Abstract
Germ cell-specific genes such as Ddx4, Dnd1, and Dazl play critical roles in the proliferation and survival of germ cells. However, the methylation state of the promoter in mature germ cells is still unknown. Here, we investigated the methylation levels of these genes and the pluripotency marker gene Nanog in chicken sperm as compared with the Alb gene in the liver. CpG islands and/or promoter motifs such as TATA box, GC box and CAAT box were found within the putative promoter regions that we identified. By using the bisulfite reaction, CpG sites in the putative promoters were converted, and they were analyzed by sequencing. The putative promoters of Ddx4, Dnd1, Dazl and Nanog showed very low methylation levels in sperm, but they were highly methylated in the liver. Conversely, the Alb gene promoter was highly methylated in sperm and hypomethylated in the liver. However, no transcripts of Ddx4, Dnd1, Dazl and Nanog were detected in sperm or the liver. Also, no transcripts of Dnmt1 and Dnmt3a were detected in sperm. Our present results may indicate that these germ cell-specific genes and the pluripotency marker gene are ready to express any time after fertilization. Our findings showing that low methylation and selective DNA methylation of specific genes are present in chicken sperm contribute to our understanding of fertilization and embryogenesis of birds.
Keywords: CpG site, DNA methylation, Gene expression, Germ cell-specific genes, Sperm
DNA methylation occurs at cytosine-phosphodiester-guanine sites (CpG sites), and its patterns are established and maintained during cell differentiation and cell division by DNA methyltransferases (DNMTs). In primordial germ cells (PGCs), the genome-wide methylation state inherited from parents is erased in a certain period, and new methylation patterns are established [1, 2]. The relationship between methylation and the regulation of imprinted genes in germ cells has recently attracted considerable attention [3,4,5,6,7]. In mammals, the methylation levels of maternal and paternal imprinted genes are different between oogenesis and spermatogenesis [8].
During gametogenesis in chickens, several germ line-specific genes such as Ddx4 [9], Dnd1 [10], and Dazl [11] are expressed in germ cells. These proteins play critical roles in the survival and differentiation of germ cells. In Xenopus laevis, PGCs are not generated after injection of the anti-Xenopus Ddx4 homologue antibody into tadpoles [12]. In mice, a defect in Ddx4 function results in the failure of differentiation and the promotion of apoptotic cell death [13]. The Ter mutation in the Dnd1 gene is known to cause marked decreases in mouse PGCs [14]. In Dazl-deficient mice, germ cells do not initiate meiosis [15].
Some studies have reported that the DNA methylation in gene promoters prevents the gene transcription by inhibiting the binding of transcription factors to some promoters [16, 17]. Although the methylation of imprinting genes plays an important role in sex determination, at the same time, that of germ cell-specific genes is also important for germ cell survival. In humans, incorrect DNA methylation of the DAZL promoter regions of sperm, identified by the presence of CpG islands, causes abnormal spermatogenesis [18]. Although the CpG island is one of the guideposts of the promoter, all promoters do not always have a CpG island. In mammals, the ratio of genes having tCpG islands around the promoter is approximately 40%. Imprinting genes are methylated with the progression of differentiation of cells, including germ cells, while a non-imprinting gene, Brahma, displays a different methylation pattern during spermatogenesis and oogenesis [19, 20]. Based on these differences in gene methylation pattern among the sexes and genes, there is a certain mechanism in mature germ cells for expression of several genes immediately after fertilization. In the zygote during cleavage, the maternal transcription factor is important for embryogenesis [21, 22]. In vertebrates including chickens, the precursors of germ cells arise in the early stage of embryogenesis. One of the germ cell-specific genes, vas, is expressed soon after fertilization in germ cells, despite the fact that the general RNA synthesis is not detected soon after fertilization in drosophila [23]. This indicates the importance of the expression of germ cell-specific genes supporting germ cell survival.
During spermatogenesis, histones are replaced by testis-specific histone variants in male germ cells. Then the variants are converted to transition proteins, which are replaced with protamine in the condensing chromatin during elongating spermatid stage in humans [24]. As a result, the gene expression activity is diminished. Based on this inactivity of transcription, in sperm, it is considered that there is an epigenetic mechanism that makes gene expression easier in the zygote. However, the methylation pattern of germ cell-specific genes in mature germ cells remains unknown. Thus, in the present study, we focused on the putative promoter region of germ cell-specific genes and the pluripotency marker gene Nanog. We first analyzed the methylation states of these genes in sperm.
