Abstract
Mitophagy (mitochondrial autophagy), which removes damaged, effete and superfluous mitochondria, has several distinct variants. In Type 1 mitophagy occurring during nutrient deprivation, preautophagic structures (PAS) grow into cup-shaped phagophores that surround and sequester individual mitochondria into mitophagosomes, a process requiring phosphatidylinositol-3-kinase (PI3K) and often occurring in coordination with mitochondrial fission. After sequestration, the outer compartment of the mitophagosome acidifies, followed by mitochondrial depolarization and ultimately hydrolytic digestion in lysosomes. Mitochondrial damage stimulates Type 2 mitophagy. After photodamage to single mitochondria, depolarization occurs followed by decoration and then coalescence of autophagic LC3-containing structures on mitochondrial surfaces. Vesicular acidification then occurs. By contrast to Type 1 mitophagy, PI3K inhibition does not block Type 2 mitophagy. Further, Type 2 mitophagy is not associated with phagophore formation or mitochondrial fission. A third form of self-eating of mitochondria is formation of mitochondria-derived vesicles (MDVs) enriched in oxidized mitochondrial proteins that bud off and transit into multivesicular bodies. Topologically, the internalization of MDV by invagination of the surface of multivesicular bodies followed by vesicle scission into the lumen is a form of microautophagy, or micromitophagy (Type 3 mitophagy). Cell biological distinctions are the basis for these three types of mitophagy. Future studies are needed to better characterize the molecular and biochemical differences between Types 1, 2 and 3 mitophagy.
Abbreviations: ΔΨ, membrane potential; Drp1, dynamin-related protein-1; GFP, green fluorescent protein; LC3, microtubule-associated protein-1 light chain-3; LTR, LysoTracker Red; 3 MA, 3-methyladenine; MDV, mitochondria-derived vesicle; MFFR, MitoFluor Far Red; mtDNA, mitochondrial DNA; MV633, MitoView 633; PAS, preautophagic structure; PI3K, phosphatidylinositol 3-kinase; TMRM, tetramethyrhodamine methyester; TOM20, transporter of the outer membrane-20
Keywords: Micromitophagy, Mitochondria-derived vesicles, Mitophagy, Nutrient deprivation, Photodamage, Preautophagic structure
Graphical abstract
Highlights
-
•
Mitophagy (mitochondrial autophagy) removes damaged and superfluous mitochondria.
-
•
In Type 1 mitophagy, cup-shaped phagophores engulf mitochondria during starvation.
-
•
In Type 2 mitophagy, autophagic membranes coalesce around damaged mitochondria.
-
•
Type 1 but not Type 2 mitophagy requires PI3K and occurs with mitochondrial fission.
-
•
Multivesicular bodies engulf mitochondria-derived vesicles in Type 3 micromitophagy.
Introduction
During fasting, pancreatic islets release glucagon, which promotes gluconeogenesis and autophagy in the liver [1,2]. Refeeding leads to pancreatic insulin release, which suppresses hepatic autophagy. Such autophagy during fasting and other forms of nutrient deprivation furnishes amino acids and fatty acids to maintain cellular metabolism. Mitochondria are particularly rich sources of protein and lipid, and during nutrient deprivation to cultured hepatocytes about 85% of autophagic events involve mitochondria [3]. This process of mitochondrial autophagy is termed mitophagy [3–5].
In healthy liver, as well as in other organs like heart, brain and kidney, cell proliferation is minimal. Nonetheless, individual mitochondria turn over with a half-life of 10–25 days as mitophagy removes worn out mitochondria in balance with biogenesis of new mitochondria [6,7]. Mitophagy protects against release of pro-apoptotic proteins, generation of toxic reactive oxygen species (ROS) and futile hydrolysis of ATP by aged, damaged and depolarized mitochondria [4,5,8–10]. Mitophagy also eliminates mitochondria during cytoplasmic remodeling and degrades mitochondrial DNA (mtDNA), including damaged and mutated mtDNA promoting mitochondrial dysfunction and disease [3,11,12]. A proper balance of mitophagy to mitochondrial biogenesis seems essential for cellular well-being, since inadequate and excess mitophagy both can promote cell injury and death [8,13,14].
