Abstract
Cells have evolved rather sophisticated mechanisms to deal with stress positively and efficiently. Accumulation of reactive oxygen species (ROS), release of damage-associated molecular pattern molecule (DAMPs), and autophagy induction, are three inter-related processes occurring during most if not all cellular adaptations to stress. They influence each other reciprocally, initiating individual pathways, mediating and/or inducing effector mechanisms and modifying cellular function. High-mobility group box 1 (HMGB1), is a prototypic DAMP molecule, with various roles depending on its compartmental localization (nuclear, cytosolic, extracellular), well-defined but rather promiscuous binding partners, and the redox status within or without the cell. Typically, HMGB1 serves as a redox sensor, where redox modification also defines its translocation, release and activity, illustrative of the coordinate and multiply determined paths involved in the response to cell stress. Since DAMPs, redox and autophagy are essential and multifaceted in their roles in host defense, inflammation, and homeostasis, understanding how they interact and coordinate various signaling pathways to adjust to the stressful environment is important in the development of various potential therapeutic strategies, including application to patients with cancer.
Introduction
Danger clearly comes in threes [1]. Stressors induce a wide range of responses to varying environmental conditions or internal stimuli. In acute settings, following disruption of equilibrium, internal homeostasis is rapidly regained following coordinate interactions of the nervous, endocrine, and immune systems. In the setting of chronic disorders such as cancer, perpetuated responses to stress become fully engaged. Various evolutionary strategies, so-called ‘cellular adaptation’ processes, are used by cells to cope with diverse physiological or pathological stimuli [2]. The initial response to a stressor coordinately increases pathways associated with apoptosis and autophagy, and the subsequent response, either cell death or adaptation is dictated by other signals, allowing a new altered metastable state to be achieved. With persistence of the stressor, cells reorient in a variety of ways, often recruiting stroma that allows sequestration of the local process as well as recruiting inflammatory and immune cells to promote and enable recovery of organ function and the barrier function central to epithelial defenses.
Among numerous cellular strategies, autophagy is a highly conserved common catabolic process that facilitates cellular homeostasis following response to a wide spectrum of cellular stressors, including nutrient starvation, hypoxia, macromolecular or organelle damage, development of protein aggregates, radiation, chemotherapy and pathogenic infection [3]. Literally ‘self-eating’, in most cases, autophagy serves as a stress survival adaptation that prevents cell death, whereas under certain circumstances, it constitutes an alternative, albeit rare, route to cell death [2].
Cell fate critically depends on the signals to promote cellular and/or immune responses towards stressful stimuli. A series of cooperative signals of immune responses in succession comprise recruiting inflammatory cells (Signal 0), switching from innate to adaptive immune responses (Signals 1–4) via dendritic cells, and a subsequent integrated inflammatory response with resolution, recovery, and tissue regeneration at the stressed site (Signal 5’s) [4]. Deeper understanding of the underlying mechanisms of autophagy and its implications in pathological conditions have led to widespread acceptance of the notion that pathogen-associated molecular pattern molecules (PAMPs) and damage-associated molecular pattern molecules (DAMPs) function as ‘Signal 0s’ to promote autophagy and immunity via binding specific innate receptors [5]. Reciprocally, several studies suggest that autophagy itself can regulate release and degradation of DAMPs [6], which functions as an endogenous ‘danger’ signal that elicits inflammation/ immune responses once released from dead or stressed cells in the setting of sterile inflammation [7,8].
Redox (reduction-oxidation) reactions are fundamental chemical switches, by which biological energy is frequently stored and released, as perfectly exemplified in the classic processes of photosynthesis and cellular respiration. Subcellular compartments and the extracellular milieu have dramatically different redox potential, which partially determines the basic properties of reactions, the functions and/or interactions of proteins in a location-dependent manner, including DAMPs. In pathological conditions, oxidative stress, an imbalance of reactive oxygen species (ROS), serves as not only a cellular or environmental stressor but a common complication accompanying a wide range of diseases, including cancer, type II diabetes, chronic inflammatory processes, arteriosclerosis, ischemia/reperfusion injury, and neurodegenerative diseases [9]. Oxidative stress facilitates autophagy with redox sensitive DAMPs playing a modulatory role. Autophagy in turn regulates translocation/release of DAMPs in a redox-dependent manner and determines the final cellular destiny, either confining or releasing redox components, such as oxidoreductases and nonprotein thiols, thereby modifying the extracellular redox state. Also, DAMP release or secretion is associated with a subsequent series of inflammatory responses and reactions, which, in turn, can induce production of reactive oxidative intermediates.
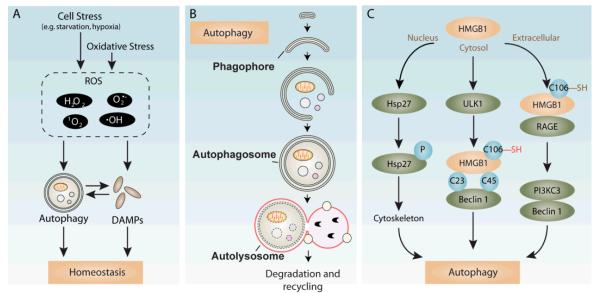
Here, we discuss the three elemental relationships linking DAMPs, redox and autophagy in response to stressful stimuli (Figure 1) with specific emphasis on one of the major and best-studied DAMP molecules, high mobility group box 1 (HMGB1) protein, along with its location and redox status in the modulation of autophagy. We thendiscuss these three elements in the setting of cancer.
Figure 1. The three elemental relationships linking autophagy, DAMPs (HMGB1) and redox.
Oxidative stress or ROS production following other cellular stress facilitates autophagy and modifies HMGB1 release and its activity. Autophagy regulates translocation/release of DAMPs in a redox-dependent manner. HMGB1 in turn promotes autophagy in individual compartments (nuclear, cytosolic, extracellular). Autophagy also confines reduced constituents intracellularly, such as nonprotein thiols, by sequestering damaged organelles and/or macromolecules. Additionally, DAMPs released or secreted in their reduced state are associated with a subsequent series of inflammatory responses, which, in turn, modulates the production of ROS.
HMGB1 Biology
As our understanding of the multifaceted role of DAMPs evolves, so too does the number of nominal DAMP family members. The majority of DAMPs are nuclear and cytosolic proteins, including high mobility group box 1 (HMGB1), constituents of exosomes, heat shock proteins (HSPs)], the S100 family of calcium-binding proteins, histone, interleukin 1(IL1) family members and plasma components such as complement (C3a, C4a, and C5a). Non-protein counterparts consist of nucleotides (adenosine triphosphate (ATP), DNA, RNA), their metabolites such as uric acid, and extracellular substances including hyaluronan and heparin sulfate [5]. Mitochondria, central in bioenergetics and, have recently emerged t as the key source of DAMPs including mitochondrial DNA, formylated peptides, and transcription factor A of mitochondria (TFAM) [10].
Although various DAMPs have been identified, HMGB1 remains the best characterized and prototypic DAMP [11]. HMGB1, a 25-kDa protein, with two DNA-binding domains, 80 amino acids homologous regions, termed the A and B boxes, and a C-terminal domain consisting largely of aspartic and glutamic acid residues. There are two nuclear localization signals (NLS) in the proximal part of the A and B boxes, respectively, important for transport via binding to nuclear exportin CRM1. Three cysteines are encoded at positions 23, 45 and 106, making HMGB1 a redox sensitive molecule [12]. Its translocation, release and activity are tightly controlled by redox modification.
HMGB1 is an evolutionarily ancient protein with multiple functions (summarized in Table 1) ranging from its central role as a chromatin-associated protein first identified more than three decades ago [13] to an autophagy promoter and to a cytokine or a DAMP. Which function prevails chiefly depends on its redox status, subcellular location and which of its synergizing partners is available. The remarkable importance of HMGB1 for ex utero growth is illustrated by the lethality of HMGB1 global knockout in mice due to deficient glucocorticoid receptor function [14] and potentially impaired autophagy. Additionally, during early brain development, a complex temporal and spatial distribution pattern of HMGB1 has been found in the central nervous system where it enhances neurite outgrowth and cell migration, essential in brain developmental programs [15]. HMGB1 is constitutively expressed in nuclei of almost all cell types and presents primarily inside nuclei in normal resting cells with some exceptions including eosinophils, whose HMGB1 resides mostly in the cytoplasm [16]. Normally, within the nucleus, HMGB1 serves as a DNA chaperone that maintains nucleosome stability, and promotes access to replication and transcription with specific gene targets [17,18]. HMGB1 binding to chromatin itself is not sequence specific although it has heightened binding to CpG motifs. In response to stress, HMGB1 shoulders the responsibility to protect DNA from oxidative damage and regulate transcription of autophagy-related targets, including HSPB1/ HSP27[12,19]. Work performed by our group shows that once translocated into the cytosol, HMGB1 occupies a regulatory role in promoting and sustaining autophagy [20–22], a conserved programmed survival pathway utilized following environmental and intracellular stresses. When released into the extracellular milieu, HMGB1 promotes a host inflammatory response to sterile (DAMP) and infectious (PAMP) signals [23], acting as an irreplaceable coordinator/integrator of immunity and the ‘universal’ biosensor for nucleic acids [24,25].
Table 1.