Materials and Methods
Chicken samples
Chicken sperm and livers were collected from White Leghorn chickens. The semen was separated into 90% and 45% layered Percoll solutions (MP Biomedicals, Solon, OH, USA) by centrifuging for 15 min at 1000 g and 4 C. All procedures were performed under the control of the Guidelines for Animal Experiments of the Faculty of Medicine, Kyushu University, and in compliance with Law No. 105 and Notification No. 6 of the Government of Japan.
Genomic DNA extraction and bisulfite reaction
Genomic DNA was isolated using an AllPrep DNA/RNA Micro Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocol. Unmethylated cytosine in the genomic DNAs isolated from each sample was converted to uracil using a MethylEasy Xceed Rapid DNA Bisulfite Modification Kit (Human Genetic Signatures Pty, Randwick, Australia) according to the manufacturer’s protocol.
Genomic DNA subcloning and sequencing
Genomic DNA sequences, CpG islands, and transcription factor binding sites were identified using the NCBI Entrez database (http://www.ncbi.nlm.nih.gov/Entrez/), MethPrimer (http://www.urogene.org/methprimer/), and TFBIND (http://tfbind.hgc.jp/), respectively. The target sequences were isolated from the genomic DNAs treated with the bisulfite reaction by PCR. The primer sets used for the PCR reaction are listed in Supplemental Table 1 (on-line only). The PCR reaction was performed in a total volume of 10 µl containing 40 ng genomic DNA, 1 µl of 10 × PCR buffer, 0.2 mM of dNTP, 0.25 units TaKaRa Taq HS (Takara Bio, Otsu, Japan) and 250 nM of each specific primer. The target sequences were ligated with T-Vector pMD20 (Takara Bio) using a DNA Ligation Kit, Mighty Mix (Takara Bio), and the construct was transformed into E. coli HST08 (Takara Bio) for sequencing.
Total RNA extraction and reverse transcription
Total RNA was isolated using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. RNA samples were treated with RNase-free DNase (Wako, Osaka, Japan). cDNA was synthesized with oligo (dT)15 and random primers using a GoTaq 2-Step RT-qPCR System (Promega, Madison, WI, USA).
RT-PCR
RT-PCR was performed in a 10-μl volume containing 45 ng cDNA, 1 µl of 10 × PCR buffer, 0.2 mM of dNTP, 0.25 units TaKaRa Taq HS (Takara Bio), and 250 nM of each specific primer (Supplemental Table 2: on-line only). All primer pairs were designed to span introns to prevent amplification of products from genomic DNA. PCR products were electrophoresed using 1.5% agarose gel and were detected by Midori Green DNA Stain (Nippon Genetics, Tokyo, Japan).
Results
Searching for promoter motifs upstream of the first exon in the chicken
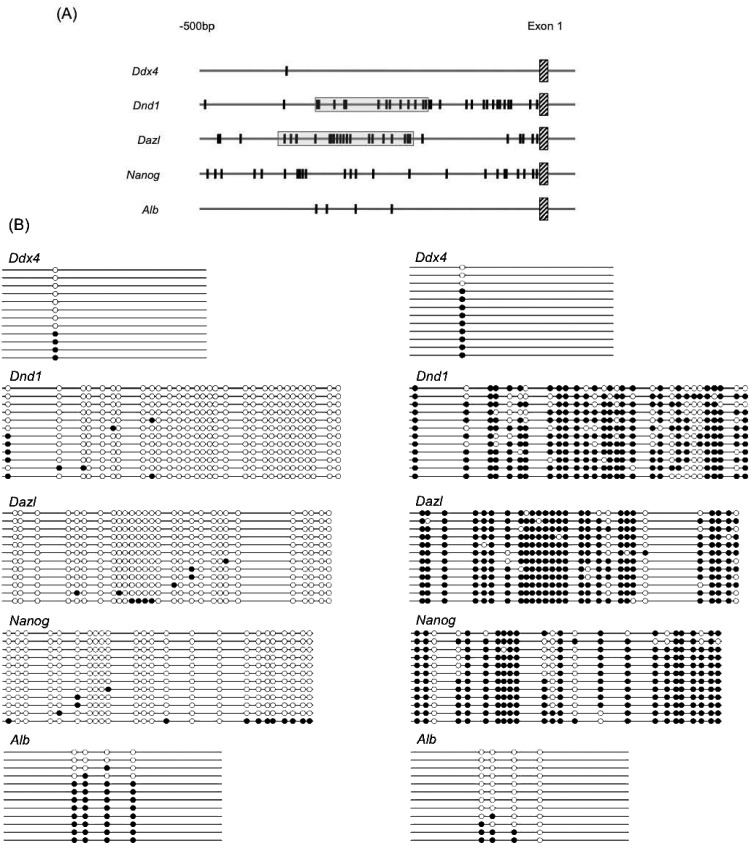
Since the chicken genomic database still does not contain sufficient data to identify the promoter, we searched for promoter motifs such as TATA box, GC box, and CAAT box upstream of their first exon as compared with the mouse genomic database. The structures of chicken Ddx4, Dnd1, Dazl and Nanog were investigated. The Alb gene was used as a reference. Since each chicken gene was constructed by several exons, we focused on a region upstream of the first exon of each gene as a putative promoter. A CpG island was present near the first exon of Dnd1 and Dazl in the chicken. Chicken Ddx4, Nanog and Alb lack CpG islands within the investigated region. However, highly homologous sequences of TATA box, GC box, and CAAT box were located near the first exon of each chicken gene (Fig. 1). Consequently, the region approximately 500 bp upstream from the first exon may function as a promoter.
