression. Journal of Molecular Biology. 2009;387:162–174. doi: 10.1016/j.jmb.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 13.Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 14.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–9. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 15.Hori T, Osaka F, Chiba T, Miyamoto C, Okabayashi K, Shimbara N, Kato S, Tanaka K. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene. 1999;18:6829–34. doi: 10.1038/sj.onc.1203093. [DOI] [PubMed] [Google Scholar]
- 16.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuang Z, Lewis RS, Curtis JM, Zhan Y, Saunders BM, Babon JJ, Kolesnik TB, Low A, Masters SL, Willson TA, Kedzierski L, Yao S, Handman E, Norton RS, Nicholson SE. The SPRY domain-containing SOCS box protein SPSB2 targets iNOS for proteasomal degradation. J Cell Biol. 2010;190:129–41. doi: 10.1083/jcb.200912087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamizono S, Hanada T, Yasukawa H, Minoguchi S, Kato R, Minoguchi M, Hattori K, Hatakeyama S, Yada M, Morita S, Kitamura T, Kato H, Nakayama K, Yoshimura A. The SOCS box of SOCS-1 accelerates ubiquitin-dependent proteolysis of TEL-JAK2. J Biol Chem. 2001;276:12530–8. doi: 10.1074/jbc.M010074200. [DOI] [PubMed] [Google Scholar]
- 19.De Sepulveda P, Ilangumaran S, Rottapel R. Suppressor of cytokine signaling-1 inhibits VAV function through protein degradation. J Biol Chem. 2000;275:14005–8. doi: 10.1074/jbc.c000106200. [DOI] [PubMed] [Google Scholar]
- 20.Rui LY, Yuan MS, Frantz D, Shoelson S, White MF. SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. Journal of Biological Chemistry. 2002;277:42394–42398. doi: 10.1074/jbc.C200444200. [DOI] [PubMed] [Google Scholar]
- 21.Croker BA, Krebs DL, Zhang JG, Wormald S, Willson TA, Stanley EG, Robb L, Greenhalgh CJ, Forster I, Clausen BE, Nicola NA, Metcalf D, Hilton DJ, Roberts AW, Alexander WS. SOCS3 negatively regulates IL-6 signaling in vivo. Nature Immunology. 2003;4:540–545. doi: 10.1038/ni931. [DOI] [PubMed] [Google Scholar]
- 22.Croker BA, Metcalf D, Robb L, Wei W, Mifsud S, DiRago L, Cluse LA, Sutherland KD, Hartley L, Williams E, Zhang JG, Hilton DJ, Nicola NA, Alexander WS, Roberts AW. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity. 2004;20:153–165. doi: 10.1016/s1074-7613(04)00022-6. [DOI] [PubMed] [Google Scholar]
- 23.Roberts AW, Robb L, Rakar S, Hartley L, Cluse L, Nicola NA, Metcalf D, Hilton DJ, Alexander WS. Placental defects and embryonic lethality in mice lacking suppressor of cytokine signaling 3. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9324–9329. doi: 10.1073/pnas.161271798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori H, Hanada R, Hanada T, Aki D, Mashima R, Nishinakamura H, Torisu T, Chien KR, Yasukawa H, Yoshimura A. Socs3 deficiency in the brain elevates leptin sensitivity and confers resistance to diet-induced obesity. Nature Medicine. 2004;10:739–743. doi: 10.1038/nm1071. [DOI] [PubMed] [Google Scholar]
- 25.Babon JJ, Yao S, DeSouza DP, Harrison CF, Fabri LJ, Liepinsh E, Scrofani SD, Baca M, Norton RS. Secondary structure assignment of mouse SOCS3 by NMR defines the domain boundaries and identifies an unstructured insertion in the SH2 domain. Febs J. 2005;272:6120–30. doi: 10.1111/j.1742-4658.2005.05010.x. [DOI] [PubMed] [Google Scholar]
- 26.Babon JJ, McManus EJ, Yao S, DeSouza DP, Mielke LA, Sprigg NS, Willson TA, Hilton DJ, Nicola NA, Baca M, Nicholson SE, Norton RS. The structure of SOCS3 reveals the basis of the extended SH2 domain function and identifies an unstructured insertion that regulates stability. Mol Cell. 2006;22:205–16. doi: 10.1016/j.molcel.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 27.Lucet IS, Fantino E, Styles M, Bamert R, Patel O, Broughton SE, Walter M, Burns CJ, Treutlein H, Wilks AF, Rossjohn J. The structural basis of Janus kinase 2 inhibition by a potent and specific pan-Janus kinase inhibitor. Blood. 2006;107:176–83. doi: 10.1182/blood-2005-06-2413. [DOI] [PubMed] [Google Scholar]
- 28.Kershaw NJ, Murphy JM, Liau NPD, Varghese LN, Laktyushin A, Whitlock EL, Lucet IS, Nicola NA, Babon JJ. SOCS3 binds specific receptor-JAK complexes to control cytokine signaling by direct kinase inhibition. Nature Structural & Molecular Biology. 2013;20:469. doi: 10.1038/nsmb.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–86. doi: 10.1074/jbc.M609659200. Epub 2007 Apr 10. [DOI] [PubMed] [Google Scholar]
- 30.Boyle K, Egan P, Rakar S, Willson TA, Wicks IP, Metcalf D, Hilton DJ, Nicola NA, Alexander WS, Roberts AW, Robb L. The SOCS box of suppressor of cytokine signaling-3 contributes to the control of G-CSF responsiveness in vivo. Blood. 2007;110:1466–1474. doi: 10.1182/blood-2007-03-079178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muniz JR, Guo K, Kershaw NJ, Ayinampudi V, von Delft F, Babon JJ, Bullock AN. Molecular Architecture of the Ankyrin SOCS Box Family of Cul5-Dependent E3 Ubiquitin Ligases. J Mol Biol. 2013;425:3166–77. doi: 10.1016/j.jmb.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, Schulman BA. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483–95. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wenzel DM, Stoll KE, Klevit RE. E2s: structurally economical and functionally replete. Biochemical Journal. 2011;433:31–42. doi: 10.1042/BJ20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brzovic PS, Klevit RE. Ubiquitin transfer from the E2 perspective - Why is UbcH5 so promiscuous? Cell Cycle. 2006;5:2867–2873. doi: 10.4161/cc.5.24.3592. [DOI] [PubMed] [Google Scholar]
- 36.Nicholson SE, De Souza D, Fabri LJ, Corbin J, Willson TA, Zhang JG, Silva A, Asimakis M, Farley A, Nash AD, Metcalf D, Hilton DJ, Nicola NA, Baca M. Suppressor of cytokine signaling-3 preferentially binds to the SHP-2-binding site on the shared cytokine receptor subunit gp130. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6493–6498. doi: 10.1073/pnas.100135197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bullock AN, Debreczeni JE, Edwards AM, Sundstrom M, Knapp S. Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase. Proc Natl Acad Sci U S A. 2006;103:7637–42. doi: 10.1073/pnas.0601638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bullock AN, Rodriguez MC, Debreczeni JE, Songyang Z, Knapp S. Structure of the SOCS4-ElonginB/C Complex Reveals a Distinct SOCS Box Interface and the Molecular Basis for SOCS-Dependent EGFR Degradation. Structure. 2007;15:1493–504. doi: 10.1016/j.str.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–60. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]