Highlights
-
•
Transcriptional directionality is controlled by premature transcription termination.
-
•
Transcriptional directionality is enforced by gene looping.
-
•
mRNA-specific termination signals and factors are required for gene looping.
Keywords: bidirectional promoters, transcriptional termination, gene loops
Abstract
Bidirectional promoters are a common feature of many eukaryotic organisms from yeast to humans. RNA Polymerase II that is recruited to this type of promoter can start transcribing in either direction using alternative DNA strands as the template. Such promiscuous transcription can lead to the synthesis of unwanted transcripts that may have negative effects on gene expression. Recent studies have identified transcription termination and gene looping as critical players in the enforcement of promoter directionality. Interestingly, both mechanisms share key components. Here, we focus on recent findings relating to the transcriptional output of bidirectional promoters.
Transcription from bidirectional promoters
Many promoters for RNA polymerase II (Pol II) are bidirectional, as found in a wide range of organisms including humans, mice, Arabidopsis thaliana, and Saccharomyces cerevisiae [1–8]. Pol II recruited to such promoters is directionally unbiased and can transcribe DNA in both directions. Divergent transcription can produce either two mRNA (head-to-head genes) or a single mRNA and a corresponding upstream noncoding RNA (ncRNA) [4–8]. The presence of head-to-head genes increases with decreasing genome length. Thus, for the compressed S. cerevisiae genome, half of mRNA coding genes are divergent, whereas in humans, only 11% of genes are so organized [9]. The vast majority of bidirectional promoters produce only one mRNA together with a divergent, usually nonfunctional, ncRNA (Box 1).
Box 1. Promoter associated ncRNA species.
In animals, promoter-associated ncRNA (upstream antisense RNA, uasRNA) are classified by their length. Short ncRNAs (<100 nt) are referred to as transcription start site-associated RNA (TSSa-RNA) [7]. TSSa-RNA are the products of 5′→3′ degradation of the nascent transcript protected by paused Pol II [93]. ncRNA >100–200 nt are termed lncRNA and are produced divergently from the coding sequence by Pol II that has escaped from pausing [94]. A prominent group of this type is called PROMPTs. PROMPTs are 5′ capped and 3′ adenylated, but in contrast to mRNA, undergo quick degradation by the nuclear exosome [6,48]. lncRNA considerably longer than PROMPTs derived from enhancers are named enhancer RNA (eRNA) (Box 2) [32–34,94]. Other lncRNA that are transcribed from intergenic regions (devoid of protein-coding genes) and are called long intergenic ncRNA (lincRNA) [95,96].
In yeast, promoter-associated ncRNA are more extensively characterized. Similar to animals, the vast majority of ncRNA are detectable only in RNA-degradation-deficient mutants as they undergo rapid degradation in wild type cells. Cryptic unstable transcripts (CUTs) include ncRNA accumulating in 3′→5′ exonucleolytic mutants of Rrp6 and exosome [4,5,97]. Their number varies from 925 when only RRP6 is deleted, to ∼1600 when both exosome and its cofactor Rrp6 are inactive. Stable unannotated transcripts (SUTs; 847 detected), are longer (median length is 761 nt compared to 440 nt for CUTs) and more resistant to exonucleolytic digestion [5]. CUTs and SUTs are transcribed mostly from divergent promoters, followed by terminators and least frequently from intragenic regions. Both classes are predominantly terminated by the NRD complex (68% of CUTs and 58% of SUTs) and have been proposed to be organized into a holo-class called Nrd1-dependent unterminated transcripts (NUTs) [40]. A total of 1526 NUTs were identified by deep sequencing after Nrd1 depletion. However, only 213 are novel ncRNA and these do not overlap with any specific genomic features. Another major class of ncRNA includes 605 Ssu72-restricted transcripts (SRTs) [73]. SRTs accumulate independently of the exonucleolytic machinery in an ssu72-2 mutant which has lost gene looping, and are directly associated with promoter directionality. The most abundant group of ncRNA (1658 detected) accumulate in a Xrn1 5′→3′ exonuclease mutant, and are called Xrn1-dependent unstable transcripts (XUTs) [98]. Most XUTs are expressed from terminators, as 66% are antisense to their ORFs. The lines of distinction between these different ncRNA classes are blurred, for example, elongated CUTs, (eCUTs), which have escaped their primary transcription termination pathway, are terminated at the nearest PAS and may become stable SUTs [51]. Moreover, 20% of CUTs and 75% of SUTs have been classified as XUTs [98].
The reason transcription initiates in both directions from a promoter region appears to be dictated by the chromatin structure. In general, Pol II promoters are nucleosome-free regions (NFRs) [10–14]. The lack of DNA packaging facilitates recruitment of transcriptional machinery, unwinding of DNA strands, and as a consequence, initiation of transcription. NFRs are partially determined by intrinsic promoter sequences, which disfavor nucleosome assembly and permit access to transcription factors (TFs) [15]. In yeast, this role is played by poly(A-T) tracts frequently occurring in promoter sequences [16–18]. In higher eukaryotes, CpG islands, which are present in more than half of human and mouse promoters, are involved in regulation of nucleosome assembly [15]. Although CpG islands support nucleosome formation in vitro [19], they appear nucleosome depleted in vivo [20]. It has been suggested that CpG islands are resistant to higher-order chromatin compaction by linker histones such as H1, which is known to have a binding preference for A-T-rich DNA [15,21].
It appears that more complex mechanisms are responsible for keeping promoters in a nucleosome-free state. For instance, several groups have reported that NFRs over promoters result from competition for DNA binding between TFs and nucleosomes [22–25]. In particular, TFs bound to promoters introduce a barrier for chromatin organization. Promoters with paused Pol II have also been shown to restrict nucleosome formation [26]. A proposed model describes stretches of poly(A-T) tracts or CpG islands that act to maintain chromatin in a state that allows transient TF access. The consequent recruitment of TFs and then Pol II results in the expansion of accessible regions and thus maintenance of NFRs [15].