Progression of mitophagy
Microtubule-associated protein-1 light chain-3 (LC3) is the mammalian ortholog of the yeast autophagy-associated protein Atg8. During autophagy, cytosolic LC3 (LC3-I) becomes conjugated to phosphatidylethanolamine to form an LC3-phosphatidylethanolamine conjugate (LC3-II), which is recruited to forming and newly formed autophagosomal membranes [15]. The fusion protein, green fluorescent protein-LC3 (GFP-LC3), is thus a convenient fluorescent marker of autophagic events [16]. GFP-LC3 is mostly diffuse in the cytosol under nutrient-replete conditions. However, some GFP-LC3 resides in small (0.2–0.3 µm) preautophagic structures (PAS) close to mitochondria [3]. During nutrient deprivation, PAS grow into cup-shaped isolation membranes or phagophores that envelop and sequester individual mitochondria into autophagosomes (mitophagosomes) (Fig. 1). Mitochondrial fission frequently occurs in coordination with sequestration of mitophagosomes. Once initiated, sequestration is complete within 6–7 min. When mitophagy is stimulated by nutrient deprivation, mitochondria maintain their membrane potential (ΔΨ) during sequestration, and depolarization does not occur until after sequestration is complete, as indicated by loss of ΔΨ-indicating fluorophores like tetramethyrhodamine methyester (TMRM) (Fig. 1). After sequestration, mitophagosomes acidify and fuse with lysosomes (or with late endosomes that then fuse with lysosomes). Mitochondrial contents are then digested after about 10 min [3,17]. Phosphatidylinositol 3-kinase (PI3K) inhibitors, wortmannin and 3-methyladenine, block mitophagic sequestration virtually completely during nutrient deprivation, signifying involvement of the classical beclin-1/Vps34 (class III PI3K) autophagic pathway [3,18].
Fig 1.
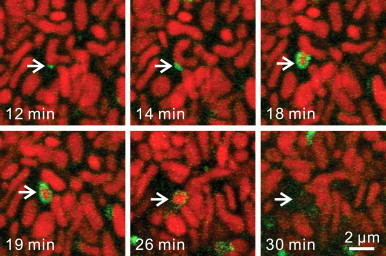
Mitochondrial fission, autophagic sequestration and depolarization during Type 1 mitophagy. Hepatocytes from GFP-LC3 transgenic hepatocytes were loaded with ΔΨ-indicating TMRM and incubated in nutrient-free Krebs-Ringer–HEPES buffer (KRH) containing 1 µM glucagon as confocal images were collected every minute. Note association of a PAS with a U-shaped mitochondrion (12 min of nutrient deprivation), which grew into a cup-shaped phagophore (14 and 18 min) that enveloped and then sequestered the middle part of the mitochondrion coordinately with mitochondrial fission (19 min). The sequestered mitochondrial fragment remained polarized for several minutes, as shown by retention of red TMRM fluorescence (26 min) before depolarizing (30 min).
Adapted from [3].
Depolarization-induced mitophagy
Global mitochondrial damage caused by mitochondrial uncoupling (depolarization) and oxidative stress induces a robust autophagic response and ultimately cell death [19–22]. The question remains, however, whether such mitophagy is actually due directly to mitochondrial damage and depolarization, since global uncoupling also causes a profound bioenergetic deficit that creates a nutrient deprivation-like and autophagy-stimulating metabolic state. Evidence that depolarization of single mitochondria can induce selective mitophagy comes from photodamage experiments in which small groups of mitochondria are exposed to bright 488-nm laser light, which excites mitochondrial flavins, leading to ROS production and mitochondrial injury [5,23–25]. At lower cumulative exposure, this light depolarizes mitochondria transiently, but greater light exposure leads to sustained and irreversible depolarization, which is accompanied by inner membrane permeabilization akin to the mitochondrial permeability transition (MPT).