Fig. 1.
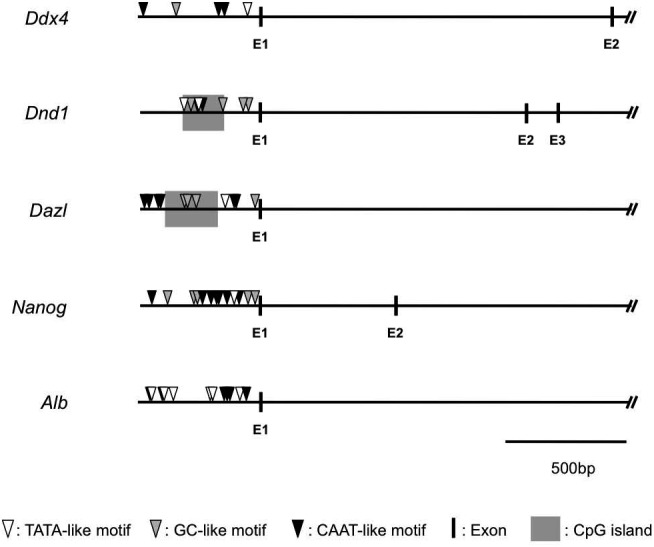
Structure of Ddx4, Dnd1, Dazl, Nanog, and Alb in the 2000-bp region including the downstream regions of the putative promoter regions in the chicken. Vertical line: exon. Shadow: CpG island. White triangle: TATA and TATA-like motifs. Gray triangle: GC and GC-like motifs. Black triangle: CAAT and CAAT-like motifs.
Expression of DNA methyltransferases in sperm and the liver
To further confirm whether the methylation level results on the expression levels of the Dnmt family, the transcripts of Dnmt1, Dnmt3a and Dnmt3b were analyzed in sperm and liver samples. As shown in Fig. 2, the transcripts of Dnmt1 and Dnmt3a were detected in liver samples, but not in sperm.
Fig. 2.
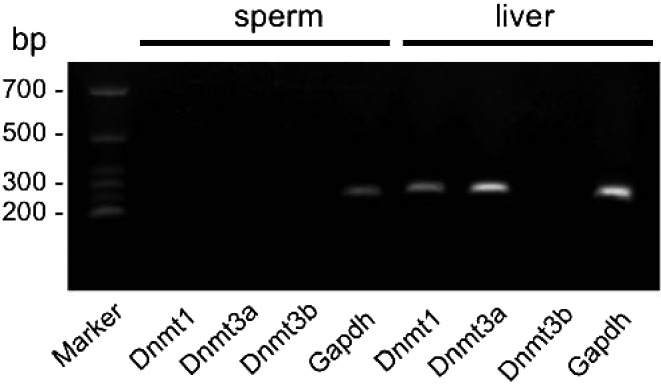
Analyses of the expression of Dnmts in chicken sperm and liver samples. Sperm and liver mRNA samples were subjected to RT-PCR for expression of Dnmt1, Dnmt3a, and Dnmt3b. Gapdh was used as an internal control.