Compartment Specific Functions of HMGB1.
| NUCLEAR | DNA chaperone |
| Modulation of transcriptional activity of various genes [11,117] | |
| Participation of DNA repair pathways [47] | |
| Maintenance of telomere and chromosomal stability [50,51] | |
| Transcription-dependent regulation of autophagy and apoptosis [19] | |
|
| |
| CYTOSOLIC | Redox sensor [12] |
| Transcription-independent regulation of autophagy [20] | |
| Mitochondrial quality control [19] | |
| Universal biosensor of nucleic acid [103] | |
| Recruitment of MyD88 to TLR9 [119] | |
| Vesicular exocytosis for release [95] | |
|
| |
| EXTRACELLULAR | DAMP signaling [8] |
| All thiol form: chemotactic activity [106] | |
| Dithiol form: TLR4/MD-2 interaction ,cytokine role [106,107] | |
| Oxidized form: immunological tolerance, apoptosis induction[107] | |
| Synergizition with other cytokines to modulate cell functions [103] | |
| Increase of IFN-γ secretion in macrophage-stimulated NK cells [120] | |
| Regulation of autophagy via RAGE [110] | |
|
| |
| CELL SURFACE | Promotion of axonal sprouting and neurite outgrowth [121] |
| Platelet activation [122] | |
HMGB1 has been implicated in various pathological states, where it can be passively released from necrotic cells and/or actively secreted by inflammatory cells, including monocytes/macrophages, dendritic cells, neutrophils, eosinophils, natural killer cells and platelets, binding with high affinity to several receptors including the receptor for advanced glycation end products (RAGE), Toll-like receptors (TLR)-2, TLR-4, TLR-9, chemokine CXC receptor (CXCR) 4 and negative signaling molecules, CD24CD24-Siglec G/10 and TIM-3,. Overwhelming evidence indicates that neutralizing or eliminating HMGB1 significantly limits activation of immune responses and tissue damage [8,23,26]. HMGB1 is tightly linked to the key hallmarks of cancer including proliferative immortality, insensitivity of growth suppressors, evasion of apoptosis, angiogenesis, invasion and metastasis [26]. Elevated serum HMGB1 has been detected in cancer patients and mice bearing tumors.
Compartment One: The Nucleus
HMGB1 was initially identified more than three decades ago as a chromatin-associated protein with two unique DNA-binding domains, termed the A and B box [27]. In terms of its binding affinity, DNA mini-circles, four-way junctions, looped structures, hemi-catenated and triplex DNA are preferential targets, in addition to supercoiled, single-stranded, B- and Z-DNA [12]. As one of the major non-histone proteins, HMGB1 acts as a DNA chaperone, mediating multiple important processes within chromatin including replication, transcription, recombination, chromatin remodeling, integration of transposons, V(D)J recombination, DNA repair and genomic stability. This is largely due to its ability to bend/distort DNA and to bind preferentially to distorted DNA structures [26,28]. The acidic C-terminus tail plays a modulatory role in its DNA-binding properties and subsequent biological processes. Apart from HMGB1 binding, other proteins also play a crucial role in promoting nucleoprotein complex formation. These amicable partners of HMGB1 include plentiful proteins, such as other transcription factors (TFs), DNA repair proteins, silencing complexes (transcriptional activators/repressors, co-repressors), and nuclear importing proteins and histones [28].
HMGs and Transcription
High Mobility Group (HMG) family members possess a common transcription-friendly activity, but they also modulate the expression of specific genes. HMGB1 facilitates binding and thus enhances the activities of numerous transcription factors, in particular members of the nuclear hormone-receptor family [29–31], p53/p73 transcriptional complexes [32–34], members of the Rel/NF-κB family [35] and octamer transcription factors [36]. HMGB1 global knockout is associated with lethal hypoglycemia, runting, and early mortality after birth [14] in outbred animals and late prenatal when backcrossed onto inbred strains. As such, HMGB1 can modulate immune/ endocrine responses following stress and mediate survival or cell death in a transcription-dependent fashion.
It is worth noting that HMGA’s, members of the unrelated but eponymous high mobility family of proteins, are key architectural transcription factors mediating enhanceosome formation. The best-studied enhanceosome is located on the interferon β (IFN- β) promoter [37]. Furthermore, several genes are regulated by HMGA1, validated by microarray analysis in embryonic stem (ES) cells bearing one or two disrupted hmga1 alleles [38]. This was further studied in other cell types, including those encoding the interferon γ, interleukin-2 receptor α, E-selectin and insulin receptor [39–42]. Given that HMGs appear to compete with each other [43,44], HMGB1 may also interact with other binding proteins at these sites, competing to fluidize and open the chromatin.
HMGB1, DNA Damage Repair and Chromatin Stability
DNA is always subject to damage induced by plentiful chemicals and environmental agents, requiring a constant and dynamic process of repair. Repair of DNA damage is indispensable for sustaining genomic integrity and cell survival, as confirmed by permanent cell-cycle arrest or cell death with irreparable DNA lesions [45].
HMGB1 binds to platinated-DNA as revealed by precipitation technique, inaugurating an era in which HMGB1 involvement in DNA repair was first revealed. Cells depleted of HMGB1 are sensitive to DNA damage with increased mutagenesis and diminished survival [46]. HMGB1 participates in both single-strand DNA damage repair (nucleotide excision repair (NER), mismatch repair (MMR) and base excision repair (BER)), as well as DNA double-strand break repair (DSBR) by targeting individual regulatory proteins [47]. When binding to DNA lesions, HMGB1 either blocks access of repair machinery (repair shielding) or facilitates the recognition and processing of DNA repair (repair enhancing).
Another mechanism for maintaining chromosomal stability is by telomere maintenance, the specific nucleoprotein structures located at chromosome ends protecting them from recombination and degradation [48]. Telomeres shorten with every cell division as a result of completing DNA replication using the terminal telomere to prime the DNA polymerase, and tend to be uncapped with loss of this protective function. This can be replenished by telomerase, a nucleoprotein complex with reverse-transcriptase activity. The presence of telomerase enables continued cell division, thus playing a significant role in immortalization and tumorigenesis, as supported by the fact that up-regulation or reactivation of telomerase occurs in almost 90% of all human cancers [49]. Ablation of HMGB1 results in hyper sensitivity to UV-irradiated damage, numerous chromosomal aberrations, shortening of telomere lengths and lower telomerase activity, as demonstrated in murine embryonic fibroblasts (MEFs) [50,51]. The underlying mechanism by which HMGB1 modulates telomerase activity has yet to be deciphered. In vitro pull-down and TRAP experiments suggest that HMGB1 binds to human TERT (telomerase reverse transcriptase, a core constituents in telomerase), which are not detectable by western blotting [50]. Two groups recently provided evidence of subtle genetic changes in the promoter region lying upstream of the gene encoding the telomerase subunit TERT, serving as driver mutations in melanoma tumorigenesis, and possibly in other types of cancer [52,53]. Given that overexpression of HMGB1 is found in most cancers and autophagy predominates as a survival advantage during the late stages of tumor development, another possible explanation for HMGB1-driven telomerase activation in addition to physical binding and activation, would be that HMGB1 or other autophagy-active factors could limit mutagenesis of the TERT promoter, thereby promoting cancer development.
Nuclear HMGB1 regulates autophagy/apoptosis
There is an intimate and sophisticated relationship between apoptosis and autophagy [54,55], with both processes increasing during periods of cellular stress. Through mutual inhibition, the predominant one usually determines the cell fate and tumor progression differentially with pronounced apoptosis early in the development of cancer under stress and pronounced obligate unbalanced autophagy late in tumor development.
Our group proposes that HMGB1 can in part promote autophagy in a transcription-dependent fashion [19] (Figure 2C). HMGB1 facilitates access to transcription of the heat shock protein beta-1 (HSPB1/Hsp27), which promotes autophagy by modulating the actin cytoskeletal response following mitochondrial injury [56,57], further regulating mitochondrial dynamics and quality. Hmgb1−/− cells have reduced level of HSPB1 expression, with mitochondrial dysfunction associated with fragmented morphology, respiratory deficits and loss of membrane potential, which are all rescued by insertion of Hsp27 cDNA [19]. Additionally, Hsp27 inhibits apoptosis via physical binding and inhibition of key components of the apoptotic signaling pathway including caspase-3 and cytochrome-c [58] or of p53-mediated induction of p21 [59]. With anti-apoptotic and cyto-protective properties against heat shock, oxidative stress and ischemia, Hsp27 is a potential target for inactivation and cell death induction [58]. HMGB1 is also a unique activator of p53 [33,34], a common tumor suppressor that controls cell cycle progression and apoptosis. Stress signals such as starvation and DNA damage trigger interactions between HMGB1 and p53 within the nucleus and cytoplasm, as suggested by our studies [60]. The ultimate cell destiny is largely due to their relative amount. Loss of p53 allows cytosolic HMGB1 accumulation, followed by increased binding to BECN1 of HMGB1, thereafter prompting autophagy and decreasing apoptosis, as further discussed below. In contrast, depletion of HMGB1 increases cytosolic p53 and apoptosis, and autophagy is thus inhibited [61].
Figure 2. Cellular response to stress and HMGB1-mediated autophagy pathways. A) Autophagy and DAMP release in response to stress.
ROS (reactive oxygen species) acts either as a prime cause (oxidative stress) for damage/death or as the secondary signals to recruit and activate inflammatory cells. When faced with such stress, by inducing autophagy and releasing DAMPs, cells undergo adaptation, thus maintaining homeostasis and promoting survival. B) Essential mechanisms of autophagy. During autophagy, intact organelles and cytosolic components are sequestered into a emergent double-membrane vesicle, named the phagophore. Subsequently, the completed autophagosome matures through fusion with endosomes and/or lysosomes, thereby forming an autolysosome for further degrading and recycling. C) Multiple roles of HMGB1 in autophagy induction. Nuclear HMGB1 modulates the expression of heat shock protein beta-1 (HSPB1/Hsp27). Phosphorylated Hsp27/HSPB1 modulates the actin cytoskeleton, followed by mitophagy and/or autophagy promotion. Cytoplasmic HMGB1 with C23-C45 binding and displacing Beclin-1 from Bcl2/Bclxl, further sustaining autophagosome formation . Extracellular HMGB1 also triggers autophagy via interaction with the receptor glycation end product (RAGE), which initiates Beclin 1- PtdIns3KC3 complex formation.