After laser-induced photodamage to mitochondria in nutrient-replete hepatocytes, GFP-LC3 fluorescence decorates the edges of individual depolarized mitochondria beginning after about 30 min and continuing for up to at least 60 min (Fig. 2A and B) [25]. Importantly, decoration with GFP-LC3 only occurs when laser-induced mitochondrial depolarization is sustained. If a mitochondrion initially depolarizes after light exposure but subsequently repolarizes, labeling by GFP-LC3 does not ensue. At first, GFP-LC3 labeling of the surfaces of individual depolarized mitochondria is discontinuous. Over time, however, GFP-LC3 fluorescence coalesces into a thin continuous ring, and the individual mitophagosomes acidify (Fig. 2B). Notably, mitophagy does not occur outside the region of photodamage, and the phototoxic stress does not lead to cell death. Additionally, autophagic sequestration of individual mitochondria after photodamage appears to occur without mitochondrial fission or involvement of PAS. Unexpectedly and in marked contrast to nutrient deprivation-induced mitophagy, PI3K inhibitors do not stop photodamage-induced mitophagy (Fig. 3). If anything, PI3K inhibitors promote greater accumulation of mitophagosomes after photodamage, possibly by preventing subsequent PI3K-dependent fusion with lysosomes or lysosomal precursors [25].
Fig 2.
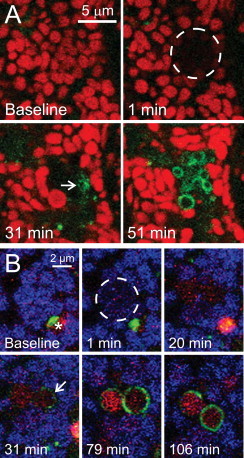
Type 2 mitophagy after selective photodamage. In (A), a TMRM-loaded GFP-LC3 hepatocyte was exposed to photodamaging 488-nm laser light within the area indicated by the circle. Note mitochondrial depolarization at 1 min after photoirradiation, followed by decoration of the depolarized mitochondria with GFP-LC3 (31 min, arrow). GFP-LC3 subsequently formed complete rings around the damaged mitochondria (51 min). In (B), a GFP-LC3 hepatocyte was loaded with ΔΨ-indicating MitoFluor Far Red (MFFR) and LysoTracker Red (LTR) for 30 min and then exposed to photodamaging 488-nm laser light within the area indicated by the circle. Note mitochondrial depolarization after photoirradiation, as indicated by loss of blue pseudo-colored MFFR fluorescence (1 min). GFP-LC3 subsequently began to decorate the depolarized mitochondria (31 min, arrow) to form mitophagosomes, which acidified as indicated by uptake of red LTR fluorescence (79 and 106 min). A pre-existing autophagosome was also present (baseline, asterisk), which matured into a red-fluorescing autolysosome and moved out of the field during the experiment.
Adapted from [25].
Fig. 3.
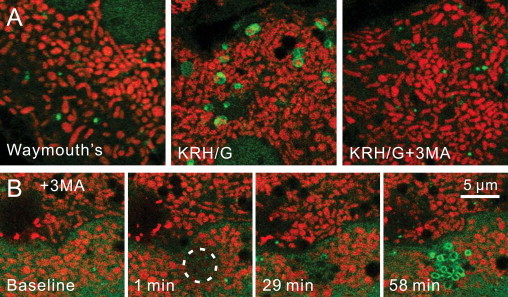
Inhibition of Type 1 but not Type 2 mitophagy by 3-methyladenine. In (A), TMRM-loaded GFP-LC3 transgenic hepatocytes were incubated 120 min in serum-containing Waymouth׳s growth medium (left panel), KRG plus glucagon (KRH/G), or KRH/G plus 10 mM 3-methyladenine (3 MA), a PI3K inhibitor and classical autophagy blocker. In comparison to Waymouth׳s medium alone, incubation in KRH/G caused abundant formation of GFP-LC3-tagged autophagosomes, many of which contained TMRM-labeled polarized mitochondria (middle panel). 3 MA treatment prevented autophagosomal formation virtually completely (right panel). In (B), GFP-LC3 hepatocytes were subjected to 488-nm photoirradiation (circle), as described in Fig. 2, in the presence of 3 MA. Note that 3 MA did not prevent formation of mitophagosomes after photodamage (58 min, compare to Fig. 2). Wortmannin (100 nM), another PI3K inhibitor, similarly failed to prevent mitophagosome formation after photodamage (not shown). Adapted from [25].