Identification of methylated CpG sites in the putative promoters
To identify the number of CpG sites in the putative promoters of Ddx4, Dnd1, Dazl, Nanog and Alb, we searched for the contiguous sequence of cytosine and guanine in the region 500 bp upstream from the first exon. The results are shown in Fig. 3A. The number of CpG sites was diverse among these five genes, and one CpG island existed in the 5′UTR of Dnd1 (–168 bp to –324 bp) and Dazl (–185 bp to –388 bp). Genomic DNA samples were prepared from sperm and liver samples to analyze their methylation states. After applying the bisulfite reaction to the target sequences, 12 clones were obtained from each gene. The methylation levels of CpG sites in all genes examined were very low in sperm samples, while Alb was highly methylated. Conversely, most CpG sites in Ddx4, Dnd1, Dazl and Nanog were highly methylated in liver samples, while the methylation level of Alb was low (Fig. 3B, Table 1).
Fig. 3.
Methylation of CpG sites within the putative promoter region containing the region 500 bp upstream from the first exon. (A) Localization of CpG sites and a CpG island within the putative promoter region. Ddx4, Nanog and Alb lack CpG islands. Vertical line: CpG site. Shadow: CpG island. (B) Methylated CpG sites in the putative promoter regions of Ddx4, Dnd1, Dazl, Nanog and Alb in chicken sperm and liver samples. Left: sperm; right: liver. Filled circles and open circles represent the methylated and unmethylated CpG sites, respectively. Each line represents an examined clone. The localizations of the CpG sites in each gene shown in A (vertical line) and B (circle) represent the same positions.
Table 1. Methylation levels in the promoters of the germ cell-specific genes, the pluripotency marker gene and the albumin gene in chicken sperm and liver samples.
| Genes | No. of CpG sites | Methylation level (%) |
|
| Sperm | Liver | ||
| Ddx4 | 1 | 33 | 75 |
| Dnd1 | 30 | 3 | 75 |
| Dazl | 28 | 3 | 88 |
| Nanog | 24 | 5 | 82 |
| Alb | 4 | 71 | 17 |
Expression of the target genes in sperm and the liver
To further investigate the relationship between gene transcription and methylation in sperm and the liver, mRNA samples were subjected to RT-PCR. No transcripts of Ddx4, Dnd1, Dazl or Nanog were detected in either sperm or liver samples (Fig. 4). However, the transcript of liver Alb, which was hypomethylated in the putative promoter, was detected in liver samples.
Fig. 4.
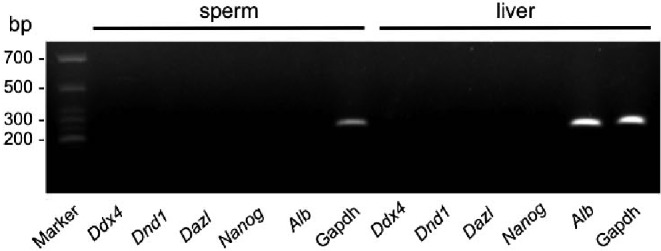
Analyses of the expression of Ddx4, Dnd1, Dazl, Nanog and Alb in chicken sperm and liver samples. Sperm and liver mRNA samples were subjected to RT-PCR for expression of Ddx4, Dnd1, Dazl, Nanog and Alb. Gapdh was used as an internal control.
Discussion
Germ cell-specific genes such as Ddx4, Dnd1, and Dazl play critical roles in the proliferation and survival of germ cells. These proteins are all classified in the RNA binding protein. Ddx4 plays a role in germ cell proliferation as meiotic progression [25]. Dnd1 is associated with germ cell survival and the mitotic arrest of male PGCs [26, 27]. Dazl-defective mice are infertile and exhibit a decrease in germ cells [15]. In oligoasthenoteratozoospermic men, incorrect methylation in the Dazl promoter is a cause of infertility [18]. In the chicken, the expression of the germ cell-specific proteins dynamically changes during germ cell differentiation [11]. This suggests that the expression of genes coding germ cell-specific proteins is also changed during gametogenesis by a certain mechanism. Epigenetic mechanisms contribute to the regulation of imprinted gene transcription [28,29,30,31]. Methylation at CpG sites plays a principal role in gene silencing [32,33,34,35,36,37]. In the present study, we investigated the methylation states of putative promoter regions in non-imprinted germ cell-specific genes. The genes Ddx4, Dnd1, Dazl and Nanog are different in terms of their numbers of CpG sites within the 500–bp putative promoters. Specifically, Dnd1, Dazl and Nanog have many CpG sites, while Ddx4 has only one. In a recent study using mouse spermatogenic cells, it was reported that the number of CpG sites affects the silencing of intronless genes [38]. The fact that the methylation site of the Ddx4 promoter region is only slightly silenced may indicate that the Ddx4 protein is expressed nearly until terminal differentiation of the germ cells.