Compartment Two: The Cytosol
The cytosol is highly reducing thanks to a group of thiol-regulating enzymatic systems, including the thioredoxin reductase and glutaredoxin systems [7] as well as the presence of abundant protein thiols, consisting ofHMGB1. Nonprotein thiols, such as amino acid cysteine and the small cysteine-containing tripeptide glutathione, which assist thiol-regulating enzymes, are mainly reduced within the cytosol and predominate in the disulfide form extracellularly. Stress can trigger the production of reactive oxygen species (ROS), mostly from mitochondria but also generated by NADPH oxidases. Thus, ROS serve as molecular switches within the cell and their balance with the extracellular environment changes and sets the stage and determines individual cell fates, including autophagy, apoptosis, and necrosis [12] (Figure 3). Autophagy is a conserved mechanism of cell survival under mild stress. Cells undergo complex and highly regulated events, orchestrated by cytosolic ROS levels and redox-sensitive HMGB1. Once translocated within the cytosol, HMGB1, with its disulfide link, plays a major role in autophagy induction [20]. Mitochondria- generated ROS is also involved in the release of cytochrome c, followed by caspase activation and apoptosis [62].
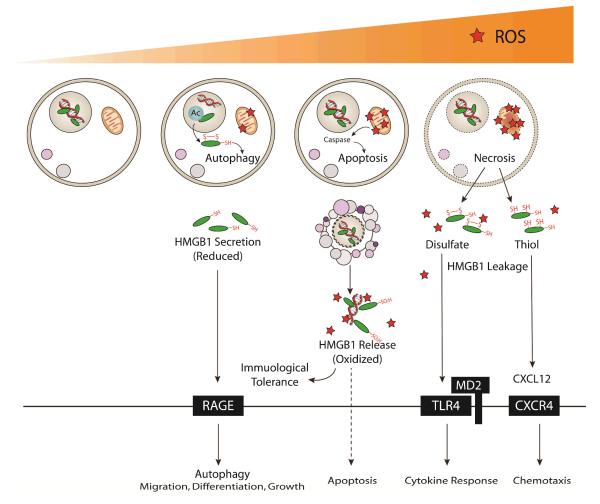
Figure 3. Pathways of HMGB1 secretion/release and activities in response to different degree of oxidative stress.
Under physiological conditions, HMGB1 is predominantly within the nucleus functioning as a DNA guardian and transcription regulator. When low oxidative stress occurs, autophagy and HMGB1 (DAMP) release reciprocally assist cell adaptation. Acetylated HMGB1 can be translocated out of the nucleus, followed by autophagy induction. Autophagy, in turn, is required for HMGB1 secretion via vesicular exocytosis. Reduced HMGB1 in the extracellular milieu could bind to the receptor for advanced glycation end products (RAGE) to further induce autophagy in surrounding cells or modulate cell migration, growth and protein expression. As oxidative damage increases, molecules such as cytochrome c could be released from redox-sensitive mitochondria, which further activates the caspase cascade and triggers apoptosis. In the early stage of apoptosis, HMGB1 bound with DNA and nucleosome, is confined within two-membrane vesicles, while in the late stage, oxidized HMGB1 is released, acting as a potentially immunological inactive molecule and possibly apoptosis inducer. At the extreme, oxidative stress causes severe mitochondrial permeability transition (MPT) or even the rupture of the mitochondrial membrane, promoting collapse. HMGB1 is leaked out of the cells, basically in two forms: 1) Disulfate HMGB1 contributes remarkably to cytokine production via TLR4/MD2 interaction while 2) All thiol HMGB1 leads to chemotaxis through induction of release and subsequent binding to CXCL12 and CXCR4.
ROS and Oxidative Homeostasis
Reactive oxygen species (ROS), free radicals containing an oxygen atom, are highly reactive and toxic resulting from the presence of unpaired valence shell electrons. ROS collectively comprise superoxide anions (O2−), hydroxyl radicals (−OH), hydrogen peroxide (H2O2), superoxide radicals (O2−), singlet oxygen (1O2) and ozone (O3). As a metabolic byproduct of electron transfer to oxygen, ROS exert important physiological effects on signal transduction and eukaryotic homeostatic maintenance [63]. Excess ROS remarkably impair mitochondrial function and influence the state of health of the organelle. Depending on the physiologic insult, cells can either clear protein aggregates/debris and repair the damage via autophagy or activate pathways triggering cellular suicide/apoptosis [62]. With profound stress, the mitochondrial permeability transition (MPT) occurs, resulting in necrosis due to ATP depletion (Figure 3).
The principle cellular source of ROS is generated by mitochondrial respiration processes and nicotinamide adenine dinucleotide phosphate reduced (NADPH) oxidases. During oxidative phosphorylation, O2 serves as the final electron acceptor of the respiratory chain, converted to H2O eventually with four-election reduction, catalyzed by cytochrome-c oxidase, a key component of the mitochondrial enzymatic complex. Incomplete reduction results in ROS generation as an inevitable byproduct of the election transfer reaction [64]. Actually, this seemingly “accidental” production of free radicals in the mitochondria accounts for 1–2% of total oxygen consumption. NADPH oxidases (NOX’s) are membrane-bound enzyme complexes that catalyze superoxide generation from oxygen, using NADPH as electron donor. Nox’s can be activated by insulin, platelet-derived growth factor, fibroblast growth factor, GM-CSF and tumor necrosis factor (TNF). This mechanism is primarily utilized by phagocytic leukocytes to clear pathogens [65]. Other less abundant sources of ROS come from the endoplasmic reticulum (flavoenzyme Ero1) and peroxisome (peroxidase) [64].
To achieve intracellular homeostasis against the potentially damaging effects of ROS in excess, cells possess a broad detoxification system consisting of several anti-oxidant enzymatic scavengers such as superoxide dismutase (SOD), catalase, glutathione peroxidase (Gpx), peroxiredoxin (Prx) and others. Mitochondrial SOD2 (Mn-SOD) is responsible for converting O2− to H2O2, which can diffuse out of the mitochondria into the cytoplasm to be further metabolized by cytosolic Gpx1 or decomposed into molecular oxygen and water by catalase and peroxiredoxin (Prx1), released from peroxisomes of nearly all aerobic cells [62,66]. Cytosolic CuZn-SOD (SOD1) catalyzes the same reaction within the cytoplasm. Another key component in the system is tripeptide glutathione, GSH (L-γ -glutamyl-L-cysteinylglycine), a substrate or cofactor for multiple GSH-linked antioxidant enzymes due to its active thiol (a cysteine residue). GSH can be converted to GSSG directly after it is oxidized by ROS or indirectly during glutathione peroxidase (GPX)-catalyzed reactions. In addition to antioxidant defense systems, GSH also has a complicated pattern of involvement in diverse biological processes, including many experimental and clinical interventions [67]. In particular, catalyzed by glutaredoxin (Grx), one of the thiol–disulfide oxidoreductases, GSH participates in thiol–disulfide exchange termed as S-glutathionylation, a process which is essential in disulfide bond breakage of proteins, maintaining the essential thiol status and further determining their location and activity [68]. A large number of potential substrates are structural proteins, molecular chaperones, and members of mitochondrial respiration complexes. Among them, HMGB1, containing three cysteines, is an abundant component susceptible to oxidation, reversible by Grx [68].
The prominent targets of ROS are exceptionally diverse. Many cysteine-dependent serine/threonine phosphatases (such as MAPK, ERK and JNK) are subject to reversible oxidative inactivation, as are caspases, with suppression of apoptosis and limiting production of IL-1 family members [69,70]. Transcription factors and nuclear/mitochondrial DNA are also susceptible to regulation by ROS. Thus, ROS, together with reactive nitrogen species (RNS), play diverse roles in host defense, inflammation, and homeostasis.
Autophagy and Oxidative Stress
Autophagy is the cyclical process by which cells engulf and break down damaged or unnecessary components following fusion with the lysosome and repurpose the constituents for new biosynthesis/anabolism (Figure 2B). It occurs continuously under homeostatic conditions to prevent toxic accumulation of protein aggregates and disordered organelles and recycle constituents to sustain metabolic homoeostasis as a method of quality control [5].
Autophagy is, however, more than just a homeostasis guardian, but also a key responder to cellular stress (Figure 2A), including starvation, hypoxia, infection and oxidative stress [71]. For example, under conditions of starvation, autophagy recycles biologic substrates and generates ATP, coping with externally limited nutrients. Cells initiate programmed survival or autophagy to survive during moderate stress, and can promote cell death, under rare conditions [72]. A hitherto unappreciated role of autophagy in promoting unconventional vesicular biogenesis and exocytosis has been uncovered recently, expanding its functional role beyond autodigestion and quality control roles in mammals [73], suggesting a broader role in tissue homeostasis.
The term autophagy encompasses three major intracellular pathways in all eukaryotic cells, macroautophagy (hereafter referred to as autophagy), microautophagy, and chaperone- mediated autophagy (CMA), with a shared destiny of lysosomal degradation but varied mechanisms employed for delivery of contents to the lysosome. During the process of macroautophagy (Figure 2B), intact organelles (such as mitochondria-mitophagy and peroxisomes-pexophagy) and portions of the cytosol are sequestered into a double-membrane vesicle, the autophagosome. Subsequently, the completed autophagosome matures through fusion with endosomes and/or lysosome, thereby forming an autolysosome. The entire process is coordinated by a series of linked proteins including the E3 ubiquitin-like ligase Atg12-5-16L1 complex, termed autophagic-related (Atg) proteins [74]. After breakdown within the autolysosome, the decomposed products can either be transported back into the cytosol for reuse or prematurely undergo autophagic vesicular exocytosis [75] or to MHC class II loading compartments for antigen presentation within dendritic cells [76]. In contrast, microautophagy involves direct delivery of largely denatured components of the cytoplasm to the lysosome surface. This is mediated following starvation by formation of autophagic tubes, which promote within the lysosomal lumen, invagination and vesicle scission, enclosing part of the cytosol. Selective microautophagy includes micropexophagy, piecemeal nuclear microautophagy, and micromitophagy. CMA, on the other hand, transfers unfolded, individual soluble proteins directly across the lysosomal membrane.