Type 1 and Type 2 mitophagy
These observations indicate that sequestration of mitochondria into autophagosomes occurs by different cellular mechanisms, which can be termed Type 1 and Type 2 mitophagy (Fig. 4). Nutrient deprivation-induced mitophagy typifies Type 1 mitophagy in which PAS enlarge to surround and sequester individual mitochondria into mitophagosomes, often in coordination with mitochondrial fission. Photodamage-induced mitophagy exemplifies Type 2 mitophagy. In Type 2 mitophagy. GFP-LC3 decorates the surfaces of individual depolarized mitochondria. This surface labeling first occurs in small aggregates of GFP-LC3. The aggregates subsequently coalesce and fuse into complete rings segregating each damaged mitochondrion into a mitophagosome. Unlike Type 1 mitophagy, cup-shaped phagophores do not form during Type 2 mitophagy, and mitochondrial fission is not evident. Nonetheless after formation by either a Type 1 or Type 2 mechanism, the mitophagosomes then acidify and their contents become degraded. A major biochemical distinction between Type 1 and Type 2 mitophagy is that PI3K inhibition with 3-methyladenine or wortmannin blocks Type 1 but not Type 2 mitophagic sequestration, implying that Type 2 mitophagy is independent of beclin-1 (Fig. 4). Beclin-1 and PI3K-independent initiation of damage-induced mitophagy has also been described in SH‑SY5Y human neuroblastoma cells and primary dopaminergic neurons exposed to the parkinsonian neurotoxin 1-methyl‑4-phenylpyridinium [26,27].
Fig. 4.
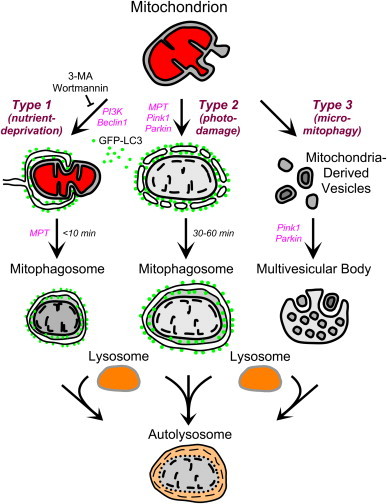
Scheme of Types 1, 2 and 3 mitophagy. In Type 1 mitophagy induced by nutrient deprivation, activation of beclin1/PI3K leads to formation of an LC3-GFP-labeled phagophore that sequesters a mitochondrion into a mitophagosome often in coordination with mitochondrial fission. Mitochondrial depolarization then occurs after sequestration due to onset of the MPT. Subsequently, the mitophagosome fuses with lysosomes, and hydrolytic digestion of the entrapped mitochondrion occurs. In Type 2 mitophagy, mitochondrial injury by photodamage or other injurious stress causes MPT onset and sustained mitochondrial depolarization with swelling of the inner membrane–matrix compartment. In PI3K- and beclin1-independent fashion, GFP-LC3-labeled membrane vesicles attach to the depolarized mitochondrion and coalesce to form a mitophagosome. Further mitophagosomal processing occurs as in Type 1 mitophagy. In Type 3 mitophagy, or micromitophagy, MDV containing oxidized mitochondrial proteins bud off from mitochondria and then become internalized into multivesicular bodies in a pink1/parkin-dependent fashion. Multivesicular bodies subsequently fuse with lysosomes to complete hydrolytic degradation of the mitochondrial fragments. In the scheme, red denotes mitochondrial polarization.
Mitochondria-derived vesicle formation (micromitophagy or Type 3 mitophagy)
Another mechanism for targeted removal of damaged mitochondrial components is formation of mitochondria-derived vesicles (MDV) that bud off and then transit to lysosomes, as shown in elegant recent experiments [28,29]. MDV are cargo-selective vesicles released from mitochondria independently of the mitochondrial fission machinery. Oxidative stress stimulates MDV formation, and the MDVs themselves are enriched in oxidized mitochondrial proteins [29]. Unlike Type 2 mitophagy, MDV formation does not require mitochondrial depolarization. Although MDV formation and transit to lysosomes occurs independently of the autophagic proteins ATG5 and LC3, this form of self-eating of mitochondria does require PTEN-induced putative kinase-1 (pink1) and parkin, proteins well known to be associated with mitophagy [30]. Interestingly, electron microscopy reveals that MDVs end up as vesicles within multivesicular bodies, a form of late endosome, as shown by immunostaining for transporter of the outer membrane-20 (TOM20) [28]. Such findings suggest that MDV incorporation occurs by invagination of the surface of multivesicular bodies followed by vesicle scission into the lumen. Afterwards, the multivesicular bodies fuse with lysosomes to complete the hydrolytic degradation of the MDV. Topologically, the transit of MDV into multivesicular bodies and then lysosomes is a form of microautophagy, or micromitophagy, termed here as Type 3 mitophagy (Fig. 4). Micromitophagy is well characterized in yeast but differs from micromitophagy (Type 3 mitophagy) in mammalian cells in that digestive vacuoles (lysosomes) of yeast engulf whole mitochondria, whereas multivesicular bodies of mammalian cells swallow up small mitochondrial MDV fragments [31,32]. Overall, micromitophagy facilitates selective removal of damaged and oxidized mitochondrial components as a form of mitochondrial quality control that occurs without mitochondrial depolarization or overt functional impairment.