Certain motifs such as TATA box, GC box and CAAT box appear in promoter regions of eukaryotes. Transcriptions of most genes including imprinted genes are controlled by these elements and epigenetic mechanisms [39,40,41,42,43,44]. At the TATA box, TFIIB and TFIIF, the basic transcription factors, promote gene transcription. One of the well-known transcriptional factors, Sp1, promotes gene transcription via its binding to the GC box in the promoter [45]. DNA methylation can potentially inhibit the binding of transcription factors and repress gene transcription [46]. The methylation patterns of some genes are different among organisms and between sexes. In mice, methylation in the promoter of Brahma decreases during spermatogenesis but increases during oocyte maturation [20]. In the present study, we identified the highly homologous promoter motifs including TATA box. The presence of the highly homologous TATA box suggests the possibility of the combination of basic transcription factors, and the region works as a promoter.
In contrast to the histone acetylation involved in histone acetyltransferase and histone deacetylase [47,48,49], DNA methylation is a strict modification with a process that is regulated by the Dnmt family, including Dnmt1, Dnmt3a, Dnmt3b and Dnmt3L. Of the Dnmt family, Dnmt3L is lacking in chickens [50]. Dnmt1 maintains DNA methylation patterns from parental cells to daughter cells [51,52,53]. Dnmt3a and Dnmt3b are involved in de novo methylation, and they work during early development and gametogenesis in mammals [54, 55]. De novo methylation occurs primarily in germ cells and pluripotent cells for the establishment of individual methylation patterns [56]. The above studies strongly suggest a relationship between methylation of the promoter and the expression of Dnmts in germ cells. In the present study, transcripts of Dnmt1 and Dnmt3a, but not Dnmt3b, were detected in liver samples but were not expressed in sperm. The lack of gene expression of Dnmts in sperm indicates that the activity of the DNA methylation is low in sperm, while it is high in the liver. We also found that methylation levels in the putative promoters of germ cell-specific genes and Nanog were low in sperm, indicating that these genes may be easily expressed in sperm. However, no transcripts were detected in sperm. Conversely, it is very reasonable that the putative promoters of germ cell-specific genes are highly methylated in the liver, where they are not expressed. Interestingly, the methylation level of the Alb promoter was very low in liver samples, where the gene is constantly expressed, while it was very high in sperm. Thus, genes are selectively methylated by unidentified mechanisms. Our present study supports a recent report demonstrating that the promoter CpG islands of germ cell-specific genes are hypermethylated in mouse somatic tissues [57].
One of the prospective reasons for this selectivity is that the genes having a hypomethylated putative promoter are necessary for the development of germ cells and embryos in early embryogenesis. Some studies have indicated the importance of the maternal factors stored in the egg for the zygotic gene activation in zygotes [21, 22]. Additionally, in the embryo of most vertebrates, the precursors of germ cells arise soon after fertilization [9, 23, 58]. Thus, since germ-cell specific genes such as Ddx4, Dnd1 and Dazl and pluripotency marker genes such as Nanog and Oct-4 need to be expressed soon, these genes might be maintained in a state with a low methylation level in their promoters.
In the process of producing sperm and eggs, the germ cells differentiate several times and undergo meiosis. This is the most distinctive event for germ cells, and many genes are involved in this process. Elucidation of the relationship between methylation and expression of functional genes is important for clarification of the mechanism of reproduction. In conclusion, the present study provides novel evidence that the putative promoters of the germ cell-specific genes Ddx4, Dnd1 and Dazl and the pluripotent marker gene Nanog are hypomethylated in chicken sperm. These genes, which have critical roles in the proliferation and survival of germ cells and the pluripotency of embryonic cells, may be ready to be expressed soon after fertilization. Conversely, the promoter of the Alb gene, which is not essential for their proliferation and survival, is highly methylated in chicken sperm. Our findings showing that low methylation and selective DNA methylation of specific genes are present in chicken sperm contribute to our understanding of fertilization and embryogenesis of birds, but the molecular mechanism(s) remains to be clarified in the future.
Conflict of interest: The authors declare that no conflicts of interest exist that would prejudice the impartiality of this study.
Supplementary
Acknowledgments
This work was supported in part by a Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Sciences (JSPS No. 19658099, 24658246) (to M-A Hattori). Gakushi Kito was supported by a Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists (Grant No. 23–1262).