Oxidative stress (also termed ROS chaos), as a consequence, reflects an imbalance between the production of reactive oxygen species (ROS) and the utilization/detoxification of excessive reactive intermediates [77]. Oxidative stress and the following ROS pathways can be stimulated by various types of stress(starvation, hypoxia, subcellular damage, protein aggregates, radiation, chemotherapy and pathogen infection) [78] and correlate with a wide variety of pathological conditions, including neurodegenerative diseases, diabetes, cancer and ageing [79]. However, whether ROS is the accompanying effect for alarm or the prime cause for damage/death is an area of debate [80]. Generally, multiple stimuli, inducing or followed by ROS production, can trigger autophagy (Figure 2A). In this regard, ROS, function as either stressors or intermediate signaling stimuli [20,81,82]. Specifically, mitochondria, prone to oxidative stress, are one of the two principle sites where ROS are generated and involved in the processes of autophagy and mitophagy (selective autophagy of mitochondria) [83,84], used as a means of mitochondrial quality [85]. Additionally, mild oxidative stress may bring about protein degradation as a means of CMA [12]. ROS trigger autophagy through a couple of distinct mechanisms, including autophagy-related (Atg) gene 4 (Atg4) activation (triggering Atg8/LC3 activation), catalase production, and disturbances in the mitochondrial electron transport chain. Knockdown of SOD1 in fibroblasts is associated with increased autophagic flux, consistent with results from cells treated with exogenous hydrogen peroxide(H2O2) [86]. Interestingly, in the program of so-called autophagic cell death, selective autophagic degradation of catalase results from caspase inhibition, leading to abnormal ROS accumulation and subsequently triggering this unusual mechanism of cell death. Collectively, ROS occupy a central role in inducing or orchestrating autophagy.
Conversely, deficient autophagy preferentially accumulates ROS, accompanied by retention of damaged mitochondria, ER stress, and accumulation of genomic damage in tumor cells [87,88], associated with apparent defects in management of protein turnover. Thus accumulated “garbage”, as a potential source of additional stressors, may contribute to cell injury and death. Therefore, previous work highlighting the pivotal role of autophagy (mitophagy) in the context of homeostatic ROS responses and/or elimination, likely understated the importance of the cascade of events underlying adaptation or apoptosis.
Redox Buffering of HMGB1 and Autophagy
Cysteines (Cys) are modified by diverse redox signals with oxidation of thiol side-chains (-SH) to several reversible redox states such as disulphide (R-S-S-R); sulphenic acid (R-SOH); and sulfonate (R-SO3H) moieties, which in turn regulate the secondary structure of proteins. Within proteins, cysteines are unequally accessible and reactive depending on their neighboring residue and protein structure. For example, in the context of mild oxidative stress, HMGB1 can form a Cys23-Cys45 intermolecular disulfide bond, while the Cys106 remains in the reduced form. During apoptosis, Cys106 is further oxidized by ROS arising from mitochondria, thus limiting its immunological stimulatory properties [89] (Figure 3).
Following stress, such as oxidative stress, starvation and chemotherapy, HMGB1 translocates from the nucleus to the cytoplasm. Although acetylation (weakening affinity binding of chromatin) and expression of CMR1/exportin (exporting HMGB1 from the nucleus) have been considered to be critical in nucleocytoplamsic shuttling of HMGB1 [28], Cys redox regulation also plays a modulatory role, as shown by serine replacement of Cys106 (nonoxidizable), but not Cys23 (C23S) and/or Cys45(C45S) altering its nuclear distribution, allowing HMGB1 entry into the cytosol [68]. Additionally, elevated endogenous and exogenous ROS supports HMGB1 translocation and release [86,90].
Present in the cytoplasm, redox-sensitive HMGB1 can also modify autophagy (Figure 2C), in addition to its transcription-dependent modulation of autophagy/mitophagy involving Hsp27/HSPB1, as discussed above. Cytosolic HMGB1 binds Beclin-1, further sustaining autophagosome formation during autophagy initiation [20,86]. The C23-C45 intramolecular disulfide bond is required for this binding and sustaining of autophagy. C106S mutation not only favors cytoplasmic translocation, but facilitates autophagy. Thus, HMGB1, regulated by post-translational redox modifications, plays an important role in overall sensing stress, ROS, and promoting autophagy.
Compartment Three: Extracellular
In healthy cells, the cytosol itself is highly reduced, while the extracellular environment is substantially oxidizing, with the exception of the Redox gradient within the endoplasmic reticulum which is increasingly oxidized to allow folding and export of membrane and secreted proteins. Following damage or injury, however, oxidoreductases and nonprotein thiols, usually confined within cells, can be released into tissues, thereby promoting an extracellular local reduction.
Endogenous HMGB1 can be passively released or actively secreted into the environment, serving as a ’Damage Associated Molecular Pattern molecule, DAMP’ to alert adjacent cells that damage has occurred [23]. As a leaderless protein, HMGB1 undergoes unconventional lysosome-mediated exocytosis, which is facilitated by autophagy (Figure 3). Following stress, depending on stress strength and cellular status, HMGB1 can be released in various redox forms, reflecting the strength of inflammation, loss of the normal balance between the intracellular/reduced and extracellular/oxidized formats, induction of autophagy, and promoting the programmed cell survival or death of neighboring cells. Further modulated by environmental conditions, individual redox states of HMGB1 exert widely divergent activities, either promoting recruitment or activation of immune cells.
Outside of the cell, HMGB1 can either bind to specific receptors itself, or with high affinity to DNA, nucleosomes, IL-1β, CXCL12, sRAGE, lipopolysaccharide, and lipoteichoic acid to mediate responses in specific physiological or pathological conditions. Currently identified receptors include TLR2, TLR4, TLR9, the receptor for advanced glycation end products (RAGE), CD24-Siglec G/10, chemokine CXC receptor 4, TIM-3 and others [8,24].
Autophagy and/or ROS-mediated HMGB1 Release
HMGB1 was first identified as a late mediator of inflammation in response to endotoxin, liberated from macrophages, following earlier responses with transient IL-1β and TNF-α release [91]. Delivery of HMGB1 into the extracellular milieu occurs in two principle ways: either passively released from necrotic cells and/or actively secreted by inflammatory cells (monocytes/macrophages, dendritic cells, natural killer cells, neutrophils, eosinophils and platelets), therefore evoking immune response or spurring autophagy as so-called ‘Signal 0’s’ following stress (infection, injury or inflammation) [5].
Active secretion is a broadly used mechanism for cells to initiate interactions with the environment. Classical protein export route in eukaryotic cells is through the endoplasmic reticulum (ER)/Golgi apparatus [92]. Unlike those proteins typically containing N-terminal signal peptides, most DAMPs, however, are devoid of a leader signal sequence, and follow non-classical transport pathways including release from secretory lysosomes, endosome, autophagosome, membrane blebbing, multivesicular body-derived exosome and ABC transport [93,94]. Among them, HMGB1,IL-1β, and ATP undergo secretory lysosome-mediated exocytosis [95], termed as vesicular exocytosis when stress exists. Several factors contribute to the process, including autophagy, post-translational modification (acetylation, phosphorylation, methylation, ADP ribosylation and oxidation) and signal transduction pathways [6]. Autophagy is a conserved pathway responsible for delivery of cytoplasmic material into the lysosomal degradation pathway, thus potentially allowing vesicular exocytosis. Our group and others have reported that autophagy facilitates exit from cells of HMGB1 in fibroblasts, macrophages and cancer cells following starvation, cytotoxic drugs and PAMPs [20,73,96]. Furthermore, cells treated with inhibitors of the mitochondrial respiratory chain or bearing superoxide dismutase (SOD) knockdown accumulate ROS and increase starvation- and rapamycin-induced autophagy, leading to extracellular delivery of HMGB1 [20]. This strongly indicates that autophagy-mediated HMGB1 secretion is dependent on ROS generation (Figure 2A and 3). Interestingly, soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) family, which are critical in membrane fusion, are also tightly linked to autophagy-related pathways, demonstrating that the fine-tuned machinery of autophagy shares components and possibly remarkably similar events with individual trafficking pathways [6].
With frank necrosis HMGB1 is passively released as cell membrane integrity is lost [97] (Figure 3). Poly(ADP)-ribose polymerase 1(PARP1) activation, triggered by DNA-alkylating damage, can destabilize HMGB1 association and induce nuclear-to-cytosolic translocation, which allows HMGB1 leakage upon subsequent ATP depletion, ROS production and plasma membrane disintegration during necrosis [98]. Additionally, p53, well known as a major mediator regulating apoptosis, also plays a role in modulating PARP enzymatic activity to in turn regulate necrosis [99]. Besides DNA-alkylating damage, we believe other stresses may also act as inducers or modulators of this event, and translocation may be the key event for cells to be susceptible to release HMGB1, alerting neighboring cells to deleterious outcome.
Apoptotic cells, however, sequester HMGB1 at early stage via firm binding to chromatin, programmed to limit its release [97] (Figure 3). Afterwards, they can release considerable quantities of immunodetectable HMGB1 at late stages in the form of nucleosomes along with DNA and histones [100]. Such HMGB1, oxidized on Cys106 by ROS in caspase-orchestrated mitochondrial production, appear to be tolerogenic rather than proinflammatory [89]. That is, it fails to considerably stimulate TNF secretion from responding macrophages in contrast to extracellular HMGB1 during necrosis, which is a potent inflammatory stimulator. The key issues within this dichotomy lies within the ROS generated by mitochondria in apoptotic cells that inhibits the inflammatory activity of HMGB1 by oxidizing the Cys 106 [89]. This mechanism at least partly, if not completely, elucidates how apoptosis tends to suppress inflammatory responses, given the fact that blocking the HMGB1 oxidation step restores its immunologically stimulating potential.
Pyroptosis, a peculiar form of programmed inflammatory cell death, occurs with inflammasome activation in macrophages, followed by caspase-1 activation [101]. In this pathway, double-stranded RNA–dependent protein kinase (PKR) has been identified as a crucial participator to trigger inflammasome activation and subsequently HMGB1 release. Regarded as a stress kinase, PKR is also a mediator of stress-induced apoptosis and autophagy, as well as proinflammatory cytokine production at the core of inflammation and immunity [102].
Altogether, among these different pathways of HMGB1 delivery, ROS seems to be the common key process, acting as either an inducer or downstream event, as further confirmed by the fact that both elevated endogenous and exogenous ROS promote HMGB1 release [90]. Once released, extracellular HMGB1 behaves differently depending on its redox state. Autophagy may also play a regulatory role, especially during vesicular exocytosis of HMGB1 and other leaderless proteins.