Future directions
Cell biological differences constitute the principal distinctions between the three types of mitophagy described here, and future studies are needed to characterize the molecular and biochemical differences between these variants of mitophagy. Models to induce mitophagy need scrutiny as well. For example, mitochondrial depolarization with an uncoupler such as dinitrophenol or FCCP is widely used to induce mitophagy, but such treatment may promote more than one type of mitophagy. One effect of uncoupling, mitochondrial depolarization, activates Type 2 mitophagy, whereas the bioenergetic deficit after uncoupling produces a nutrient deprivation-like state and hence Type 1 mitophagy. Similarly depending on severity, oxidative stress likely induces all three types of mitophagy: Type 3 mitophagy with mild oxidative stress and Types 1 and 2 mitophagy as more severe oxidative stress causes mitochondrial depolarization and ATP depletion. Thus, use of models that selectively activate specific types of mitophagy will be essential to delineate the exact molecular mechanisms underlying each form of mitophagy.
The cell biological differences between the different types of mitophagy give clues as to likely differences in their biochemistry and molecular biology. For example in Type 1 mitophagy, mitochondrial depolarization does not occur until after a mitochondrion is sequestered and captured inside a mitophagosome [3]. Indeed, our data indicates that mitochondrial depolarization does not occur until the outer compartment of the mitophagosome (space between the membranes originating from the phagophore) acidifies (Fig. 5) [33]. By contrast, sequestration in Type 2 mitophagy does not begin until well after mitochondrial depolarization [25]. Pink1 and parkin are proteins whose mutations cause familial forms of Parkinson׳s disease, and mitochondrial depolarization leads to pink1-dependent recruitment of parkin to the mitochondrial outer membrane. Parkin is an E3 ubiquitin ligase that ubiquinates outer membrane proteins to target mitochondria for mitophagy [20,34–38]. Thus, pink1 and parkin are essential for Type 2 mitophagy but may not be involved in Type 1 mitophagy, although pink1 and parkin are also involved in Type 3 mitophagy perhaps because MDV are depolarized [30]. Future work will be needed to determine the role, if any, of pink1 and parkin in Type 1 mitophagy where mitochondrial depolarization follows rather than precedes autophagic sequestration. Likewise, since PI3K inhibitors block Type 1 but not Type 2 mitophagy, the beclin1/vps34 PI3K complex would appear to play an essential role in Type 1 mitophagy but not to be involved in Type 2 mitophagy. However, these expectations require experimental testing and confirmation.
Fig . 5.
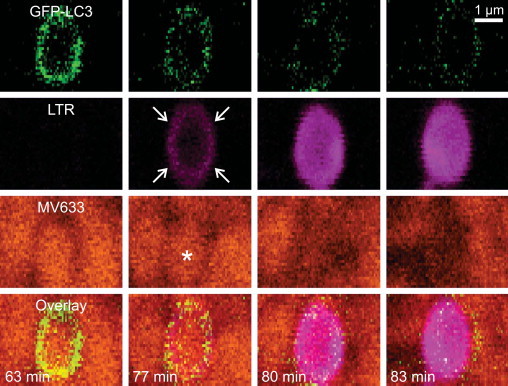
Acidification of the outer mitophagosomal compartment precedes cargo compartment acidification and mitochondrial depolarization. Rat hepatocytes were infected with an adenovirus expressing GFP-LC3 and loaded with LTR and ΔΨ-indicating MitoView 633 (MV633). The cells were then incubated in nutrient-free Krebs-Ringer–HEPES buffer (KRH) containing 1 µM glucagon as confocal images were collected every minute. After sequestration of a mitochondrion into a mitophagosome at 63 min of incubation, the compartment between the inner and outer mitophagosomal membranes acidified (77 min, arrows), as indicated by uptake of magenta-pseudocolored LTR, followed minutes later by acidification of the mitochondrion-containing cargo compartment (80 and 83 min). Cargo acidification was accompanied by loss of orange-pseudocolored MV633 fluorescence seen earlier at 63 and 77 min (asterisk), signifying mitochondrial depolarization (80 and 83 min). Acidification also quenched GFP-LC3 fluorescence internalized inside the mitophagosome. Pixelation is due to the high magnification of the image.