References
- 1.Lefèvre C, Mann JR. RNA expression microarray analysis in mouse prospermatogonia: identification of candidate epigenetic modifiers. Dev Dyn 2008; 237: 1082–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohno R, Nakayama M, Naruse C, Okashita N, Takano O, Tachibana M, Asano M, Saitou M, Seki Y. A replication-dependent passive mechanism modulates DNA demethylation in mouse primordial germ cells. Development 2013; 140: 2892–2903 [DOI] [PubMed] [Google Scholar]
- 3.Obata Y, Kono T. Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J Biol Chem 2002; 277: 5285–5289 [DOI] [PubMed] [Google Scholar]
- 4.Lucifero D, Mann MR, Bartolomei MS, Trasler JM. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet 2004; 13: 839–849 [DOI] [PubMed] [Google Scholar]
- 5.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development 1987; 99: 371–382 [DOI] [PubMed] [Google Scholar]
- 6.Davis TL, Yang GJ, McCarrey JR, Bartolomei MS. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet 2000; 9: 2885–2894 [DOI] [PubMed] [Google Scholar]
- 7.Ueda T, Abe K, Miura A, Yuzuriha M, Zubair M, Noguchi M, Niwa K, Kawase Y, Kono T, Matsuda Y, Fujimoto H, Shibata H, Hayashizaki Y, Sasaki H. The paternal methylation imprint of the mouse H19 locus is acquired in the gonocyte stage during foetal testis development. Genes Cells 2000; 5: 649–659 [DOI] [PubMed] [Google Scholar]
- 8.Sanford JP, Clark HJ, Chapman VM, Rossant J. Differences in DNA methylation during oogenesis and spermatogenesis and their persistence during early embryogenesis in the mouse. Genes Dev 1987; 1: 1039–1046 [DOI] [PubMed] [Google Scholar]
- 9.Tsunekawa N, Naito M, Sakai Y, Nishida T, Noce T. Isolation of chicken vasa homolog gene and tracing the origin of primordial germ cells. Development 2000; 127: 2741–2750 [DOI] [PubMed] [Google Scholar]
- 10.Aramaki S, Kubota K, Soh T, Yamauchi N, Hattori MA. Chicken dead end homologue protein is a nucleoprotein of germ cells including primordial germ cells. J Reprod Dev 2009; 55: 214–218 [DOI] [PubMed] [Google Scholar]
- 11.Kito G, Aramaki S, Tanaka K, Soh T, Yamauchi N, Hattori MA. Temporal and spatial differential expression of chicken germline-specific proteins cDAZL, CDH and CVH during gametogenesis. J Reprod Dev 2010; 56: 341–346 [DOI] [PubMed] [Google Scholar]
- 12.Ikenishi K, Tanaka TS. Involvement of the protein of Xenopus vasa homolog (Xenopus vasa-like gene 1, XVLG1) in the differentiation of primordial germ cells. Dev Growth Differ 1997; 39: 625–633 [DOI] [PubMed] [Google Scholar]
- 13.Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev 2000; 14: 841–853 [PMC free article] [PubMed] [Google Scholar]
- 14.Youngren KK, Coveney D, Peng X, Bhattacharya C, Schmidt LS, Nickerson ML, Lamb BT, Deng JM, Behringer RR, Capel B, Rubin EM, Nadeau JH, Matin A. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature 2005; 435: 360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saunders PT, Turner JM, Ruggiu M, Taggart M, Burgoyne PS, Elliott D, Cooke HJ. Absence of mDazl produces a final block on germ cell development at meiosis. Reproduction 2003; 126: 589–597 [DOI] [PubMed] [Google Scholar]
- 16.Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev 1989; 3: 612–619 [DOI] [PubMed] [Google Scholar]
- 17.Comb M, Goodman HM. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res 1990; 18: 3975–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navarro-Costa P, Nogueira P, Carvalho M, Leal F, Cordeiro I, Calhaz-Jorge C, Gonçalves J, Plancha CE. Incorrect DNA methylation of the DAZL promoter CpG island associates with defective human sperm. Hum Reprod 2010; 25: 2647–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patra SK, Deb M, Patra A. Molecular marks for epigenetic identification of developmental and cancer stem cells. Clin Epigenetics 2011; 2: 27–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagrani SR, Levens ED, Baxendale V, Boucheron C, Chan WY, Rennert OM. Methylation patterns of Brahma during spermatogenesis and oogenesis: potential implications. Fertil Steril 2011; 95: 382–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leichsenring M, Maes J, Mössner R, Driever W, Onichtchouk D. Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science 2013; 341: 1005–1009 [DOI] [PubMed] [Google Scholar]