Redox HMGB1-Partner-Receptor Signaling Pathways
Extracellular HMGB1 signals via various receptors, including the receptor for advanced glycation end products (RAGE), Toll-like receptors (TLRs, including TLR2, TLR4/MD-2, and TLR9) , chemokine CXC receptor (CXCR) 4, Mac-1, syndecan-1 (CD138), CD24-Siglec-G/10, T cell immunoglobulin mucin-2 (Tim-3), thrombospondin, and others [8,24]. The number of receptors keeps on growing, so does the understanding of synergistic cooperation or feedback modulation. The intracellular signaling cascades following recognition involve two major specific adaptors, myeloid differentiation primary response protein (MyD88) and the Toll/IL-1R domain-containing adaptor-inducing interferon-β (TRIF), which are responsible for activating NF-κB and IFN regulatory factor-2 (IRF3) respectively [5,24]. In addition to binding receptors by itself, HMGB1 can also form heterocomplexes with other partners, including IL-1, CXCL12(SDF-1), DNA, RNA, nucleosomes, LPS or sRAGE, potentially defining the nature of the receptors to bind [8] to and strength of the resultant signal.
HMGB1 receptors are expressed in almost all cell types, ranging from neurons (high-differentiated) to cancer cells (low-differentiated), from immune cells (inflammatory initiator) to smooth muscle cells (late responder) [23]. Conventional wisdom is that activation of RAGE chiefly occurs in endothelial and somatic cells, whereas TLRs signals principally within myeloid cells [103]. However, the exclusive or cooperative relationship between RAGE and TLR4 is still not clear. Functional cellular responses can be summed up as stimulation of immune cells, induction of cytokine and chemokine responses, facilitation of proliferation and migration, promotion of autophagy and contribution of all the key hallmarks of cancer [11,23,26].
Although it is widely appreciated that the central role of HMGB1 in evoking immune response, whether HMGB1 per se exhibits the ability to initiate cellular signaling has been called into question, with observation of weak pro-inflammatory activity of purified HMGB1 itself [104]. Differing outcomes are likely dictated by environmental conditions (redox condition in particular), given that redox regulation of the three conserved Cys residues (Cys23, 45, and 106) modify its receptor-binding ability and biological consequences [103,105]. In other words, cytokine and chemokine-like activities are critically orchestrated by adopting mutually exclusive redox states (Figure 3).
All thiol form
chemotactic activity has been achieved in an all thiol form of HMGB1, independent from its cytokine activity [106]. Fully reduced HMGB1 functions as a chemoattractant for leukocyte migration, by the means of forming a heterocomplex with the chemokine CXCL12 (stromal cell-derived factor 1,SDF-1) and interacting with CXCR4 in a synergistic fashion. Nonoxidizable HMGB1 with serine replacement of all three cysteines acts similarly, suggesting that cysteines per se are not required for the property; on the contrary, oxidizing any cysteine dramatically ameliorates this potential.
Dithiol form
Cys 106 expressing a thiol group in B box is indispensable for both TLR4/MD-2 binding and its subsequent cytokine role. Oxidation or selective mutation abrogates interaction and its effect [107,108], since the TLR4 binding domain is adjacent. Considering that reduced HMGB1 promotes cancer cell autophagy through RAGE-Beclin 1-dependent pathways [109] and the HMGB1-RAGE axis is vital in immunity and tumorigenesis, understanding the interaction of HMGB1, RAGE, and TLR4 and how these two pathways interplay with each other during sepsis or tissue trauma are of great significance. Meanwhile, inflammation induction also requires the presence of a C23-C45 disulfide bridge, along with the thiol form of C106. Either reduction or oxidation impairs its cytokine activity [106].
Oxidized form
Consistent with apoptosis-induced immunological tolerance [89], oxidized HMGB1 (mainly released from apoptotic cells) limits inflammation [106], dependent on ROS production from mitochondria or probably ROS existing within the environment.
Further clarification to determine the binding characteristics of various redox HMGB1 to putative receptors and/or its partners is needed, which may deepen understanding of the redox status of HMGB1 and its role in signaling pathways and related pathological events.
Reduced HMGB1/RAGE-mediated Autophagy
Autophagy administers protein and organelle quality control under physiologically basal conditions and can be further activated following stress, following release of the DAMP, HMGB1. Endogenous HMGB1 mediates autophagy in both transcription-dependent and -independent manners (Figure 2C). As described below, exogenous HMGB1 can also modify the balance between apoptosis and autophagy.
We [109] have found that reduced HMGB1 (Cys106-SH) binds to RAGE, but not to Toll-like receptor 4, induces Beclin1-dependent autophagy and promotes tumor resistance to various anti-tumor drugs. Oxidized HMGB1, in contrast, induces apoptosis with activation of caspase-9/-3, thus increasing cytotoxicity of these agents. Accordingly, HMGB1 release, as well as the status of its redox state, serves as a promising target for directing cells towards heightened cell survival (autophagy) or cell death (apoptosis). Administration of ethyl pyruvate or HMGB1-neutralizing antibody, to either block release or neutralize extracellular HMGB1 release diminishes autophagic flux but promotes apoptosis in chemotherapy-treated cancer cells.
The Receptor for Advanced Glycation End-products (RAGE) is an evolutionarily more recent transmembrane receptor, a member of the immunoglobulin gene super family, and encoded within the MHC Class III region. Expressed on cancer cells and cells in the tumor microenvironment (leukocytes, endothelial cells, and fibroblasts), RAGE can recognize a diverse array of ligands, including AGEs, HMGB1, S100 family members, β-sheet fibrils and others [26]. We have previously demonstrated that RAGE act as a positive modulator of autophagy during chemotherapy and oxidative stress [22,110] and that RAGE expression level markedly correlates with enhanced levels of autophagy in pancreatic cancer, What’s more, RAGE-mediated autophagy is required for IL-6-triggered mitochondrial localization and function of signal transducers and activators of transcription 3 (STAT3) [111], revealing a novel pathway coupling autophagy and cellular energy metabolism with resultant tumor progression.
Multiple roles of HMGB1 in cancer
Tumorigenesis is a multi-step, complex process which largely depends on the crosstalk between cancer cell intrinsic factors and extrinsic immunosurveillance and inflammatory mediators. Overexpression of HMGB1 has been found in most cancers [26]. The contribution of HMGB1 to tumorigenesis is quite paradoxical and potentially redox-dependent. It can act as both an important intrinsic factor, promoting DNA damage repair and extracellular modulatory element, promoting or suppressing tumor development.
Cancers exhibit distinctive functional characteristics, concluded the “ten hallmarks” that enable tumor development and metastatic dissemination [112]. Abnormal HMGB1 is tightly related to the central hallmarks of cancer including proliferative immortality, insensitivity of growth suppressors, angiogenesis, invasion, metastasis, tumor-promoting inflammation, resistance to cell death, genome instability, dysregulation of energetics and evasion of immune destruction [26]. Additionally, as a key catabolic degradation pathway for clearance of superfluous materials or damaged organelles, perturbed and aberrant autophagy emerges as another hallmark of cancer [113]. Given that HMGB1 (disulfide or reduced form) is a major regulator of autophagy and mitophagy [20], it is even more persuasive that HMGB1 is critical in the regulation of cancer biology and tumor progression.
On the other hand, as a damage signal, HMGB1 can be released from necrotic cancer cells treated with most anti-cancer agents and triggers immunogenicity, a process called immunogenic cell death (ICD) [114]. ICD is characterized by secretion, release or surface exposure of DAMPs (ATP, HMGB1, DNA and HSPs), which are required for maturation and antigen presentation of dendritic cells via the TLR4 and Myd88 signaling pathway, thus evoking acute anti-tumor immunity. However, during chemotherapy, HMGB1 could also prompt regrowth and metastasis of the remaining cells in a RAGE-dependent manner [115], whereby blockade of HMGB1–RAGE suppresses growth/metastasis of tumor [116] and enhances the efficacy of chemotherapy [110]. One of the explanations lies in the role of HMGB1 (reduced) -RAGE-Beclin 1 in promotion of autophagy and inhibition of apoptosis (Figure 2C). Collectively, HMGB1 can bind to either TLR4 on dendritic cells, mediating ICD response, or RAGE in cancer cells, essential for cell survival after chemotherapy.
All these paradoxically diverse functions of HMGB1 could be partially, if not completely be explained by its redox modifications and accompanying varying partners/receptors. Reduced cysteines (all thiol form) make HMGB1 a chemoattractant, whereas a disulfide bond makes it a proinflammatory cytokine, and further cysteine oxidation to sulfonates mediated by ROS abrogates both activities (Figure 3). Apart from discrepancy in the setting of immune responses, these three forms of HMGB1 may also affect cancer cell fate and further influence therapeutic efficacy by possibly modulating the autophagic response following therapeutic anti-cancer stress. For example, reducible HMGB1 induces RAGE-dependent autophagy, leading to resistance to chemotherapeutic agents or radiotherapy, while oxidized HMGB1 increases the cytotoxicity of these chemotherapeutics via induction of apoptosis [109]. If the redox modifications occur in the environment following release or come from identifiable biochemical pathways in a specific case of cell death remains largely unknown. Moreover, how redox forms relate to receptor binding needs further resolution.
Conclusion
Cells have evolved multiple strategies in response to stressful conditions, including translocation and release of DAMPs (HMGB1), production of ROS ‘chaos’, and induction of elevated autophagy. The crosstalk of these three elements is essential, intimate but complicated in the context of cell stress (Figure 1 and 2A). ROS serves as not only a prime cause (oxidative stress) but also the accompanying intermediates, generated by cell stress—hypoxia, starvation, pathogen infection and growth factor stimulation. Moderate stress (including oxidative stress) promotes autophagy modulated by redox sensitive HMGB1 and other DAMPs (including ATP). The autophagic machinery in turn regulates translocation/release of DAMPs in a redox-dependent manner, contributing to inflammation and immunity, as well as determining the destiny of neighboring cellular elements to confine and respond to variable redox environments. The subsequent inflammatory response, in turn, can induce production of reactive oxidative intermediates. Stress and/or the ensuing heightened ROS levels triggers apoptosis. Conversely necrosis promotes leakage of cellular components, thereby reducing the local extracellular redox state. In this case, plentiful proteins, which would normally be eliminated extracellularly due to oxidative inactivation, are potent agents recruiting immune cells and inducing a proinflammatory response.