Although such chromatin organization of promoters may aid gene expression, its tendency to be bidirectional introduces the potential danger of deleterious transcript synthesis. Biased recruitment of the transcriptional machinery can be determined by the orientation specific binding of TATA-binding protein (TBP) to the TATA box [4,27]. Indeed, in Drosophila melanogaster, Pol II-transcribed promoters are normally unidirectional with the protein coding strand defined by prominent TATA box sequences [28]. In the absence of a TATA element, TBP is still delivered to promoters by the multi-subunit TFIID complex that lacks sequence specificity [27]. Consequently, Pol II occupation at the initiation sites of bidirectional promoters is evident in the coding and noncoding directions [7,8] (Figure 1). High-resolution chromatin immunoprecipitation preceded by exonucleolytic digestion (ChIP-exo analysis) of the preinitiation complex (PIC) in S. cerevisiae has revealed that bidirectional promoters expressing ncRNA–mRNA contain two independent PICs in inverted orientation, similar to bidirectional promoters controlling head-to-head protein coding genes [29]. However, despite the presence of separate PICs, transcription in one direction is dependent on the other. For example, the repression of mRNA synthesis by a TATA box mutation leads to an increase in transcription of antisense divergent RNA [4]. Furthermore, divergent promoters often share the same transcriptional activators, as indicated by the co-regulation of ncRNA–mRNA as well as head-to-head mRNA pairs [1,4,5,9].
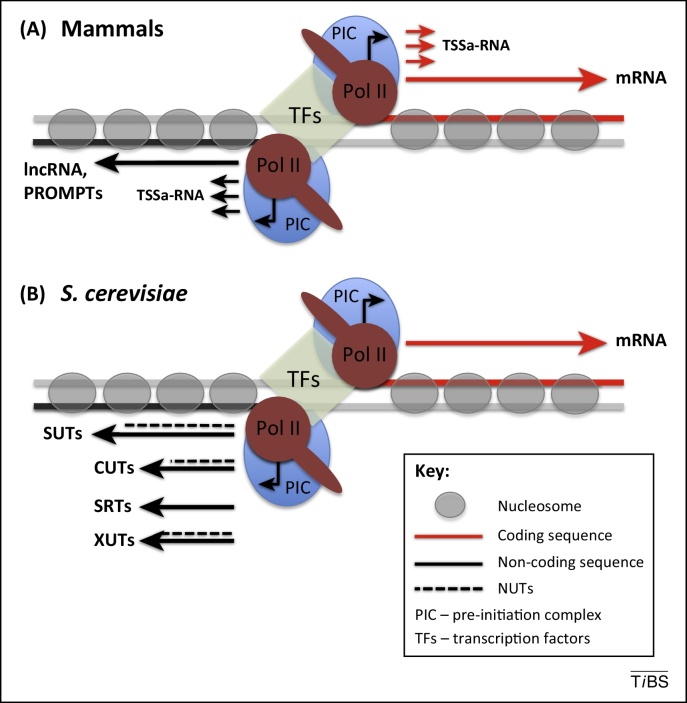
Figure 1.
Promoter-associated noncoding RNA. (A) Major classes of promoter-associated ncRNA in animals. (B) ncRNA transcribed from divergent promoters in Saccharomyces cerevisiae. NFR acts as a Pol II promoter for both the protein-coding sequence (marked as a red line) and antisense noncoding sequence (black line). The open and unbiased nature of NFR allows for formation of two independent PICs, which share TFs. Such promoter structure allows Pol II to transcribe in both directions. In mammals the upstream regions are transcribed into PROMPTs and longer ncRNA generally referred to as lncRNA. TSSa-RNA, related to Pol II pausing, are synthesized in both directions. S. cerevisiae bidirectional promoters for protein-coding genes are similarly used to initiate transcription of upstream noncoding regions. Transcribed ncRNA are classified as SUTs, CUTs, or XUTs by their susceptibility to different degrading enzymes (Box 1). SRTs are synthesized when interactions between the promoter and the sense open reading frame terminator is disrupted. All ncRNA initiated from Pol II promoters, except for SRTs, may undergo NRD dependent termination and so are classified as NUTs (marked by dotted line). The most common classes of promoter-associated ncRNA are SUTs and CUTs, whereas occurrence of SRTs and XUTs is similar. Due to the lack of Pol II promoter pausing TSSa-RNA are not present in S. cerevisiae. Abbreviations: CUT, cryptic unstable transcript; lncRNA, long noncoding RNA; ncRNA, noncoding RNA; NFR, nucleosome-free region; NRD, NRD complex; NUT, Nrd1-dependent unterminated transcript; PIC, preinitiation complex; Pol II, RNA polymerase II; PROMPT, promoter upstream transcript; SRT, Ssu72-restricted transcript; SUT, stable unannotated transcript; TF, transcription factor; TSSa-RNA, transcription start site-associated RNA; XUT, Xrn1-depednent transcript.
NFRs present elsewhere across the genome can also be used for transcription initiation. For example, NFRs over terminators of protein coding genes appear to be formed similarly to those in promoter regions. Their sequences may impede nucleosome formation [17,18,30], but transcription-dependent mechanisms may again play a role in NFR maintenance [31]. ncRNA classes initiated from terminators are identical to those expressed from the promoters [118]. However, terminator-derived transcripts may be selectively fired in the direction of the mRNA promoter, so that the ncRNA is complementary to the corresponding mRNA and may have regulatory function. Inter- or intragenically located NFRs may be also used for transcription initiation. However, a likely future focus of research will be NFRs located over enhancers which are the source of enhancer RNA (eRNA) [32–34] (Box 2). Although terminator or enhancer-associated NFRs are Pol II promoters, they appear to produce only ncRNA. In many cases it is not clear whether it is RNA synthesis or just the process of transcription itself that is relevant. Pol II initiation in these regions may also be effectively random and merely represent byproducts of NFRs. In this review we focus only on promoters engaged in protein-coding gene expression and the mechanisms that direct Pol II into the productive synthesis of mRNA.
Box 2. eRNA.
Enhancers are described as DNA elements that activate promoters from a variable distance. The ∼55 000 enhancers identified in the human genome play regulatory roles in development, differentiation, and tissue-specific gene expression [99,100]. Like promoters, enhancers comprise accessible chromatin structure bound by TFs and Pol II [32,33,101]. However, in contrast to promoters, only 5% of enhancer sequences consist of CpG islands, and enhancers have significantly lower levels of H3K4me3 marks, which are responsible for PIC assembly stimulation [84,99,102]. Instead, enhancers are generally more defined by H3K4me1 and H3K327 acetylation marks. Analysis of ncRNA in human embryonic stem cells, murine embryonic stem cells, and human endodermal cells has revealed that RNA initiated from enhancers (eRNA) comprises, respectively, 19%, 27%, and 23% of all lncRNA [34]. However, if only lincRNA are taken into account, eRNA comprise the vast majority.