- 22.Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, Giraldez AJ. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 2013; 503: 360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol 1998; 8: 243–246 [DOI] [PubMed] [Google Scholar]
- 24.Carrell DT, Emery BR, Hammoud S. Altered protamine expression and diminished spermatogenesis: what is the link? Hum Reprod Update 2007; 13: 313–327 [DOI] [PubMed] [Google Scholar]
- 25.Medrano JV, Ramathal C, Nguyen HN, Simon C, Reijo Pera RA. Divergent RNA-binding proteins, DAZL and VASA, induce meiotic progression in human germ cells derived in vitro. Stem Cells 2012; 30: 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhattacharya C, Aggarwal S, Zhu R, Kumar M, Zhao M, Meistrich ML, Matin A. The mouse dead-end gene isoform alpha is necessary for germ cell and embryonic viability. Biochem Biophys Res Commun 2007; 355: 194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cook MS, Munger SC, Nadeau JH, Capel B. Regulation of male germ cell cycle arrest and differentiation by DND1 is modulated by genetic background. Development 2011; 138: 23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onyango P, Jiang S, Uejima H, Shamblott MJ, Gearhart JD, Cui H, Feinberg AP. Monoallelic expression and methylation of imprinted genes in human and mouse embryonic germ cell lineages. Proc Natl Acad Sci USA 2002; 99: 10599–10604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, Barros A, Sousa M. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod 2008; 14: 67–74 [DOI] [PubMed] [Google Scholar]
- 30.Marques CJ, Francisco T, Sousa S, Carvalho F, Barros A, Sousa M. Methylation defects of imprinted genes in human testicular spermatozoa. Fertil Steril 2010; 94: 585–594 [DOI] [PubMed] [Google Scholar]
- 31.Park CH, Uh KJ, Mulligan BP, Jeung EB, Hyun SH, Shin T, Ka H, Lee CK. Analysis of imprinted gene expression in normal fertilized and uniparental preimplantation porcine embryos. PLoS ONE 2011; 6: e22216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mostovich LA, Prudnikova TY, Kondratov AG, Loginova D, Vavilov PV, Rykova VI, Sidorov SV, Pavlova TV, Kashuba VI, Zabarovsky ER, Grigorieva EV. Integrin alpha9 (ITGA9) expression and epigenetic silencing in human breast tumors. Cell Adh Migr 2011; 5: 395–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ristola M, Arpiainen S, Saleem MA, Holthöfer H, Lehtonen S. Transcription of nephrin-Neph3 gene pair is synergistically activated by WT1 and NF-κB and silenced by DNA methylation. Nephrol Dial Transplant 2012; 27: 1737–1745 [DOI] [PubMed] [Google Scholar]
- 34.Kocemba KA, Groen RW, van Andel H, Kersten MJ, Mahtouk K, Spaargaren M, Pals ST. Transcriptional silencing of the Wnt-antagonist DKK1 by promoter methylation is associated with enhanced Wnt signaling in advanced multiple myeloma. PLoS ONE 2012; 7: e30359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navarro A, Yin P, Monsivais D, Lin SM, Du P, Wei JJ, Bulun SE. Genome-wide DNA methylation indicates silencing of tumor suppressor genes in uterine leiomyoma. PLoS ONE 2012; 7: e33284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alvarez C, Tapia T, Cornejo V, Fernandez W, Muñoz A, Camus M, Alvarez M, Devoto L, Carvallo P. Silencing of tumor suppressor genes RASSF1A, SLIT2, and WIF1 by promoter hypermethylation in hereditary breast cancer. Mol Carcinog 2013; 52: 475–487 [DOI] [PubMed] [Google Scholar]
- 37.Gao L, Smit MA, van den Oord JJ, Goeman JJ, Verdegaal EM, van der Burg SH, Stas M, Beck S, Gruis NA, Tensen CP, Willemze R, Peeper DS, van Doorn R. Genome-wide promoter methylation analysis identifies epigenetic silencing of MAPK13 in primary cutaneous melanoma. Pigment Cell Melanoma Res 2013; 26: 542–554 [DOI] [PubMed] [Google Scholar]
- 38.Kato Y, Nozaki M. Distinct DNA methylation dynamics of spermatogenic cell-specific intronless genes is associated with CpG content. PLoS ONE 2012; 7: e43658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maity SN, Golumbek PT, Karsenty G, de Crombrugghe B. Selective activation of transcription by a novel CCAAT binding factor. Science 1988; 241: 582–585 [DOI] [PubMed] [Google Scholar]