Adapted from [33].
Mitochondrial dynamics are also different in the three types of mitophagy. In Type 1 mitophagy, mitochondria fission typically occurs in coordination with sequestration of mitophagosomes. By contrast in Type 2 mitophagy, mitochondrial fission is not observed. Instead, fusion seems required to form a continuous mitophagosomal membrane from LC3-labeled structures that initially decorate depolarized mitochondria. Indeed, LC3 membranes may also fuse with the mitochondrial outer membrane during Type 2 mitophagy, consistent with reports that mitochondrial outer membrane components incorporate into autophagosomal membranes [39,40]. Careful correlative electron microscopy will be needed to characterize more precisely the topology of membrane fusion events during Type 2 mitophagy. Lastly in Type 3 mitophagy, a fission event is required for the budding off of MDV from the mitochondrial surface. However, the fission apparatus to release MDV is different from that associated with binary fission of mitochondria, since MDV formation is not dependent on dynamin-related protein-1 (Drp1), the GTPase required for binary fission of mitochondria [28]. The molecular basis for membrane fission in Type 3 mitophagy thus also needs further study.
Autophagy after nutrient deprivation is often described as non-selective, but this assumes what we really do not know. Specifically, we do not know whether mitochondria targeted by Type 1 mitophagy carry a greater burden of damaged components, such as oxidized proteins and lipids, than mitochondria that avoid Type 1 mitophagy. Mild oxidative stress is established to stimulate Type 3 micromitophagy, but future experiments are needed to determine how mild oxidative and other stresses affect the probability of mitochondrial sequestration and degradation by Type 1 mitophagy.
Another question concerns the role of the MPT in mitophagy. Although mitochondrial depolarization after onset of MPT is sufficient to initiate Type 2 mitophagy, MPT blockers like cyclosporin A and nonimmunosuppressive N-methyl-4-isoleucine cyclosporin (NIM811) also block nutrient deprivation-induced Type 1 mitophagy, apparently by preventing mitochondrial depolarization after sequestration [11,41–44]. Factors inducing apparent MPT onset after Type 1 mitophagic sequestration as well as why MPT onset is required for further mitophagic processing remain poorly understood.
Overall, mitophagy represents an essential quality control mechanism whose disruption causes disease through failure to remove dysfunctional mitochondria that promote oxidative stress and cell death, including mitochondria with mutated mtDNA. Mitophagy is also an essential survival strategy against nutrient deprivation and starvation. Future studies should be fruitful in addressing open questions about the several variants of mitophagy.
Footnotes
This work was supported, in part, by Grants 2 R01 DK37034, 1 R01 DK073336, P30 CA138313, P20 GM103542 and C06 RR015455 from the National Institutes of Health and Grant 14.Z50.31.0028 from the Russian Federation.