Enhancer regions promoting transcription (referred as e-promoters) can be either uni- or bidirectional [32,33,103]. Genome-wide studies in mouse cells have revealed that eRNA transcribed from bidirectional e-promoters are relatively short (0.5–2 kb) and nonpolyadenylated [103]. By contrast, unidirectional e-promoters generate longer transcripts (often >4 kb) and are normally polyadenylated. Although features of Pol II transcribing long eRNA appear similar to protein-coding genes, its C-terminal domain (CTD) phosphorylation pattern differs: CTD Ser2 levels are low along the enhancer-derived transcription unit. Also, the transcribed region lacks H3K36me3 elongation marks [103]. A poly(A) tail indicates the presence of a PAS at the 3′ end of long eRNA and implies termination by CPAC. This may control directionality of transcription initiated in enhancers by the mechanisms as described in this review. Studies on the murine α-globin locus in mouse erythrocytes have shed significant light on long eRNA [104]. Transcription from intragenic α-globin enhancers results in short nonpolyadenylated eRNA in both directions. However, because these enhancers are located in the intron of a separate host gene Nprl3, they can also act as alternative promoters for Nprl3. This results in directional transcription of lncRNA referred to as multiexonic RNA (meRNA), which are spliced and polyadenylated as mRNA. Such meRNA are prevalent in the human genome [104]. This observation raises the intriguing question: how can bidirectional intergenic e-promoters convert to unidirectionality? Does such a mechanism act to enforce transcriptional directionality in a similar manner to that of protein-coding genes? Taking into account the exceptionally important role of enhancers in the regulation of basic biological processes, the better understanding of enhancer-associated transcription may significantly broaden our knowledge of how protein-coding genes are activated or silenced.
Divergent transcription from bidirectional promoters can be deleterious to cells. Transcription of noncoding regions may result in the downregulation of coexpressed mRNA and can also lead to accumulation of ncRNA. This may be harmful for the cell because such ncRNA could be translated into a toxic product or compete with mRNA for RNA-binding proteins and their regulatory factors [35]. In spite of this and for the reasons that are discussed in this review, divergent transcription is a common feature in many eukaryotes. Therefore, mechanisms have evolved to direct Pol II into the coding region and limit transcription of divergent ncRNA as described later.
Transcriptional directionality is controlled by termination
The simplest approach to block unwanted transcription is the immediate termination and degradation of newly synthesized ncRNA [36–40]. In this situation, Pol II transcribing ncRNA encounters intrinsic sequences that trigger transcription termination and subsequent degradation. Therefore, despite efficient transcription initiation in both directions, only Pol II transcribing the protein coding sequence successfully enters into a full elongation phase.
Multiple mechanisms can promote transcription termination of Pol II in eukaryotic organisms [41,42]. For protein-coding genes, the major transcription termination mechanism uses a poly(A) site (PAS) comprising a central AAUAAA sequence in humans and less conserved, degenerate Py(A)n sequence in S. cerevisiae. PAS also include 5′ positioned U-rich and 3′ positioned GU/U-rich sequences. PAS-dependent termination (Box 3) is mediated by macromolecular complexes generally referred to as the cleavage and polyadenylation complex (CPAC), which is remarkably conserved during evolution [43]. CPAC recognizes the PAS and cleaves the nascent RNA. Following release of the mRNA, Pol II transcribes further downstream, up to 200 nucleotides (nt) in yeast and 1500 nt in mammals, before being dismantled from the DNA template. A subset of human mRNA coding genes uses a variation of PAS-dependent termination for which a distal AU-rich region acts to mediate cotranscriptional cleavage (CoTC) with rapid 5′→3′ degradation of nascent RNA leading to Pol II release [44,45].
Box 3. PAS- and NRD-dependent transcription termination.
PAS-dependent transcription termination is tightly coupled with 3′ end processing. Despite the lack of PAS sequence conservation between mammals and yeast, most of mammalian termination and processing factors have homologs in S. cerevisiae. Throughout this review we generally refer to these factors as components of the CPAC. In higher eukaryotes, CPAC consists of four multi-subunit complexes: CPSF, CstF, and two cleavage factors (CF Im and CF IIm). In yeast, homologous proteins are organized into subcomplexes; CPF and CF IA and CF IB [42,105]. The eukaryotic termination and processing machinery also contains poly(A) polymerase (PAP) and poly(A) binding proteins (PABP). CPAC recognizes and binds the PAS. The CPAC component CPSF-73 (Brr5 in yeast) cleaves the nascent RNA releasing pre-mRNA [106,107], which is subsequently polyadenylated. The downstream RNA generated by the cleavage is digested by 5′→3′ exoribonuclease Xrn2 (Rat1 in yeast). This is accompanied by an RNA:DNA helicase called Senataxin which unwinds potential R-loops formed with the DNA template [108]. Xrn2 degrades RNA and together with the other CPAC components, displaces Pol II from DNA [109,110].
Recruitment of CPAC also depends on the Pol II CTD. Pcf11, a component of CF IIm in humans and CF IA in yeast, is crucial for transcription termination [111]. This protein contains a CTD interacting domain (CID) and is recruited to Ser-2 phosphorylated CTD [112], characteristic of late stages of transcription. The presence of termination factors on Pol II approaching the end of the gene enhances the termination process.
In S. cerevisiae Pol II can be also terminated by the NRD complex consisting of Nrd1, Nab3, and RNA:DNA helicase Sen1 [50,113]. Similarly to CPAC, NRD recognizes specific sequences in the nascent RNA [the strongest motifs are UGUA/GUAG and UCUU/CUUG for the RNA recognition motifs (RRMs) of Nrd1 and Nab3, respectively] and interacts with CTD via the Nrd1 CID [52,114,115]. However, in contrast to Pcf11, Nrd1 has a stronger affinity to Ser-5 phosphorylated CTD and is therefore recruited at an earlier stage of transcription [114]. Remarkably, Ser2-P marks are also present over NRD terminators [59,116] and most CPAC mutants, including Pcf11 but excluding Brr5, display a termination defect for NRD-dependent genes [58–64]. However, NRD–CPAC interplay remains poorly understood. The current model, backed by in vitro data, assumes that Sen1, delivered to the nascent RNA by NRD, translocates in a 5′→3′ direction and somehow displaces Pol II [117].