Redox-sensitive HMGB1 regulates autophagy in both transcription-dependent and transcription-independent mechanisms (Figure 2C). Nuclear HMGB1 promotes access to Hsp27 transcription, enhancing autophagy via actin cytoskeletal responses. Cytoplasmic HMGB1, through the vicinal cysteines at positions 23 and 45, binds to Beclin 1, which sustains Beclin 1- PtdIns3KC3 complex formation during autophagy initiation and sustenance. When present outside the cell, reduced HMGB1 triggers autophagy through interaction with RAGE which it also induces, creating an autocrine feed forward loop during tissue damage and injury.
Although the three-partner ‘Ménage à Trois‘ relationship appears to be integral in the response to stress, the precise reciprocal interactions and subsequent signaling pathways in specific settings remain to be completely characterized. Understanding the mechanism(s) by which DAMPs, redox and autophagy interact and are choreographed will elucidate important tissue-centric biological activities and provide key physiological and pathological insights into adaptation of cells and tissues to injury. Of particular importance, integrating all three elements, are the unusual aspects of cancer for which these elements seem both dangerous and fundamental.
Acknowledgments
This work was supported by the National Institutes of Health (R01CA160417 to D.T.).
References
- [1].Mantovani A. MSCs, macrophages, and cancer: a dangerous ménage-à-trois. Cell Stem Cell. 2012;11:730–2. doi: 10.1016/j.stem.2012.11.016. [DOI] [PubMed] [Google Scholar]
- [2].Fulda S, Gorman AM, Hori O, Samali A. Cellular stress responses: cell survival and cell death. International Journal of Cell Biology. 2010;2010:214074. doi: 10.1155/2010/214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Current Opinion in Cell Biology. 2010;22:124–31. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lotze MT, Zeh HJ, Rubartelli A, Sparvero LJ, Amoscato AA, Washburn NR, Devera ME, Liang X, Tör M, Billiar T. The grateful dead: damage-associated molecular pattern molecules and reduction/oxidation regulate immunity. Immunological Reviews. 2007;220:60–81. doi: 10.1111/j.1600-065X.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- [5].Tang D, Kang R, Coyne CB, Zeh HJ, Lotze MT. PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunological Reviews. 2012;249:158–75. doi: 10.1111/j.1600-065X.2012.01146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang Q, Kang R, Zeh HJ, Lotze MT, Tang D. DAMP and autophagy: Cellular adaptation to injury and unscheduled cell death. Autophagy. 2013;9:451–458. doi: 10.4161/auto.23691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rubartelli A, Lotze MT. Inside, outside, upside down: damage-associated molecular-pattern molecules (DAMPs) and redox. Trends in Immunology. 2007;28:429–36. doi: 10.1016/j.it.2007.08.004. [DOI] [PubMed] [Google Scholar]
- [8].Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annual Review of Immunology. 2010;28:367–88. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- [9].Dröge W. Free radicals in the physiological control of cell function. Physiological Reviews. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- [10].Krysko DV, Agostinis P, Krysko O, Garg AD, Bachert C, Lambrecht BN, Vandenabeele P. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends in Immunology. 2011;32:157–64. doi: 10.1016/j.it.2011.01.005. [DOI] [PubMed] [Google Scholar]
- [11].Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nature Reviews. Immunology. 2005;5:331–42. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- [12].Tang D, Kang R, Zeh HJ, Lotze MT. High-mobility group box 1, oxidative stress, and disease. Antioxidants & Redox Signaling. 2011;14:1315–35. doi: 10.1089/ars.2010.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Goodwin G, Rabbani A, Nicolas P, Johns E. The isolation of the high mobility group non-histone chromosomal protein HMG 14. FEBS Letters. 1977;80:413–416. doi: 10.1016/0014-5793(77)80488-2. [DOI] [PubMed] [Google Scholar]
- [14].Calogero S, Grassi F, Aguzzi a, Voigtländer T, Ferrier P, Ferrari S, Bianchi ME. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nature Genetics. 1999;22:276–80. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- [15].Fang P, Schachner M, Shen Y-Q. HMGB1 in development and diseases of the central nervous system. Molecular Neurobiology. 2012;45:499–506. doi: 10.1007/s12035-012-8264-y. [DOI] [PubMed] [Google Scholar]
- [16].Lotfi R, Lee JJ, Lotze MT. Eosinophilic granulocytes and damage-associated molecular pattern molecules (DAMPs): role in the inflammatory response within tumors. Journal of Immunotherapy. 2007;30:16–28. doi: 10.1097/01.cji.0000211324.53396.f6. [DOI] [PubMed] [Google Scholar]
- [17].Müller S, Scaffidi P, Degryse B, Bonaldi T, Ronfani L, Agresti a, Beltrame M, Bianchi ME. New EMBO members’ review: the double life of HMGB1 chromatin protein: architectural factor and extracellular signal. The EMBO Journal. 2001;20:4337–40. doi: 10.1093/emboj/20.16.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Thomas JO. HMG1 and 2: architectural DNA-binding proteins. Biochemical Society Transactions. 2001;29:395–401. doi: 10.1042/bst0290395. [DOI] [PubMed] [Google Scholar]
- [19].Tang D, Kang R, Livesey KM, Kroemer G, Billiar TR, Van Houten B, Zeh HJ, Lotze MT. High-mobility group box 1 is essential for mitochondrial quality control. Cell Metabolism. 2011;13:701–11. doi: 10.1016/j.cmet.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tang D, Kang R, Livesey KM, Cheh C-W, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ, Lotze MT. Endogenous HMGB1 regulates autophagy. The Journal of Cell Biology. 2010;190:881–92. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kang R, Livesey KM, Zeh HJ, Lotze MT, Tang D. HMGB1: a novel Beclin 1-binding protein active in autophagy. Autophagy. 2010;6:1209–11. doi: 10.4161/auto.6.8.13651. [DOI] [PubMed] [Google Scholar]
- [22].Kang R, Tang D, Schapiro NE, Patricia L, Bierhaus A, Lotze MT, Zeh HJ. The receptor for advanced glycation end products (RAGE) sustains autophagy and limits apoptosis, promoting pancreatic tumor cell survival. Cell Death & Differentiation. 2010;17:666–76. doi: 10.1038/cdd.2009.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Andersson U, Tracey KJ. HMGB1 is a therapeutic target for sterile inflammation and infection. Annual Review of Immunology. 2011;29:139–62. doi: 10.1146/annurev-immunol-030409-101323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li G, Liang X, Lotze MT. HMGB1: The Central Cytokine for All Lymphoid Cells. Frontiers in Immunology. 2013;4:68. doi: 10.3389/fimmu.2013.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yanai H, Ban T, Wang Z, Choi MK, Kawamura T, Negishi H, Nakasato M, Lu Y, Hangai S, Koshiba R, Savitsky D, Ronfani L, Akira S, Bianchi ME, Honda K, Tamura T, Kodama T, Taniguchi T. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462:99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- [26].Tang D, Kang R, Zeh HJ, Lotze MT. High-mobility group box 1 and cancer. Biochimica Et Biophysica Acta. 2010;1799:131–40. doi: 10.1016/j.bbagrm.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Goodwin GH, Johns EW. The isolation and purification of the high mobility group (HMG) nonhistone chromosomal proteins. Methods in Cell Biology. 1977;16:257–67. doi: 10.1016/s0091-679x(08)60104-1. [DOI] [PubMed] [Google Scholar]
- [28].Stros M. HMGB proteins: interactions with DNA and chromatin. Biochimica Et Biophysica Acta. 2010;1799:101–13. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- [29].Pasqualini JR, Sterner R, Mercat P, Allfrey VG. Estradiol enhanced acetylation of nuclear high mobility group proteins of the uterus of newborn guinea pigs. Biochemical and Biophysical Research Communications. 1989;161:1260–6. doi: 10.1016/0006-291x(89)91378-8. [DOI] [PubMed] [Google Scholar]
- [30].Verrier CS, Roodi N, Yee CJ, Bailey LR, Jensen RA, Bustin M, Parl FF. High-mobility group (HMG) protein HMG-1 and TATA-binding protein-associated factor TAF(II)30 affect estrogen receptor-mediated transcriptional activation. Molecular Endocrinology. 1997;11:1009–19. doi: 10.1210/mend.11.8.9962. [DOI] [PubMed] [Google Scholar]
- [31].Zhang CC, Krieg S, Shapiro DJ. HMG-1 stimulates estrogen response element binding by estrogen receptor from stably transfected HeLa cells. Molecular Endocrinology. 1999;13:632–43. doi: 10.1210/mend.13.4.0264. [DOI] [PubMed] [Google Scholar]
- [32].Stros M, Ozaki T, Bacikova A, Kageyama H, Nakagawara A. HMGB1 and HMGB2 cell-specifically down-regulate the p53- and p73-dependent sequence-specific transactivation from the human Bax gene promoter. The Journal of Biological Chemistry. 2002;277:7157–64. doi: 10.1074/jbc.M110233200. [DOI] [PubMed] [Google Scholar]
- [33].Jayaraman L, Moorthy NC, Murthy KG, Manley JL, Bustin M, Prives C. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes & Development. 1998;12:462–72. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Banerjee S, Kundu TK. The acidic C-terminal domain and A-box of HMGB-1 regulates p53-mediated transcription. Nucleic Acids Research. 2003;31:3236–47. doi: 10.1093/nar/gkg412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Agresti A, Lupo R, Bianchi ME, Müller S. HMGB1 interacts differentially with members of the Rel family of transcription factors. Biochemical and Biophysical Research Communications. 2003;302:421–6. doi: 10.1016/s0006-291x(03)00184-0. [DOI] [PubMed] [Google Scholar]