References
- 1.Yu Q.C., Marzella L. Response of autophagic protein degradation to physiologic and pathologic stimuli in rat hepatocyte monolayer cultures. Laboratory Investigation: A Journal of Technical Methods and Pathology. 1988;58:643–652. [PubMed] [Google Scholar]
- 2.Arstila A.U., Trump B.F. Studies on cellular autophagocytosis. The formation of autophagic vacuoles in the liver after glucagon administration. American Journal of Pathology. 1968;53:687–733. 4300890 [PMC free article] [PubMed] [Google Scholar]
- 3.Kim I., Lemasters J.J. Mitochondrial degradation by autophagy (mitophagy) in GFP-LC3 transgenic hepatocytes during nutrient deprivation. American Journal of Physiology: Cell Physiology. 2011;300:C308–C317. doi: 10.1152/ajpcell.00056.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemasters J.J. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Research. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. 15798367 [DOI] [PubMed] [Google Scholar]
- 5.Kim I., Rodriguez-Enriquez S., Lemasters J.J. Selective degradation of mitochondria by mitophagy. Archives of Biochemistry and Biophysics. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. 17475204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfeifer U. Inhibition by insulin of the formation of autophagic vacuoles in rat liver. A morphometric approach to the kinetics of intracellular degradation by autophagy. Journal of Cell Biology. 1978;78:152–167. doi: 10.1083/jcb.78.1.152. 670291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menzies R.A., Gold P.H. The turnover of mitochondria in a variety of tissues of young adult and aged rats. Journal of Biological Chemistry. 1971;246:2425–2429. 5553400 [PubMed] [Google Scholar]
- 8.Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. 18305538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin X.M., Ding W.X., Gao W. Autophagy in the liver. Hepatology (Baltimore, Md.) 2008;47:1773–1785. doi: 10.1002/hep.22146. 18393362 [DOI] [PubMed] [Google Scholar]
- 10.Youle R.J., Narendra D.P. Mechanisms of mitophagy. Nature Reviews. Molecular Cell Biology. 2011;12:9–14. doi: 10.1038/nrm3028. 21179058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez-Enriquez S., Kai Y., Maldonado E., Currin R.T., Lemasters J.J. Roles of mitophagy and the mitochondrial permeability transition in remodeling of cultured rat hepatocytes. Autophagy. 2009;5:1099–1106. doi: 10.4161/auto.5.8.9825. 19783904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suen D.F., Narendra D.P., Tanaka A., Manfredi G., Youle R.J. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11835–11840. doi: 10.1073/pnas.0914569107. 20547844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine B., Klionsky D.J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Developmental Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 14.Lemasters J.J. Necrapoptosis and the mitochondrial permeability transition: shared pathways to necrosis and apoptosis. American Journal of Physiology. 1999;276:G1–G6. doi: 10.1152/ajpgi.1999.276.1.G1. [DOI] [PubMed] [Google Scholar]
- 15.Tanida I., Ueno T., Kominami E. LC3 and autophagy. Methods in Molecular Biology (Clifton, N.J.) 2008;445:77–88. doi: 10.1007/978-1-59745-157-4_4. 18425443 [DOI] [PubMed] [Google Scholar]
- 16.Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. in vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Molecular Biology of the Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. 14699058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Enriquez S., Kim I., Currin R.T., Lemasters J.J. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy. 2006;2:39–46. doi: 10.4161/auto.2229. 16874071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxidants & Redox Signaling. 2011;14:2201–2214. doi: 10.1089/ars.2010.3482. 20712405 [DOI] [PubMed] [Google Scholar]
- 19.Nieminen A.L., Saylor A.K., Tesfai S.A., Herman B., Lemasters J.J. Contribution of the mitochondrial permeability transition to lethal injury after exposure of hepatocytes to t-butylhydroperoxide. Biochemical Journal. 1995;307:99–106. doi: 10.1042/bj3070099. 7718000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. Journal of Cell Biology. 2008;183:795–803. doi: 10.1083/jcb.200809125. 19029340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim E.H., Choi K.S. A critical role of superoxide anion in selenite-induced mitophagic cell death. Autophagy. 2008;4:76–78. doi: 10.4161/auto.5119. 17952022 [DOI] [PubMed] [Google Scholar]
- 22.Nieminen A.L., Saylor A.K., Herman B., Lemasters J.J. ATP depletion rather than mitochondrial depolarization mediates hepatocyte killing after metabolic inhibition. American Journal of Physiology. 1994;267:C67–C74. doi: 10.1152/ajpcell.1994.267.1.C67. 8048493 [DOI] [PubMed] [Google Scholar]
- 23.Aggarwal B.B., Quintanilha A.T., Cammack R., Packer L. Damage to mitochondrial electron transport and energy coupling by visible light. Biochimica et Biophysica Acta. 1978;502:367–382. doi: 10.1016/0005-2728(78)90057-9. 656406 [DOI] [PubMed] [Google Scholar]