Recently, a balance between PAS recognition and PAS blockage by U1 small nuclear RNA (snRNA) was predicted to regulate transcription termination and thus define the directionality of Pol II transcription in mammals [38,39]. Deep sequencing of polyadenylated RNA 3′ ends from mouse embryonic cells identified cleavage sites associated with the PAS hexamer AAUAAA or close variants, proximal to Pol II transcription start sites (TSS) and at least 5 kb away from known transcription termination sites. Upstream antisense regions contain twofold more cleavage sites, peaking at 700 bp from a TSS, compared to the protein-coding sense sequences. Consistent with this, 48% of divergent promoters produce ncRNA susceptible to PAS-dependent antisense cleavage events in proximity to the TSS. Moreover, antisense divergent PAS are bound by cleavage and polyadenylation factors in a similar manner to mRNA PAS. This suggests that mammalian promoter-associated ncRNA (in particular promoter upstream transcripts, PROMPTs; Box 1) are generally prematurely terminated by early PAS selection. However, the frequency of candidate PAS sequences in TSS proximal sense regions (up to 6 kb) is only reduced by 33% as compared to antisense regions. Such a sequence bias does not adequately account for the discrepancy in cleavage distribution. Consequently, additional elements were searched for to explain better the asymmetric pattern of cleavage events for sense and antisense transcripts. This led to finding that 5′ splice site related sequences (5′SS; U1 sites), which are recognized by U1 snRNA are more enriched in the coding sequence direction as compared to antisense divergent sequence. This provides a possible explanation for the biased PAS-mediated cleavage pattern, because the U1 small nuclear ribonucleoprotein (snRNP) complex is known to suppress cleavage and polyadenylation within a 1-kb vicinity of 5′SS, by inhibiting cleavage and polyadenylation specificity factor (CPSF) [46,47]. Indeed, downregulation of U1 increases cleavage events over coding regions, but has little impact on antisense regions [38].
Parallel studies characterizing PROMPT 3′ end formation in HeLa cells [39] have validated some of the above mechanisms. This genome-wide analysis shows that these ncRNA contain functional PAS with AWUAAA, GU/U and U-rich motifs in positions resembling mRNA 3′ ends. PROMPT PAS cloned into reporter genes generate unstable polyadenylated RNA. Mutation of the GU/U PAS motif causes significant read-through transcription of the PROMPT PAS, whereas mutation of AWUAAA hexamer completely abolishes 3′ end formation [39]. Overall, the involvement of a PAS-dependent pathway in directing expression from bidirectional promoters in mammalian cells appears unequivocal. However, a few questions remain unanswered. In contrast to protein-coding genes, PAS in a PROMPT context yield unstable RNA that is subjected to degradation [6,48]. It is therefore speculated that PAS-dependent termination close to promoters occurs in suboptimal conditions, which somehow facilitates transcript degradation. This is achieved by the recruitment of the multimeric degradation complex called exosome via interaction of the cap binding complex (CBP) with the nuclear exosome targeting complex (NEXT) [39,49]. Nonetheless, the mechanism whereby the PAS-associated degradation pathway differentiates ncRNA from mRNA remains unclear.
In S. cerevisiae, apart from PAS-dependent termination, Pol II also uses a separate NRD-dependent pathway (Box 3) for termination of short transcripts [42]. The NRD complex consists of three polypeptides (Nrd1, Nab3, and Sen1) and is recruited at early stages of transcription. Furthermore, it responds to specific short sequences in the nascent RNA (NRD binding sites, NBS) and acts to promote transcription termination in the downstream region. NRD complex interacts with CBP and recruits the exosome together with the associated exonuclease Rrp6 [50]. In contrast to mRNA, NRD-terminated transcripts are subjected to rapid 3′→5′ exonucleolytic digestion. This connection between a termination complex and RNA-degrading enzymes allows the NRD pathway to maintain proper transcriptional directionality from bidirectional promoters. Unwanted transcription is effectively restricted close to the transcription start site and its product, ncRNA, is immediately degraded.
Recent extensive genome-wide studies [40] on the function of NRD have confirmed and broadened previous findings [36,37,51,52] by demonstrating that ∼50% of known ncRNA in S. cerevisiae are controlled by NRD. Following nuclear depletion of Nrd1, RNA-seq analysis of newly synthesized RNA showed termination defects for many ncRNA, with 55% of them arising from bidirectional promoters. Lack of NRD caused a twofold increase in divergent transcription. Furthermore, photoactivable ribonucleoside enhanced crosslinking (PAR-CLIP), a method used for analysis of protein–RNA binding, has revealed relatively higher NRD binding affinity for divergent ncRNA than for other Pol II transcripts, and has identified several RNA-binding motifs including those already known (Box 3). As with human PAS distribution [38,39], NBS are rarer in protein-coding sequences than in antisense ncRNA. However, the mechanism that antagonizes NRD termination in mRNA sequences, mimicking the mammalian PAS–U1 axis, has not yet been established. Another PAR-CLIP study has revealed that a quarter of protein-coding genes in yeast are bound by NRD [53], but only 302 genes show significant concentration of crosslinked NRD at the 5′ end, suggesting the location of functional terminators. Nevertheless, even a single NBS is sufficient to trigger transcription termination [54], therefore, many genes theoretically may be susceptible to the NRD termination pathway. However, given that only 44 mRNA coding genes have been shown to be prematurely terminated by NRD [40,52,55,56], a mechanism that negatively affects NRD-dependent termination over the protein-coding sequences may exist. Possibly, suppression of NRD termination relies on phosphorylation of Pol II C-terminal domain (CTD) Tyr-1, which may impair Nrd1–CTD interaction and so encourage elongation [57]. Other mechanisms that control promoter directionality at the transcription termination level in yeast have yet to be characterized.
In higher eukaryotes no homologs of the NRD complex have been identified. However, surprisingly, yeast NRD termination also requires CPAC factors. Although the functional links between CPAC and NRD are still under investigation, it appears that many CPAC components (e.g., Pcf11, Ssu72, Glc7) are crucial for NRD termination [58–64]. We speculate that in the course of evolution, NRD has been lost and entirely replaced with CPAC to terminate ncRNA in higher organisms. Alternatively, yeast might be considered to have evolved NRD to facilitate the recruitment of CPAC close to the transcription start site.
Regardless of the organism, ncRNA are enriched in termination signals. These act to trigger recruitment of termination factors resulting in premature transcription termination and subsequent transcript degradation, thus maintaining transcriptional directionality. Remarkably, termination factors are also essential for another mechanism that acts to enforce promoter directionality: gene loops.