- [36].Butteroni C, De Felici M, Schöler HR, Pesce M. Phage display screening reveals an association between germline-specific transcription factor Oct-4 and multiple cellular proteins. Journal of Molecular Biology. 2000;304:529–40. doi: 10.1006/jmbi.2000.4238. [DOI] [PubMed] [Google Scholar]
- [37].Agresti A, Bianchi M. HMGB proteins and gene expression. Current Opinion in Genetics & Development. 2003;13:170–178. doi: 10.1016/s0959-437x(03)00023-6. [DOI] [PubMed] [Google Scholar]
- [38].Hoyos J. Martinez, Fedele M, Battista S, Pentimalli F, Kruhoffer M, Arra C, Orntoft TF, Croce CM, Fusco A. Identification of the genes up- and down-regulated by the high mobility group A1 (HMGA1) proteins: tissue specificity of the HMGA1-dependent gene regulation. Cancer Research. 2004;64:5728–35. doi: 10.1158/0008-5472.CAN-04-1410. [DOI] [PubMed] [Google Scholar]
- [39].Chau K-Y, Keane-Myers AM, Fedele M, Ikeda Y, Creusot RJ, Menozzi L, Cousins DJ, Manfioletti G, Feigenbaum L, Fusco A, Ono SJ. IFN-gamma gene expression is controlled by the architectural transcription factor HMGA1. International Immunology. 2005;17:297–306. doi: 10.1093/intimm/dxh209. [DOI] [PubMed] [Google Scholar]
- [40].Attema JL, Reeves R, Murray V, Levichkin I, Temple MD, Tremethick DJ, Shannon MF. The human IL-2 gene promoter can assemble a positioned nucleosome that becomes remodeled upon T cell activation. Journal of Immunology. 2002;169:2466–76. doi: 10.4049/jimmunol.169.5.2466. [DOI] [PubMed] [Google Scholar]
- [41].Foti D, Chiefari E, Fedele M, Iuliano R, Brunetti L, Paonessa F, Manfioletti G, Barbetti F, Brunetti A, Croce CM, Fusco A, Brunetti A. Lack of the architectural factor HMGA1 causes insulin resistance and diabetes in humans and mice. Nature Medicine. 2005;11:765–73. doi: 10.1038/nm1254. [DOI] [PubMed] [Google Scholar]
- [42].Whitley MZ, Thanos D, Read MA, Maniatis T, Collins T. A striking similarity in the organization of the E-selectin and beta interferon gene promoters. Molecular and Cellular Biology. 1994;14:6464–75. doi: 10.1128/mcb.14.10.6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Molecular and Cellular Biology. 2004;24:4321–8. doi: 10.1128/MCB.24.10.4321-4328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gerlitz G, Hock R, Ueda T, Bustin M. The dynamics of HMG protein-chromatin interactions in living cells. Biochemistry and Cell Biology = Biochimie Et Biologie Cellulaire. 2009;87:127–37. doi: 10.1139/O08-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nature Reviews. Molecular Cell Biology. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- [46].Lange SS, Mitchell DL, Vasquez KM. High mobility group protein B1 enhances DNA repair and chromatin modification after DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10320–5. doi: 10.1073/pnas.0803181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lange SS, Vasquez KM. HMGB1: the jack-of-all-trades protein is a master DNA repair mechanic. Molecular Carcinogenesis. 2009;48:571–80. doi: 10.1002/mc.20544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Štros M, Launholt D, Grasser K. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cellular and Molecular Life Sciences. 2007;64:2590–606. doi: 10.1007/s00018-007-7162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Shay JW. Telomerase in human development and cancer. Journal of Cellular Physiology. 1997;173:266–70. doi: 10.1002/(SICI)1097-4652(199711)173:2<266::AID-JCP33>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- [50].Polanská E, Dobšáková Z, Dvořáčková M, Fajkus J, Štros M. HMGB1 gene knockout in mouse embryonic fibroblasts results in reduced telomerase activity and telomere dysfunction. Chromosoma. 2012;121:419–31. doi: 10.1007/s00412-012-0373-x. [DOI] [PubMed] [Google Scholar]
- [51].Giavara S, Kosmidou E, Hande M. Yeast Nhp6A/B and mammalian Hmgb1 facilitate the maintenance of genome stability. Current Biology. 2005;15:68–72. doi: 10.1016/j.cub.2004.12.065. [DOI] [PubMed] [Google Scholar]
- [52].Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, Schadendorf D, Kumar R. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–61. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- [53].Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science (New York, N.Y.) 2013;339:957–9. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nature Reviews. Molecular Cell Biology. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- [55].Weiner L, Lotze M. Tumor-cell death, autophagy, and immunity. New England Journal of Medicine. 2012:28–30. doi: 10.1056/NEJMcibr1114526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lavoie JN, Gingras-Breton G, Tanguay RM, Landry J. Induction of Chinese hamster HSP27 gene expression in mouse cells confers resistance to heat shock HSP27 stabilization of the microfilament organization. The Journal of Biological Chemistry. 1993;268:3420–9. [PubMed] [Google Scholar]
- [57].Lavoie JN, Hickey E, Weber LA, Landry J. Modulation of actin microfilament dynamics and fluid phase pinocytosis by phosphorylation of heat shock protein 27. The Journal of Biological Chemistry. 1993;268:24210–4. [PubMed] [Google Scholar]
- [58].Acunzo J, Katsogiannou M, Rocchi P. Small heat shock proteins HSP27 (HspB1), alphaB-ccrystallin (HspB5) and HSP22 (HspB8) as regulators of cell death. The International Journal of Biochemistry & Cell Biology. 2012;44:1622–1631. doi: 10.1016/j.biocel.2012.04.002. [DOI] [PubMed] [Google Scholar]
- [59].O’Callaghan-Sunol C, Gabai VL, Sherman MY. Hsp27 modulates p53 signaling and suppresses cellular senescence. Cancer Research. 2007;67:11779–88. doi: 10.1158/0008-5472.CAN-07-2441. [DOI] [PubMed] [Google Scholar]
- [60].Livesey KM, Kang R, Vernon P, Buchser W, Loughran P, Watkins SC, Zhang L, Manfredi JJ, Zeh HJ, Li L, Lotze MT, Tang D. p53/HMGB1 complexes regulate autophagy and apoptosis. Cancer Research. 2012;72:1996–2005. doi: 10.1158/0008-5472.CAN-11-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].M.T.L., Kristen DT, Livesey M, Kang Rui, Zeh Herbert J., III Direct molecular interactions between HMGB1 and TP53 in colorectal cancer. Autophagy. 2012;8:846–848. doi: 10.4161/auto.19891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ott M, Gogvadze V, Orrenius S, Zhivotovsky B. Mitochondria, oxidative stress and cell death. Apoptosis : an International Journal on Programmed Cell Death. 2007;12:913–22. doi: 10.1007/s10495-007-0756-2. [DOI] [PubMed] [Google Scholar]
- [63].Vernon PJ, Tang D. Eat-me: autophagy, phagocytosis, and reactive oxygen species signaling. Antioxidants & Redox Signaling. 2013;18:677–91. doi: 10.1089/ars.2012.4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Thannickal V, Fanburg B. Reactive oxygen species in cell signaling. American Journal of Physiology. 2000;279:L1005–L1028. doi: 10.1152/ajplung.2000.279.6.L1005. [DOI] [PubMed] [Google Scholar]
- [65].Babior BM. NADPH oxidase. Current Opinion in Immunology. 2004;16:42–7. doi: 10.1016/j.coi.2003.12.001. [DOI] [PubMed] [Google Scholar]
- [66].Neumann CA, Cao J, Manevich Y. Peroxiredoxin 1 and its role in cell signaling. Cell Cycle. 2009;8:4072–8. doi: 10.4161/cc.8.24.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lushchak V. Glutathione homeostasis and functions: potential targets for medical interventions. Journal of Amino Acids. 2012;2012:736837. doi: 10.1155/2012/736837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hoppe G, Talcott KE, Bhattacharya SK, Crabb JW, Sears JE. Molecular basis for the redox control of nuclear transport of the structural chromatin protein Hmgb1. Experimental Cell Research. 2006;312:3526–38. doi: 10.1016/j.yexcr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- [69].Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell. 2005;121:667–70. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- [70].Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–47. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- [71].Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–35. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, Baehrecke EH, Lenardo M. Autophagic programmed cell death by selective catalase degradation. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4952–7. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. The EMBO Journal. 2011;30:4701–11. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annual Review of Cell and Developmental Biology. 2011;27:107–32. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- [75].Jiang S, Dupont N, Castillo EF, Deretic V. Secretory versus Degradative Autophagy: Unconventional Secretion of Inflammatory Mediators. Journal of Innate Immunity. 2013 doi: 10.1159/000346707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Virgin HW, Levine B. Autophagy genes in immunity. Nature Immunology. 2009;10:461–70. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Demple B, Amábile-Cuevas CF. Redox redux: the control of oxidative stress responses. Cell. 1991;67:837–9. doi: 10.1016/0092-8674(91)90355-3. [DOI] [PubMed] [Google Scholar]
- [78].Kang R, Livesey KM, Zeh HJ, Lotze MT, Tang D. HMGB1 as an autophagy sensor in oxidative stress. Autophagy. 2011;7:904–906. doi: 10.4161/auto.7.8.15704. [DOI] [PubMed] [Google Scholar]
- [79].Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Molecular Cell. 2012;48:158–67. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Naviaux RK. Oxidative shielding or oxidative stress? The Journal of Pharmacology and Experimental Therapeutics. 2012;342:608–18. doi: 10.1124/jpet.112.192120. [DOI] [PubMed] [Google Scholar]