- 24.Alexandratou E., Yova D., Handris P., Kletsas D., Loukas S. Human fibroblast alterations induced by low power laser irradiation at the single cell level using confocal microscopy. Photochemical & Photobiological Sciences: Official Journal of the European Photochemistry Association and the European Society for Photobiology. 2002;1:547–552. doi: 10.1039/b110213n. 12659495 [DOI] [PubMed] [Google Scholar]
- 25.Kim I., Lemasters J.J. Mitophagy selectively degrades individual damaged mitochondria after photoirradiation. Antioxidants & Redox Signaling. 2011;14:1919–1928. doi: 10.1089/ars.2010.3768. 21126216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J.H., Horbinski C., Guo F., Watkins S., Uchiyama Y., Chu C.T. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. American Journal of Pathology. 2007;170:75–86. doi: 10.2353/ajpath.2007.060524. 17200184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu C.T., Zhu J., Dagda R. Beclin 1-independent pathway of damage-induced mitophagy and autophagic stress: implications for neurodegeneration and cell death. Autophagy. 2007;3:663–666. doi: 10.4161/auto.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soubannier V., McLelland G.L., Zunino R., Braschi E., Rippstein P., Fon E.A. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Current Biology. 2012;22:135–141. doi: 10.1016/j.cub.2011.11.057. [DOI] [PubMed] [Google Scholar]
- 29.Soubannier V., Rippstein P., Kaufman B.A., Shoubridge E.A., McBride H.M. Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PloS One. 2012;7:e52830. doi: 10.1371/journal.pone.0052830. 23300790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLelland G.L., Soubannier V., Chen C.X., McBride H.M., Fon E.A. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO Journal. 2014;33:282–295. doi: 10.1002/embj.201385902. 24446486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shutt T.E., McBride H.M. Staying cool in difficult times: mitochondrial dynamics, quality control and the stress response. Biochimica et Biophysica Acta. 2013;1833:417–424. doi: 10.1016/j.bbamcr.2012.05.024. [DOI] [PubMed] [Google Scholar]
- 32.Li W.W., Li J., Bao J.K. Microautophagy: lesser-known self-eating. Cellular and Molecular Life Sciences. 2012;69:1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deepe R.N., Kim J.-S., Lemasters J.J. Autophagosomal acidification is associated with depolarization of mitochondria during nutrient deprivation-induced mitophagy. Hepatology. 2013;58(Suppl.):946A–947A. [Google Scholar]
- 34.Dagda R.K., Cherra S.J., III, Kulich S.M., Tandon A., Park D., Chu C.T. Loss of pink1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. Journal of Biological Chemistry. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. 19279012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tracy K., Dibling B.C., Spike B.T., Knabb J.R., Schumacker P., Macleod K.F. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Molecular and Cellular Biology. 2007;27:6229–6242. doi: 10.1128/MCB.02246-06. 17576813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J., Ney P.A. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death and Differentiation. 2009;16:939–946. doi: 10.1038/cdd.2009.16. 19229244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rikka S., Quinsay M.N., Thomas R.L., Kubli D.A., Zhang X., Murphy A.N. Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death and Differentiation. 2011;18(4):721–731. doi: 10.1038/cdd.2010.146. 21278801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuda S., Kitagishi Y., Kobayashi M. Function and characteristics of PINK1 in mitochondria. Oxidative Medicine and Cellular Longevity. 2013;2013:601587. doi: 10.1155/2013/601587. 23533695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hailey D.W., Rambold A.S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P.K. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. 20478256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. 23455425 [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez-Enriquez S., Kim I., Currin R.T., Lemasters J.J. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy. 2006;2:39–46. doi: 10.4161/auto.2229. 16874071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cui T., Fan C., Gu L., Gao H., Liu Q., Zhang T. Silencing of PINK1 induces mitophagy via mitochondrial permeability transition in dopaminergic MN9D cells. Brain Research. 2011;1394:1–13. doi: 10.1016/j.brainres.2011.01.035. 21262209 [DOI] [PubMed] [Google Scholar]
- 43.Elmore S.P., Qian T., Grissom S.F., Lemasters J.J. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. 11511528 [DOI] [PubMed] [Google Scholar]
- 44.Carreira R.S., Lee Y., Ghochani M., Gustafsson Å.B., Gottlieb R.A. Cyclophilin D is required for mitochondrial removal by autophagy in cardiac cells. Autophagy. 2010;6:462–472. doi: 10.4161/auto.6.4.11553. 20364102 [DOI] [PMC free article] [PubMed] [Google Scholar]