Gene loops enhance promoter directionality
As described above, early transcription termination and subsequent RNA degradation act to remove unwanted divergent transcription. However, it is now apparent that a specific gene looping mechanism may further select productive gene transcription by encouraging Pol II to transcribe into the sense coding sequence. The fact that functional and physical interaction between initiation and termination factors leads to the juxtaposition of promoter and terminator regions has been known for a decade [65,66]. Thus ChIP analysis in S. cerevisiae of the general transcription factor TFIIB, which functions in promoter regions, also detected it over the 3′ ends of mRNA coding genes [67]. Conversely, some CPAC components crosslinked to the 5′ ends of genes [66,68,69]. Among them, were the Pol II CTD phosphatase Ssu72, which interacts genetically with TFIIB [70] and the whole CPAC subcomplex called cleavage factor IA (CF IA) [66,68]. Formation of gene loops was also independently confirmed by chromosome conformation capture analysis (3C) and shown to be dependent on active transcription [65,66].
Although gene loop formation is transcription dependent, mRNA synthesis is not strongly affected when the loop is disrupted. However, gene loops were shown to contribute to transcriptional memory in yeast. Thus, although transcriptional reactivation of inducible GAL10 and HXK1 genes was faster than the first round of activation, this effect was lost following gene loop disruption [71,72]. Physical proximity of the promoter and termination regions may facilitate reinitiation of RNA polymerase and therefore improve gene expression [66]. However, it has not been directly proven that the same Pol II molecule released at the 3′ end of the gene is immediately transferred to the promoter. Such evidence can only be obtained from single molecule experiments.
Recently, intragenic gene loops have also been shown to enhance transcriptional directionality from bidirectional promoters in S. cerevisiae [73]. Mutations of Ssu72 affects loop formation, which in turn results in Pol II relocation on the promoter towards the production of divergent ncRNA and their consequent accumulation. ssu72-2 mutant cells show the appearance of 605 novel ncRNA called Ssu72 restricted transcripts (SRTs); the majority of which are associated with bidirectional promoters. Mutation of the TFIIB component Sua7 and other CPAC factors involved in gene looping, Pta1, Rna14 and RNA15, also causes an increase in promoter-associated ncRNA. Furthermore, replacement of the PAS with an Rnt1 cleavage signal (RCS) results in a similar phenotype. RCS can act as an alternative terminator for Pol-II-transcribed genes [74,75]. However, the lack of PAS disrupts the association of CPAC and consequently prevents gene loop formation. This results in threefold higher transcription of the noncoding divergent region compared to the gene possessing a normal PAS. Overall, gene loop formation favors transcription in the direction that forms a looped structure. It must be mentioned that Ssu72 is also essential for the NRD pathway. By controlling both processes of early transcription termination and gene looping, this protein appears to be the determinant factor for promoter directionality. The CPAC subcomplex CF IA has consistently been shown to be involved in gene loop formation, where it promotes Pol II reinitiation and transcription of the coding region [69]. Intrinsic DNA sequences can also modulate transcriptional directionality through physical properties that facilitate looping. Genes where TFIIB was detected at the 3′ end are likely to form loops [76,77]. Interestingly, their gene sequences display higher flexibility in the middle of the open reading frame (ORF) than in nonlooped genes [77].
In higher organisms the process of gene looping has not been investigated in depth. However there are strong indications that the same mechanism exists in animals and plants. Human TFIIB recruits both CPSF and CstF to the promoter at an early transcriptional stage, before Pol II enters into the elongation phase [78]. TFIIB is also necessary for CstF recruitment to the terminator region and interacts directly with human Ssu72 and CstF-64 [68,78]. Similarly, in D. melanogaster TFIIB is required for termination of the polo gene. In this case, 3C analysis shows the existence of a TFIIB-dependent gene loop between promoter and terminator region [79]. Looping of the mammalian genes CD68 and BRCA1 as well as the plant gene FLC has also been reported [80–82]. Finally, a gene loop was detected across integrated HIV-1 provirus following transcriptional activation [83].
Gene loops provide an additional mechanism to enforce promoter directionality. In contrast to transcription termination, which aborts transcription of noncoding upstream regions, gene loops direct Pol II into the protein-coding sequences. However, their genome-wide prevalence is yet to be estimated.
Chromatin marks reinforce promoter directionality
On leaving the promoter and transcribing into the gene body, Pol II encounters nucleosomes; chromatin structural elements that consist of histones. Modified histones act as modulators of transcription, including regulation of transcription from bidirectional promoters. Divergent PICs on the promoters are in general compositionally equivalent [29]. Consequently, in human cells, histone H3 lysine 4 is trimethylated (H3K4me3) on both sides of the promoter and acts to stimulate PIC formation [7,84]. By contrast, the marker of active transcription, H3 lysine 79 dimethylation (H3K79me2) is found only over the coding region [7].
In yeast, the ssu72-2 mutation results in H4 acetylation in the upstream noncoding region of the bidirectional promoters [73]. This suggests that H4 deacetylation normally acts to reinforce transcriptional directionality established by Ssu72-dependent gene loops. Deacetylated histones are thought to form more compact chromatin than when acetylated. This restricts access to transcription factors and so affects transcription [85].
Why is divergent transcription needed?
Promoter bidirectionality as a general feature of eukaryotic genes could facilitate genome evolution by providing a pool of noncoding transcripts that may potentially cause a gain of function. It has been suggested that some long ncRNA (lncRNA) might be evolutionary precursors of mRNA-coding genes [38]. Indeed, the distribution of PAS and U1 sites over mRNA–PROMPT, mRNA–lncRNA, and head to head mRNA–mRNA genes, reveals that mRNA–lncRNA have an intermediate pattern. Consistent with this, evolutionary analysis of mouse genes has uncovered that the gain of U1 sites and loss of PAS at gene 5′ ends is correlated with the age of the gene [38]. This reinforces promoter directionality towards the coding sequence and promotes the stability and inheritance of the gene. The role of divergent transcription in genome evolution has been recently reviewed [86].