- [81].Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. The EMBO Journal. 2007;26:1749–60. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Azad MB, Chen Y, Gibson SB. Regulation of autophagy by reactive oxygen species (ROS): implications for cancer progression and treatment. Antioxidants & Redox Signaling. 2009;11:777–90. doi: 10.1089/ars.2008.2270. [DOI] [PubMed] [Google Scholar]
- [83].Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends in Cell Biology. 2007;17:422–7. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- [84].Kiffin CAM, Bandyopadhyay R. Urmi. Oxidative stress and autophagy, Antioxidants & Redox Signaling. 2006;8:152–162. doi: 10.1089/ars.2006.8.152. [DOI] [PubMed] [Google Scholar]
- [85].Tatsuta T, Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. The EMBO Journal. 2008;27:306–14. doi: 10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Tang D, Kang R, Livesey KM, Zeh HJ, Lotze MT. High mobility group box 1 (HMGB1) activates an autophagic response to oxidative stress. Antioxidants & Redox Signaling. 2011;15:2185–95. doi: 10.1089/ars.2010.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen H-Y, Bray K, Reddy A, Bhanot G, Gelinas C, Dipaola RS, Karantza-Wadsworth V, White E. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2770–5. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kazama H, Ricci J-E, Herndon JM, Hoppe G, Green DR, Ferguson TA. Induction of immunological tolerance by apoptotic cells requires caspase-dependent oxidation of high-mobility group box-1 protein. Immunity. 2008;29:21–32. doi: 10.1016/j.immuni.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Tang D, Shi Y, Kang R, Li T, Xiao W, Wang H, Xiao X. Hydrogen peroxide stimulates macrophages and monocytes to actively release HMGB1. Journal of Leukocyte Biology. 2007;81:741–7. doi: 10.1189/jlb.0806540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Wang H. HMG-1 as a Late Mediator of Endotoxin Lethality in Mice. Science. 1999;285:248–251. doi: 10.1126/science.285.5425.248. [DOI] [PubMed] [Google Scholar]
- [92].Walter P, Gilmore R, Blobel G. Protein translocation across the endoplasmic reticulum. Cell. 1984;38:5–8. doi: 10.1016/0092-8674(84)90520-8. [DOI] [PubMed] [Google Scholar]
- [93].Nickel W, Rabouille C. Mechanisms of regulated unconventional protein secretion. Nature Reviews. Molecular Cell Biology. 2009;10:148–55. doi: 10.1038/nrm2617. [DOI] [PubMed] [Google Scholar]
- [94].Nickel W. Pathways of unconventional protein secretion. Current Opinion in Biotechnology. 2010;21:621–6. doi: 10.1016/j.copbio.2010.06.004. [DOI] [PubMed] [Google Scholar]
- [95].Gardella S, Andrei C, Ferrera D, Lotti L. The nuclear protein HMGB1 is secreted by monocytes via a non-classical, vesicle-mediated secretory pathway. EMBO Reports. 2002;3:995–1001. doi: 10.1093/embo-reports/kvf198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Thorburn J, Horita H, Redzic J, Hansen K, Frankel a E., Thorburn a. Autophagy regulates selective HMGB1 release in tumor cells that are destined to die. Cell Death and Differentiation. 2009;16:175–83. doi: 10.1038/cdd.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–5. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- [98].Ditsworth D, Zong W-X, Thompson CB. Activation of poly(ADP)-ribose polymerase (PARP-1) induces release of the pro-inflammatory mediator HMGB1 from the nucleus. The Journal of Biological Chemistry. 2007;282:17845–54. doi: 10.1074/jbc.M701465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Montero J, Dutta C, van Bodegom D, Weinstock D, Letai A. p53 regulates a non-apoptotic death induced by ROS. Cell Death and Differentiation. 2013 doi: 10.1038/cdd.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Bell CW, Jiang W, Reich CF, Pisetsky DS. The extracellular release of HMGB1 during apoptotic cell death. American Journal of Physiology. Cell Physiology. 2006;291:C1318–25. doi: 10.1152/ajpcell.00616.2005. [DOI] [PubMed] [Google Scholar]
- [101].Lu B, Nakamura T, Inouye K, Li J, Tang Y, Lundbäck P, Valdes-Ferrer SI, Olofsson PS, Kalb T, Roth J, Zou Y, Erlandsson-Harris H, Yang H, Ting JP-Y, Wang H, Andersson U, Antoine DJ, Chavan SS, Hotamisligil GS, Tracey KJ. Novel role of PKR in inflammasome activation and HMGB1 release. Nature. 2012;488:670–4. doi: 10.1038/nature11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Kang R, Tang D. PKR-Dependent Inflammatory Signals. Science Signaling. 2012;5:1–3. doi: 10.1126/scisignal.2003511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Yanai H, Ban T, Taniguchi T. High-mobility group box family of proteins: ligand and sensor for innate immunity. Trends in Immunology. 2012;33:633–40. doi: 10.1016/j.it.2012.10.005. [DOI] [PubMed] [Google Scholar]
- [104].Rouhiainen A, Tumova S, Valmu L, Kalkkinen N, Rauvala H. Pivotal advance: analysis of proinflammatory activity of highly purified eukaryotic recombinant HMGB1 (amphoterin) Journal of Leukocyte Biology. 2007;81:49–58. doi: 10.1189/jlb.0306200. [DOI] [PubMed] [Google Scholar]
- [105].Yang H, Antoine DJ, Andersson U, Tracey KJ. The many faces of HMGB1: molecular structure-functional activity in inflammation, apoptosis, and chemotaxis. Journal of Leukocyte Biology. 2013;93:1–9. doi: 10.1189/jlb.1212662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Venereau E, Casalgrandi M, Schiraldi M, Antoine DJ, Cattaneo A, De Marchis F, Liu J, Antonelli A, Preti A, Raeli L, Shams SS, Yang H, Varani L, Andersson U, Tracey KJ, Bachi A, Uguccioni M, Bianchi ME. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. The Journal of Experimental Medicine. 2012;209:1519–28. doi: 10.1084/jem.20120189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Yang H, Lundbäck P, Ottosson L, Erlandsson-Harris H, Venereau E, Bianchi ME, Al-Abed Y, Andersson U, Tracey KJ, Antoine DJ. Redox modification of cysteine residues regulates the cytokine activity of high mobility group box-1 (HMGB1) Molecular Medicine. 2012;18:250–9. doi: 10.2119/molmed.2011.00389. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [108].Yang H, Hreggvidsdottir HS, Palmblad K, Wang H, Ochani M, Li J, Lu B, Chavan S, Rosas-Ballina M, Al-Abed Y, Akira S, Bierhaus A, Erlandsson-Harris H, Andersson U, Tracey KJ. A critical cysteine is required for HMGB1 binding to Toll-like receptor 4 and activation of macrophage cytokine release. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11942–7. doi: 10.1073/pnas.1003893107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Tang D, Kang R, Cheh CW, Livesey KM, Liang X, Schapiro NE, Benschop R, Sparvero LJ, Amoscato a a, Tracey KJ, Zeh HJ, Lotze MT. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene. 2010;29:5299–310. doi: 10.1038/onc.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Kang R, Tang D, Livesey KM, Schapiro NE, Lotze MT, Zeh HJ. The Receptor for Advanced Glycation End-products (RAGE) protects pancreatic tumor cells against oxidative injury. Antioxidants & Redox Signaling. 2011;15:2175–84. doi: 10.1089/ars.2010.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Kang R, Loux T, Tang D, Schapiro NE, Vernon P, Livesey KM, Krasinskas A, Lotze MT, Zeh HJ. The expression of the receptor for advanced glycation endproducts (RAGE) is permissive for early pancreatic neoplasia. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7031–6. doi: 10.1073/pnas.1113865109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- [113].Kimmelman AC. The dynamic nature of autophagy in cancer. Genes & Development. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Krysko O, Aaes T. Løve, Bachert C, Vandenabeele P, Krysko DV. Many faces of DAMPs in cancer therapy. Cell Death & Disease. 2013;4:e631. doi: 10.1038/cddis.2013.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Luo Y, Chihara Y, Fujimoto K, Sasahira T, Kuwada M, Fujiwara R, Fujii K, Ohmori H, Kuniyasu H. High mobility group box 1 released from necrotic cells enhances regrowth and metastasis of cancer cells that have survived chemotherapy. European Journal of Cancer. 2013;49:741–51. doi: 10.1016/j.ejca.2012.09.016. [DOI] [PubMed] [Google Scholar]
- [116].Taguchi A, Blood DC, del Toro G, Canet A, Lee DC, Qu W, Tanji N, Lu Y, Lalla E, Fu C, Hofmann M. a, Kislinger T, Ingram M, Lu A, Tanaka H, Hori O, Ogawa S, Stern DM, Schmidt a M. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature. 2000;405:354–60. doi: 10.1038/35012626. [DOI] [PubMed] [Google Scholar]
- [117].Harris H. Erlandsson, Andersson U. Mini-review: The nuclear protein HMGB1 as a proinflammatory mediator. European Journal of Immunology. 2004;34:1503–12. doi: 10.1002/eji.200424916. [DOI] [PubMed] [Google Scholar]
- [118].Ivanov S, Dragoi A-M, Wang X, Dallacosta C, Louten J, Musco G, Sitia G, Yap GS, Wan Y, Biron CA, Bianchi ME, Wang H, Chu W-M. A novel role for HMGB1 in TLR9-mediated inflammatory responses to CpG-DNA. Blood. 2007;110:1970–81. doi: 10.1182/blood-2006-09-044776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].DeMarco RA, Fink MP, Lotze MT. Monocytes promote natural killer cell interferon gamma production in response to the endogenous danger signal HMGB1. Molecular Immunology. 2005;42:433–44. doi: 10.1016/j.molimm.2004.07.023. [DOI] [PubMed] [Google Scholar]
- [120].Merenmies J, Pihlaskari R, Laitinen J, Wartiovaara J, Rauvala H. 30-kDa heparin-binding protein of brain (amphoterin) involved in neurite outgrowth Amino acid sequence and localization in the filopodia of the advancing plasma membrane. The Journal of Biological Chemistry. 1991;266:16722–9. [PubMed] [Google Scholar]
- [121].Rouhiainen A, Imai S, Rauvala H, Parkkinen J. Occurrence of amphoterin (HMG1) as an endogenous protein of human platelets that is exported to the cell surface upon platelet activation. Thrombosis and Haemostasis. 2000;84:1087–94. [PubMed] [Google Scholar]