- 40.Ragimov N, Krauskopf A, Navot N, Rotter V, Oren M, Aloni Y. Wild-type but not mutant p53 can repress transcription initiation in vitro by interfering with the binding of basal transcription factors to the TATA motif. Oncogene 1993; 8: 1183–1193 [PubMed] [Google Scholar]
- 41.Kim EC, Lau JS, Rawlings S, Lee AS. Positive and negative regulation of the human thymidine kinase promoter mediated by CCAAT binding transcription factors NF-Y/CBF, dbpA, and CDP/cut. Cell Growth Differ 1997; 8: 1329–1338 [PubMed] [Google Scholar]
- 42.Eggen BJ, Benus GF, Folkertsma S, Jonk LJ, Kruijer W. TAK1 activation of the mouse JunB promoter is mediated through a CCAAT box and NF-Y. FEBS Lett 2001; 506: 267–271 [DOI] [PubMed] [Google Scholar]
- 43.Bernadt CT, Rizzino A. Roles of the conserved CCAAT and GC boxes of the human and mouse type II transforming growth factor-beta receptor genes. Mol Reprod Dev 2003; 65: 353–365 [DOI] [PubMed] [Google Scholar]
- 44.Schiavoni G, Bennati AM, Castelli M, Fazia MA, Beccari T, Servillo G, Roberti R. Activation of TM7SF2 promoter by SREBP-2 depends on a new sterol regulatory element, a GC-box, and an inverted CCAAT-box. Biochim Biophys Acta 2010; 1801: 587–592 [DOI] [PubMed] [Google Scholar]
- 45.Letovsky J, Dynan WS. Measurement of the binding of transcription factor Sp1 to a single GC box recognition sequence. Nucleic Acids Res 1989; 17: 2639–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oda M, Kumaki Y, Shigeta M, Jakt LM, Matsuoka C, Yamagiwa A, Niwa H, Okano M. DNA methylation restricts lineage-specific functions of transcription factor Gata4 during embryonic stem cell differentiation. PLoS Genet 2013; 9: e1003574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love IM, Sekaric P, Shi D, Grossman SR, Androphy EJ. The histone acetyltransferase PCAF regulates p21 transcription through stress-induced acetylation of histone H3. Cell Cycle 2012; 11: 2458–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao J, Zhang H, Xing L, Xu S, Liu H, Chong K, Xu Y. Requirement of histone acetyltransferases HAM1 and HAM2 for epigenetic modification of FLC in regulating flowering in Arabidopsis. J Plant Physiol 2013; 170: 444–451 [DOI] [PubMed] [Google Scholar]
- 49.Lopez-Atalaya JP, Ito S, Valor LM, Benito E, Barco A. Genomic targets, and histone acetylation and gene expression profiling of neural HDAC inhibition. Nucleic Acids Res 2013; 41: 8072–8084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yokomine T, Hata K, Tsudzuki M, Sasaki H. Evolution of the vertebrate DNMT3 gene family: a possible link between existence of DNMT3L and genomic imprinting. Cytogenet Genome Res 2006; 113: 75–80 [DOI] [PubMed] [Google Scholar]
- 51.Okuwaki M, Verreault A. Maintenance DNA methylation of nucleosome core particles. J Biol Chem 2004; 279: 2904–2912 [DOI] [PubMed] [Google Scholar]
- 52.Estève PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev 2006; 20: 3089–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spada F, Haemmer A, Kuch D, Rothbauer U, Schermelleh L, Kremmer E, Carell T, Längst G, Leonhardt H. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J Cell Biol 2007; 176: 565–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999; 99: 247–257 [DOI] [PubMed] [Google Scholar]
- 55.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol 2003; 23: 5594–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, Laird PW, Jones PA. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol 2002; 22: 480–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki M, Sato S, Arai Y, Shinohara T, Tanaka S, Greally JM, Hattori N, Shiota K. A new class of tissue-specifically methylated regions involving entire CpG islands in the mouse. Genes Cells 2007; 12: 1305–1314 [DOI] [PubMed] [Google Scholar]
- 58.Lawson KA, Hage WJ. Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Found Symp 1994; 182: 68–84, discussion :84–91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.