Bidirectional promoters offer an additional possibility to control gene expression. The negative impact on expression of protein-coding genes by competition for TFs between sense and antisense PICs [4] was mentioned above. However, divergent transcription can also positively affect gene expression. PICs formed in a noncoding direction may help to maintain NFRs and locally provide a pool of available TFs, which may then be used to enhance transcription of the coding region, if so needed. Negative supercoils generated behind actively transcribing Pol II may create tension that helps unwind the DNA duplex in promoters and so increases the efficiency of transcription initiation in either direction [87]. Finally, head-to-head protein-coding genes often encode proteins involved in the same biological process. This arrangement appears to enhance their co-regulation and gives an advantage in rapid response to different stimuli [88]. The best examples are genes encoding DNA repair enzymes, where bidirectional promoters, both expressing proteins respond to DNA damage, are five times more represented than unidirectional [1].
Concluding remarks
Transcriptional initiation de novo appears to be directionally unbiased. Pol II starts transcribing in both directions making either mRNA or ncRNA. Uncontrolled transcription of noncoding regions may lead to downregulation of divergent mRNA and potentially toxic accumulation of ncRNA. However conditions that facilitate transcription initiation of ncRNA also promote mRNA synthesis. Finally, divergent transcription provides the opportunity for the evolution of new genes. Therefore eukaryotes have retained mechanisms that allow for the existence of bidirectional ncRNA–mRNA promoters, by restricting the synthesis of unwanted ncRNA (Figure 2).
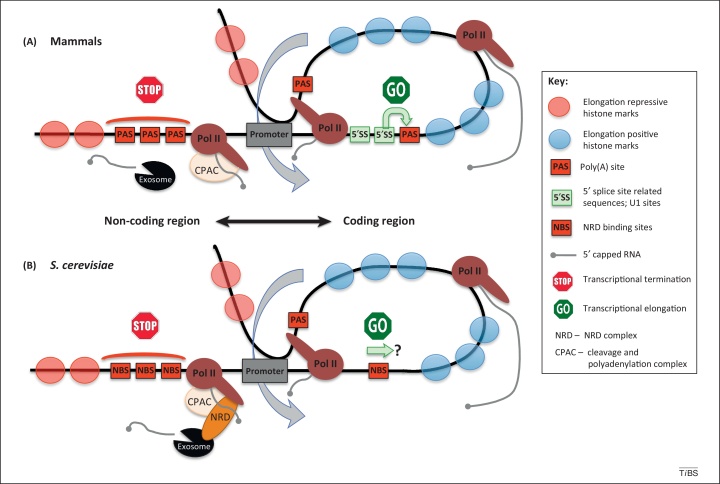
Figure 2.
Mechanisms enforcing promoter directionality. (A) In mammalian cells; (B) in Saccharomyces cerevisiae. Transcription of the upstream noncoding region is restricted by over-representation of termination signals (STOP sign, red rectangles), PAS in mammals or NBS in S. cerevisiae. NBS trigger the NRD termination pathway, which depends on both the NRD complex and CPAC. In contrast PAS-dependent termination is mediated by CPAC only. In both cases released ncRNA is degraded by the exosome. PAS and NBS are depleted over the coding regions. Moreover, in mammalian cells, the coding sequence is enriched in 5′ splice-site-related sequences recognized by U1 small nuclear RNA (snRNA), which inhibits PAS-dependent transcription termination (GO sign). A similar mechanism in yeast is yet to be discovered. Transcriptional directionality is also reinforced by chromatin modification. Nucleosomes with transcription-positive histone marks are presented as blue semitransparent circles, whereas nucleosomes with negative marks are shown in red. A pioneer round of transcription of the coding region establishes a gene loop. Termination factors recruited to Pol II transcribing in the vicinity of the terminator region interacts with initiation factors on the promoter and these juxtapose both regions. The formed loop enhances transcriptional reinitiation into the coding sequence (denoted by bent blue arrows). Abbreviations: CPAC, cleavage and polyadenylation complex; NBS, NRD-binding sites; ncRNA, noncoding RNA; NRD, NRD complex; PAS, poly(A) site; Pol II, RNA polymerase II.
The first line of defense against pervasive transcription is early transcription termination of ncRNA. This is mediated by PAS- and NRD-dependent pathways in mammals and yeast, respectively. In both cases termination signals are unequally positioned in each direction. Protein-coding sequences are depleted of PAS in human or NBS in yeast, whereas divergent noncoding regions are enriched in these sequences. Conversely, in mammals there is an opposite distribution of U1 sites that inhibit premature PAS recognition in protein-coding sequences. In addition, the first round of transcription of the coding sequence assists in establishing chromatin marks that promote efficient elongation. Finally, when transcribing Pol II approaches the 3′ end of the coding sequence, CPAC is recruited. This establishes interactions with TFIIB on the promoter and so creates a gene loop, which in turn strengthens transcriptional directionality. Taken together, bidirectional promoters are controlled at two stages. First, Pol II elongation directionality is regulated by transcriptional termination, and second, gene loops selectively enhance Pol II transcription on coding sequences.
Many questions concerning transcriptional directionality remain unanswered. How is it regulated in the case of head-to-head protein-coding genes when both divergent genes form loops? Can the promoter and PAS of ncRNA juxtapose? How widely distributed are gene loops in yeast and higher eukaryotes? Although the mammalian CoTC-type termination mechanism is as yet poorly understood, it will be interesting to investigate whether this type of PAS-dependent transcription termination is also involved in promoter-dependent antisense termination.
Transcriptional directionality can also potentially be regulated by RNA editing. Adenosine deaminase acting on RNA (ADAR) proteins can convert adenosine to inosine (reviewed in [89]). Inosine is then recognized as guanosine by the splicing and translation machineries. ADAR has been shown to introduce alternative 5′ (U1 site) and 3′ splicing sites [90,91]. Similarly, recently described R-loop-related editing of nascent transcripts frequently introduces U/A-to-G changes [92], which could also preferentially alter RNA sequence. It is therefore tempting to speculate that such RNA editing contributes to transcriptional directionality by creating U1 sites or by destroying PAS sequences, thus selectively enhancing mRNA synthesis.
Directing Pol II towards the coding region and restricting transcription of noncoding regions ensures proper gene expression. Moreover, bidirectional promoters offer an opportunity to create additional layers of either positive or negative regulation on protein-coding genes. Immediate transcription termination in the noncoding regions and subsequent degradation of unwanted RNA reduces the potential selective pressure to convert bidirectional into unidirectional promoters. This may in turn facilitate the origin of new genes and thus evolutionary progress.
Acknowledgments
The NJP laboratory is supported by a Wellcome Trust Programme Grant (091805/Z/10/Z) and a European Research Council Advanced Grant (339270, polyloop).
References
- 1.Trinklein N.D. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koyanagi K.O. Comparative genomics of bidirectional gene pairs and its implications for the evolution of a transcriptional regulation system. Gene. 2005;353:169–176. doi: 10.1016/j.gene.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q. Searching for bidirectional promoters in Arabidopsis thaliana. BMC Bioinform. 2009;10(Suppl. 1):S29. doi: 10.1186/1471-2105-10-S1-S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neil H. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 5.Xu Z. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Preker P. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 7.Seila A.C. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Core L.J. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu C. The preservation of bidirectional promoter architecture in eukaryotes: what is the driving force? BMC Syst. Biol. 2012;6(Suppl. 1):S21. doi: 10.1186/1752-0509-6-S1-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan G.C. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 11.Albert I. Translational and rotational settings of H2A.Z. nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 12.Mito Y. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 13.Ozsolak F. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- 14.Mavrich T.N. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guertin M.J., Lis J.T. Mechanisms by which transcription factors gain access to target sequence elements in chromatin. Curr. Opin. Genet. Dev. 2013;23:116–123. doi: 10.1016/j.gde.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekinger E.A. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Segal E. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan N. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valouev A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui F., Zhurkin V.B. Distinctive sequence patterns in metazoan and yeast nucleosomes: implications for linker histone binding to AT-rich and methylated DNA. Nucleic Acids Res. 2009;37:2818–2829. doi: 10.1093/nar/gkp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Floer M. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010;141:407–418. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai L. Multiple sequence-specific factors generate the nucleosome-depleted region on CLN2 promoter. Mol. Cell. 2011;42:465–476. doi: 10.1016/j.molcel.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozonov E.A., van Nimwegen E. Nucleosome free regions in yeast promoters result from competitive binding of transcription factors that interact with chromatin modifiers. PLoS Comput. Biol. 2013;9:e1003181. doi: 10.1371/journal.pcbi.1003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat. Struct. Mol. Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilchrist D.A. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papai G. New insights into the function of transcription factor TFIID from recent structural studies. Curr. Opin. Genet. Dev. 2011;21:219–224. doi: 10.1016/j.gde.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Core L.J. Defining the status of RNA polymerase at promoters. Cell Rep. 2012;2:1025–1035. doi: 10.1016/j.celrep.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhee H.S., Pugh B.F. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaplan N. Nucleosome sequence preferences influence in vivo nucleosome organization. Nat. Struct. Mol. Biol. 2010;17:918–920. doi: 10.1038/nsmb0810-918. author reply 920–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan X. Nucleosome depletion at yeast terminators is not intrinsic and can occur by a transcriptional mechanism linked to 3′-end formation. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17945–17950. doi: 10.1073/pnas.1012674107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim T.K. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Santa F. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sigova A.A. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen T.H. Dealing with pervasive transcription. Mol. Cell. 2013;52:473–484. doi: 10.1016/j.molcel.2013.10.032. [DOI] [PubMed] [Google Scholar]
- 36.Arigo J.T. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 37.Thiebaut M. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol. Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 38.Almada A.E. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–363. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ntini E. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat. Struct. Mol. Biol. 2013;20:923–928. doi: 10.1038/nsmb.2640. [DOI] [PubMed] [Google Scholar]
- 40.Schulz D. Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell. 2013;155:1075–1087. doi: 10.1016/j.cell.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 41.Richard P., Manley J.L. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mischo H.E., Proudfoot N.J. Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast. Biochim. Biophys. Acta. 2013;1829:174–185. doi: 10.1016/j.bbagrm.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Proudfoot N.J. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White E. AT-rich sequence elements promote nascent transcript cleavage leading to RNA polymerase II termination. Nucleic Acids Res. 2012;41:1797–1806. doi: 10.1093/nar/gks1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nojima T. Definition of RNA polymerase II CoTC terminator elements in the human genome. Cell Rep. 2013;3:1080–1092. doi: 10.1016/j.celrep.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaida D. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berg M.G. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Preker P. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011;39:7179–7193. doi: 10.1093/nar/gkr370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andersen P.R. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat. Struct. Mol. Biol. 2013;20:1367–1376. doi: 10.1038/nsmb.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vasiljeva L., Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol. Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 51.Marquardt S. Distinct RNA degradation pathways and 3′ extensions of yeast non-coding RNA species. Transcription. 2011;2:145–154. doi: 10.4161/trns.2.3.16298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Creamer T.J. Transcriptome-wide binding sites for components of the Saccharomyces cerevisiae non-poly(A) termination pathway: Nrd1, Nab3, and Sen1. PLoS Genet. 2011;7:e1002329. doi: 10.1371/journal.pgen.1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Webb S. PAR-CLIP data indicate that Nrd1-Nab3-dependent transcription termination regulates expression of hundreds of protein coding genes in yeast. Genome Biol. 2014;15:R8. doi: 10.1186/gb-2014-15-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim K.Y., Levin D.E. Mpk1 MAPK association with the Paf1 complex blocks Sen1-mediated premature transcription termination. Cell. 2011;144:745–756. doi: 10.1016/j.cell.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuehner J.N., Brow D.A. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol. Cell. 2008;31:201–211. doi: 10.1016/j.molcel.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 56.Thiebaut M. Futile cycle of transcription initiation and termination modulates the response to nucleotide shortage in S. cerevisiae. Mol. Cell. 2008;31:671–682. doi: 10.1016/j.molcel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 57.Mayer A. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336:1723–1725. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- 58.Al Husini N. A role for CF1A 3′ end processing complex in promoter-associated transcription. PLoS Genet. 2013;9:e1003722. doi: 10.1371/journal.pgen.1003722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim H. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat. Struct. Mol. Biol. 2010;17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim M. Distinct pathways for snoRNA and mRNA termination. Mol. Cell. 2006;24:723–734. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 61.Singh N. The Ess1 prolyl isomerase is required for transcription termination of small noncoding RNAs via the Nrd1 pathway. Mol. Cell. 2009;36:255–266. doi: 10.1016/j.molcel.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steinmetz E.J., Brow D.A. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol. Cell. Biol. 2003;23:6339–6349. doi: 10.1128/MCB.23.18.6339-6349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nedea E. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol. Cell. 2008;29:577–587. doi: 10.1016/j.molcel.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 64.Lenstra T.L. The role of Ctk1 kinase in termination of small non-coding RNAs. PLoS ONE. 2014;8:e80495. doi: 10.1371/journal.pone.0080495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O'Sullivan J.M. Gene loops juxtapose promoters and terminators in yeast. Nat. Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 66.Ansari A., Hampsey M. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh B.N., Hampsey M. A transcription-independent role for TFIIB in gene looping. Mol. Cell. 2007;27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 68.Calvo O., Manley J.L. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell. 2001;7:1013–1023. doi: 10.1016/s1097-2765(01)00236-2. [DOI] [PubMed] [Google Scholar]
- 69.Medler S. Evidence for a complex of transcription factor IIB with poly(A) polymerase and cleavage factor 1 subunits required for gene looping. J. Biol. Chem. 2011;286:33709–33718. doi: 10.1074/jbc.M110.193870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun Z.W., Hampsey M. Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol. Cell. Biol. 1996;16:1557–1566. doi: 10.1128/mcb.16.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laine J.P. A physiological role for gene loops in yeast. Genes Dev. 2009;23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan-Wong S.M. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tan-Wong S.M. Gene loops enhance transcriptional directionality. Science. 2012;338:671–675. doi: 10.1126/science.1224350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rondon A.G. Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol. Cell. 2009;36:88–98. doi: 10.1016/j.molcel.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ghazal G. Yeast RNase III triggers polyadenylation-independent transcription termination. Mol. Cell. 2009;36:99–109. doi: 10.1016/j.molcel.2009.07.029. [DOI] [PubMed] [Google Scholar]
- 76.Mavrich T.N. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dai Z. The pattern and evolution of looped gene bendability. Mol. Biol. Evol. 2014;31:319–329. doi: 10.1093/molbev/mst188. [DOI] [PubMed] [Google Scholar]
- 78.Wang Y. Phosphorylation of TFIIB links transcription initiation and termination. Curr. Biol. 2010;20:548–553. doi: 10.1016/j.cub.2010.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Henriques T. Transcription termination between polo and snap, two closely spaced tandem genes of D. melanogaster. Transcription. 2012;3:198–212. doi: 10.4161/trns.21967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tan-Wong S.M. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5160–5165. doi: 10.1073/pnas.0801048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O’Reilly D., Greaves D.R. Cell-type-specific expression of the human CD68 gene is associated with changes in Pol II phosphorylation and short-range intrachromosomal gene looping. Genomics. 2007;90:407–415. doi: 10.1016/j.ygeno.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 82.Crevillen P. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 2013;32:140–148. doi: 10.1038/emboj.2012.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Perkins K.J. Transcription-dependent gene looping of the HIV-1 provirus is dictated by recognition of pre-mRNA processing signals. Mol. Cell. 2008;29:56–68. doi: 10.1016/j.molcel.2007.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lauberth S.M. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152:1021–1036. doi: 10.1016/j.cell.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shahbazian M.D., Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 86.Wu X., Sharp P.A. Divergent transcription: a driving force for new gene origination? Cell. 2013;155:990–996. doi: 10.1016/j.cell.2013.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Parvin J.D., Sharp P.A. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 88.Li Y.Y. Systematic analysis of head-to-head gene organization: evolutionary conservation and potential biological relevance. PLoS Comput. Biol. 2006;2:e74. doi: 10.1371/journal.pcbi.0020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mallela A., Nishikura K. A-to-I editing of protein coding and noncoding RNAs. Crit. Rev. Biochem. Mol. Biol. 2012;47:493–501. doi: 10.3109/10409238.2012.714350. [DOI] [PubMed] [Google Scholar]
- 90.Rueter S.M. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 91.Solomon O. Global regulation of alternative splicing by adenosine deaminase acting on RNA (ADAR) RNA. 2013;19:591–604. doi: 10.1261/rna.038042.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang I.X. RNA-DNA differences are generated in human cells within seconds after RNA exits polymerase II. Cell Rep. 2014;6:906–915. doi: 10.1016/j.celrep.2014.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Valen E. Biogenic mechanisms and utilization of small RNAs derived from human protein-coding genes. Nat. Struct. Mol. Biol. 2011;18:1075–1082. doi: 10.1038/nsmb.2091. [DOI] [PubMed] [Google Scholar]
- 94.Kapranov P. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 95.Guttman M. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cabili M.N. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gudipati R.K. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol. Cell. 2012;48:409–421. doi: 10.1016/j.molcel.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van Dijk E.L. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 99.Heintzman N.D. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Levine M. Transcriptional enhancers in animal development and evolution. Curr. Biol. 2010;20:R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Orom U.A., Shiekhattar R. Long non-coding RNAs and enhancers. Curr. Opin. Genet. Dev. 2011;21:194–198. doi: 10.1016/j.gde.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Natoli G., Andrau J.C. Noncoding transcription at enhancers: general principles and functional models. Annu. Rev. Genet. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- 103.Koch F. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat. Struct. Mol. Biol. 2011;18:956–963. doi: 10.1038/nsmb.2085. [DOI] [PubMed] [Google Scholar]
- 104.Kowalczyk M.S. Intragenic enhancers act as alternative promoters. Mol. Cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 105.Chan S. Pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip Rev. RNA. 2011;2:321–335. doi: 10.1002/wrna.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chanfreau G. Essential yeast protein with unexpected similarity to subunits of mammalian cleavage and polyadenylation specificity factor (CPSF) Science. 1996;274:1511–1514. doi: 10.1126/science.274.5292.1511. [DOI] [PubMed] [Google Scholar]
- 107.Mandel C.R. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Skourti-Stathaki K. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.West S. Human 5′→3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 110.Kim M. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 111.Sadowski M. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 2003;22:2167–2177. doi: 10.1093/emboj/cdg200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meinhart A., Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- 113.Steinmetz E.J. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- 114.Vasiljeva L. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Carroll K.L. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol. Cell. Biol. 2004;24:6241–6252. doi: 10.1128/MCB.24.14.6241-6252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mayer A. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 117.Porrua O., Libri D. A bacterial-like mechanism for transcription termination by the Sen1p helicase in budding yeast. Nat. Struct. Mol. Biol. 2013;20:884–891. doi: 10.1038/nsmb.2592. [DOI] [PubMed] [Google Scholar]
- 118.Murray S.C. A pre-initiation complex at the 3′-end of genes drives antisense transcription independent of divergent sense transcription. Nucleic Acids Res. 2012;40:2432–2444. doi: 10.1093/nar/gkr1121. [DOI] [PMC free article] [PubMed] [Google Scholar]